Abstract
Double-stranded RNA has been shown to induce gene silencing in diverse eukaryotes and by a variety of pathways. We have examined the taxonomic distribution and the phylogenetic relationship of key components of the RNA interference (RNAi) machinery in members of five eukaryotic supergroups. On the basis of the parsimony principle, our analyses suggest that a relatively complex RNAi machinery was already present in the last common ancestor of eukaryotes and consisted, at a minimum, of one Argonaute-like polypeptide, one Piwi-like protein, one Dicer, and one RNA-dependent RNA polymerase. As proposed before, the ancestral (but non-essential) role of these components may have been in defense responses against genomic parasites such as transposable elements and viruses. From a mechanistic perspective, the RNAi machinery in the eukaryotic ancestor may have been capable of both small-RNA-guided transcript degradation as well as transcriptional repression, most likely through histone modifications. Both roles appear to be widespread among living eukaryotes and this diversification of function could account for the evolutionary conservation of duplicated Argonaute-Piwi proteins. In contrast, additional RNAi-mediated pathways such as RNA-directed DNA methylation, programmed genome rearrangements, meiotic silencing by unpaired DNA, and miRNA-mediated gene regulation may have evolved independently in specific lineages.
Keywords: RNA interference, Transposon silencing, Heterochromatin, RNAi phylogenetics
Introduction
RNA-mediated silencing is an evolutionarily conserved mechanism(s) through which double-stranded RNA (dsRNA) induces the inactivation of cognate sequences. The role of dsRNA in triggering repression was initially characterized in Caenorhabditis elegans and termed RNA interference (Fire et al. 1998). However, silencing phenomena had already been described in a number of eukaryotes and the connection to dsRNA helped to unify several, apparently disparate, processes involving post-transcriptional RNA degradation, transcriptional gene silencing via heterochromatin formation and/or DNA methylation, DNA elimination, or meiotic silencing by unpaired DNA (Baulcombe 2004; Matzke and Birchler 2005; Meister and Tuschl 2004; Ullu et al. 2004; Zamore and Haley 2005). In plants and animals, the RNAi machinery is also involved in the production of microRNAs (miRNAs), by the processing of genome encoded imperfect RNA hairpins, which play a role in developmental regulation (Bartel 2004; Chen 2005; Wienholds and Plasterk 2005; Zamore and Haley 2005).
Currently, the most extensively characterized dsRNA-mediated mechanism is targeted mRNA degradation guided by small interfering RNAs (siRNAs). Genetic and biochemical studies from diverse species have revealed that long dsRNAs are processed into siRNAs by an RNaseIII-like endonuclease, named Dicer (Bernstein et al. 2001; Meister and Tuschl 2004; Sontheimer 2005; Tomari and Zamore 2005). siRNAs are then incorporated into a multiprotein complex, the RNA-induced silencing complex (RISC) (Pham et al. 2004; Tomari et al. 2004). Members of the Argonaute-Piwi (Ago-Piwi) family of proteins are core components of the RISC and some of these polypeptides function as siRNA-guided endonucleases (Baumberger and Baulcombe 2005; Hammond et al. 2001; Liu et al. 2004a; Meister and Tuschl 2004; Qi et al. 2005; Tomari and Zamore 2005). Recent evidence suggests that a siRNA duplex may be loaded into RISC and then Ago cleaves one of the siRNA strands (the passenger strand) triggering its dissociation from the complex (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005). Activated RISC then functions as a multiple-turnover enzyme that recognizes and cleaves RNA molecules complementary to the incorporated single-stranded guide siRNA (Meister and Tuschl 2004; Sontheimer 2005; Tomari and Zamore 2005).
Members of the Argonaute-Piwi family fall into two main classes, one named after Arabidopsis thaliana Argonaute and the other after Drosophila melanogaster Piwi (Carmell et al. 2002). These proteins are highly basic, approximately 100-kD in size, and contain two conserved motifs, the PAZ (after Piwi/Argonaute/Zwille) and the PIWI domains (Cerutti et al. 2000; Lingel et al. 2004; Ma et al. 2004; Song et al. 2004; Yuan et al. 2005). A number of experiments have implicated certain argonautes, such as human Ago2, as the catalytic unit (‘slicer’) of the RISC (Liu et al. 2004a; Okamura et al. 2004; Rivas et al. 2005). However, several other Ago paralogs are not endonucleolytically active (Liu et al. 2004a; Meister et al. 2004; Rivas et al. 2005). In fact, in several species, there is evidence for functional specialization of Ago-Piwi proteins (Grishok et al. 2001; Lee et al. 2003; Matzke and Birchler 2005; Okamura et al. 2004).
RNA-dependent RNA polymerases (RdRPs) also play an important role in RNAi in some eukaryotes (Wassenegger and Krczal 2006). For instance, putative homologs of a tomato RdRP are required for post-transcriptional gene silencing (PTGS) triggered by sense transgenes in A. thaliana, for quelling (a phenomenon similar to PTGS) and for meiotic silencing by unpaired DNA in Neurospora crassa, as well as for RNAi in C. elegans and Dictyostelium discoideum (Baulcombe 2004; Cogoni and Macino 2000; Martens et al. 2002; Shiu et al. 2001; Sijen et al. 2001; Wassenegger and Krczal 2006). It has been proposed that RdRPs generate dsRNA from single-stranded transcripts either by de novo, primer independent second-strand synthesis (utilizing as template ‘aberrant’ RNAs, presumably lacking normal processing signals such as a 5′ cap or a polyA tail) or by using siRNAs as primers to synthesize RNA complementary to the target mRNA (Baulcombe 2004; Sijen et al. 2001; Wassenegger and Krczal 2006). Thus, RdRP activity may initiate RNAi (by producing the trigger dsRNA) or dramatically enhance the RNAi response (by amplifying the amount of dsRNA) (Baulcombe 2004). However, dsRNA-induced RNAi can occur in the absence of RdRP activity (Schwarz et al. 2002; Stein et al. 2003).
The biochemical and genetic studies briefly summarized above have led to the identification of three key components of the RNAi machinery, namely Dicer, Argonaute-Piwi, and RdRP. However, the taxonomic distribution of these proteins and their ancestry have not been explored in detail. In this review we have examined the phylogenetic relationship of Ago-Piwi, Dicer-like, and RdRP proteins present in members of five eukaryotic supergroups. On the basis of the parsimony principle we have attempted to infer the composition and function(s) of the RNAi machinery in the last common ancestor of eukaryotes. We have also assessed putatively derived RNAi functions that might have evolved in specific lineages. Our findings provide a framework for predicting the existence of RNAi-related mechanisms in uncharacterized eukaryotes.
Distribution of Argonaute-Piwi, Dicer-like, and RdRP proteins in eukaryotes
Based on morphological, biochemical, and molecular phylogenetic approaches, eukaryotes have recently been classified into six supergroups: the Opisthokonta, including animals and fungi; the Amoebozoa, including most traditional amoebae and slime moulds; the Excavata, grouping diplomonads, several genera of heterotrophic flagellates, and possibly the Euglenozoa; the Rhizaria, including the Foraminifera and the Cercozoa; the Archaeplastida, grouping red algae, green algae, and plants; and the Chromalveolata, including dinoflagellates, apicomplexan parasites, and the Stramenopiles (brown algae, diatoms, and many zoosporic fungi) (Adl et al. 2005; Medina 2005). In order to evaluate the phyletic distribution of the RNAi machinery components, we have surveyed 25 complete or near-complete genomes that belong to five eukaryotic supergroups (with only Rhizaria remaining unsampled). Proteins containing conserved Argonaute-Piwi, Dicer, or RdRP domains were identified by either BLAST or PSI-BLAST searches of protein and/or translated genomic DNA databases. Since several of the examined genomes are in draft stage, an important caveat in our analyses is that some proteins may be missing from the databases whereas others may have errors in the predicted gene structure. However, we only considered as potential homologs proteins that exhibited enough sequence similarity to be aligned and used for phylogenetic tree construction.
Argonaute-Piwi, Dicer-like, and RdRP proteins occur in members of all the eukaryotic supergroups examined (Table 1). This widespread taxonomic distribution, as well as the direct demonstration of RNAi related phenomena in most of these organisms (Table 2), suggests that the main components of the RNAi machinery were already present in the last common ancestor of eukaryotes. However, Ago-Piwi, Dicer-like, and RdRP polypeptides (or a subset of these proteins) also appear to have been lost from specific lineages. The RNAi machinery seems to be entirely absent in Saccharomyces cerevisiae (Opisthokonta), Trypanosoma cruzi and Leishmania major (Excavata), Cyanidioschyzon merolae (Archaeplastida), and Plasmodium falciparum (Chromalveolata) (Table 1). For some of these organisms there is also convincing evidence that they are unable to utilize dsRNA to trigger degradation of target RNA (DaRocha et al. 2004; Robinson and Beverley 2003; Ullu et al. 2004). Thus, the RNAi mechanism appears to have been lost independently several times during eukaryotic evolution.
Table 1.
Distribution of RNAi machinery components in eukaryotes
| Species | Genomea | Argonaute-Piwi | Dicer-like | RdRP | Role in virus, transposon, or repetitive DNA silencingb |
|---|---|---|---|---|---|
| Excavata | |||||
| Giardia intestinalis | Assembly | + | + | + | ? |
| Trypanosoma brucei | Assembly | + | − | − | + |
| Trypanosoma cruzi | Complete | − | − | − | NA |
| Leishmania major | Assembly | − | − | − | NA |
| Chromalveolata | |||||
| Paramecium tetraurelia | In progress | + | + | + | ? |
| Tetrahymena thermophila | In progress | + | + | + | + |
| Plasmodium falciparum | Assembly | − | − | − | NA |
| Phytophthora sojae | In progress | + | + | + | ? |
| Thalassiosira pseudonana | Assembly | + | − | − | ? |
| Rhizaria | |||||
| Data not available | |||||
| Archaeplastida | |||||
| Cyanidioschyzon merolae | Complete | − | − | − | NA |
| Chlamydomonas reinhardtii | In progress | + | + | − | + |
| Arabidopsis thaliana | Complete | + | + | + | + |
| Oryza sativa (japonica) | Complete | + | + | + | + |
| Amoebozoa | |||||
| Dictyostelium discoideum | Assembly | + | + | + | + |
| Entamoeba histolytica | Complete | + | − | + | ? |
| Opisthokonta | |||||
| Saccharomyces cerevisiae | Complete | − | − | − | NA |
| Schizosaccharomyces pombe | Complete | + | + | + | + |
| Neurospora crassa | Complete | + | + | + | + |
| Aspergillus nidulans | Assembly | + | + | + | ? |
| Caenorhabditis elegans | Complete | + | + | + | + |
| Drosophila melanogaster | Complete | + | + | − | + |
| Anopheles gambiae | Assembly | + | + | − | + |
| Strongylocentrotus purpuratus | In progress | + | + | − | ? |
| Ciona intestinalis | Assembly | + | + | − | ? |
| Homo sapiens | Complete | + | + | − | + |
Status of genome sequencing projects taken from http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi
Inferred from the phenotype of mutant or RNAi knock-down strains defective in at least one of the RNAi effectors (?, lack of evidence; NA, not applicable)
Table 2.
Taxonomic distribution of RNAi-mediated silencing pathways in eukaryotes
Representative publications are indicated whenever there is experimental evidence for a silencing pathway in a particular species
Indirect evidence such as transcription repression and/or chromosome segregation defects in RNAi mutants
Direct evidence such as RNAi-induced H3K9 methylation
The greatest conservation among the examined polypeptides corresponded to Ago-Piwi proteins, which are clearly identifiable in all species where RNAi-related phenomena have been experimentally demonstrated (Tables 1, 2). Moreover, the dual domain structure of the Ago-Piwi polypeptides, namely a PAZ domain followed by a PIWI domain, has also been well conserved. The only exception among the RNAi-positive organisms listed in Table 1 is Giardia intestinalis that encodes a protein with a well-defined PIWI domain fused to a highly divergent PAZ domain. However, Giardia protein-encoding genes are notoriously fast-evolving compared with those of most other eukaryotes (Richards and Cavalier-Smith 2005), which might explain the poor conservation of the PAZ domain. Several organisms, including a few archaea and eubacteria, encode proteins with a single PIWI domain recognizable by primary sequence comparisons (Anantharaman et al. 2002; Cerutti et al. 2000; Ullu et al. 2004). Though, the crystal structures of the Pyrococcus furiosus and the Aquifex aeolicus Ago-like polypeptides revealed that they also contain somewhat variant PAZ-like domains (Song et al. 2004; Yuan et al. 2005). Yet, since similar proteins are also present in species such as T. cruzi and L. major that are RNAi negative (Ullu et al. 2004) and the Trypanosoma brucei single PIWI polypeptide is not required for RNAi (Durand-Dubief and Bastin 2003), their functional role(s) is uncertain and they were not included in our analyses. Intriguingly, recent findings have indicated that the Archaeoglobus fulgidus Piwi and the A. aeolicus Ago bind ssDNA with greater affinity than ssRNA (Ma et al. 2005; Yuan et al. 2005) and the A. aeolicus protein has been postulated to function as a DNA-guided site-specific endoribonuclease (Yuan et al. 2005).
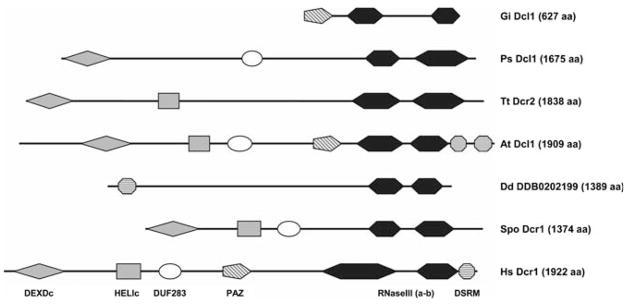
Dicer-like proteins are relatively well conserved among organisms that have retained the RNAi pathway (Table 1), albeit with significant variability in their primary sequence and domain organization (Fig. 1). The Dicer enzymes initially characterized in D. melanogaster and humans (Bernstein et al. 2001; Zhang et al. 2004) are multidomain proteins consisting of a SFII RNA helicase domain, a domain of unknown function (DUF283), a PAZ domain, two RNaseIII catalytic domains (RNaseIIIa and RNaseIIIb), and a dsRNA binding domain (DSRM) (Bernstein et al. 2001; Meister and Tuschl 2004). This overall organization is maintained in Dicer-like proteins from animals (H. sapiens), fungi (Schyzosaccharomyces pombe), and plants (A. thaliana) (Fig. 1) with the greatest variability associated with the presence or absence of the DSRM and/or of the PAZ domains. In Tetrahymena thermophila there are three Dicer-like sequences: Dcr2, which contains only the helicase and the two RNaseIII domains (Fig. 1), and Dcr1 and Dcl1, which are more divergent (Lee and Collins 2006; Mochizuki and Gorovsky 2005). Indeed, Dcl1 only includes the RNaseIII motifs and a C-terminal DSRM. In the incomplete genome of Phytophthora sojae the only recognizable Dicer-like sequence consists of a poorly conserved helicase domain, the DUF283 motif, and the two RNaseIII domains (Fig. 1). In D. discoideum only the RNaseIII domains have been conserved, in association with a DSRM fused at the N-terminal end of the two Dicer-like proteins (Fig. 1). Interestingly, in this organism the RdRPs now contain SFII RNA helicase motifs homologous to that of Dicer (Martens et al. 2002). In G. intestinalis the sole Dicer-like protein is characterized by a PAZ domain and the RNaseIII motifs (Fig. 1), yet this polypeptide has recently been shown to work as a fully functional enzyme (MacRae et al. 2006). Thus, the only Dicer domains that appear to be predominantly conserved as a fusion across the eukaryotic spectrum are the two RNaseIII catalytic motifs (Fig. 1). Intriguingly, both T. brucei and Entamoeba histolytica, species with demonstrated RNAi (Djikeng et al. 2001; Kaur and Lohia 2004), appear to encode only proteins with single RNaseIII domains (Abed and Ankri 2005). These enzymes may perhaps act as dimers to assume a catalytic core similar to that of Dicer (Zhang et al. 2004; MacRae et al. 2006). Alternatively, the T. brucei and E. histolytica genomes might be incomplete or the corresponding Dicer-like sequences might be so divergent that they are no longer recognizable by primary sequence searches.
Fig. 1.
Domain architecture of Dicer-like proteins from species belonging to five eukaryotic supergroups. Protein sequences were examined for the presence of conserved domains by comparison with the SMART and PFAM databases and their diagrams are shown to scale. DEXDc DEAD-like helicase domain; HELIc helicase C-terminal domain; DUF283 DUF283 domain; PAZ PAZ domain; RNAseIII (a-b) RNaseIII domains a and b, DSRM Double-stranded RNA binding domain. The DEXDc and HELIc motifs are referred to as the SFII RNA helicase domain in the text. At Dcl1 A. thaliana Dcl1 (NP_171612); Dd DDB0202199 D. discoideum DDB0202199 (EAL73658); Gi Dcl1 G. intestinalis Dcl1 (AAO17549); Hs Dcr1 H. sapiens Dcr1 (NP_803187); Ps Dcl1 P. sojae Dcl1 (v1_C_860007 at http://www.genome.jgi-psf.org/sojae1/sojae1.home.html); Spo Dcr1 S. pombe Dcr1 (Q09884); Tt Dcr2 T. thermophila Dcr2 (BAD34723)
RdRPs are not as widely distributed among eukaryotes as Ago-Piwi and Dicer-like proteins (Table 1). Even though RNAi occurs in animals (Schwarz et al. 2002; Stein et al. 2003), Chlamydomonas reinhardtii (a green alga) (Rohr et al. 2004; Schroda 2006), and T. brucei (Ullu et al. 2004), RdRPs were not detected in their genomes (Table 1), with the exception of C. elegans and Branchiostoma floridae (AAQ10792). In Aspergillus nidulans it has been experimentally demonstrated that RNAi induced by an inverted repeat transgene does not require any of the two RdRPs encoded in the genome (Hammond and Keller 2005). This is consistent with the postulated ancillary roles of RdRPs in generating the dsRNA trigger and/or in amplifying siRNA levels (Baulcombe 2004; Sijen et al. 2001; Wassenegger and Krczal 2006). If enough dsRNA is produced by other means RdRPs might not be needed for the degradative RNAi pathway, explaining their more widespread loss from specific eukaryotic lineages. Conversely, processes that depend on RdRPs such as transitive RNAi (i.e., the spreading of silencing to regions outside that initially targeted by dsRNA) and the generation of transacting siRNAs will be limited to eukaryotes encoding these enzymes in their genomes (Allen et al. 2005; Baulcombe 2004; Sijen et al. 2001; Yoshikawa et al. 2005).
Phylogeny of Argonaute-Piwi, Dicer-like, and RdRP proteins
The phylogeny of eukaryotic organisms has been difficult to resolve. The relationship among the supergroups, the potential lack of monophyly of Chromalveolata and Excavata, and the placement of the root of the tree have remained contentious (Adl et al. 2005; Simpson and Roger 2004). Earlier phylogenetic analyses indicated that diplomonads (including G. intestinalis) are among the deepest divergences in the eukaryotic lineage and the tree was ‘rooted’ with these mitochondrion-lacking unicellular eukaryotes (Sogin et al. 1989; Sogin 1991). Recent studies have suggested that this rooting may have been patterned by methodological artefacts (Philippe et al. 2000). Arisue et al. (2004) have argued that two possibilities seem to exist for the root of the eukaryotic tree, namely the branch leading to Opisthokonta (animals and fungi) or that leading to the common ancestor of Diplomonadida/Parabasalia (within Excavata). Moreover, combined protein phylogenies strongly suggest that Opisthokonta are most closely related to Amoebozoa (Richards and Cavalier-Smith 2005; Simpson and Roger 2004). This grouping has been called ‘unikonts’ (ancestrally monociliate). Based on similar evidence all the other major groups of eukaryotes (Archaeplastida, Chromalveolata, Rhizaria, and Excavata) might be related to each other and have been called ‘bikonts’ (ancestrally biciliate) (Richards and Cavalier-Smith 2005; Simpson and Roger 2004). Thus, an emerging hypothesis is that the earliest evolutionary divergence within eukaryotes (and the root of the eukaryotic tree) falls between unikonts and bikonts (Richards and Cavalier-Smith 2005; Simpson and Roger 2004; Stechmann and Cavalier-Smith 2003). If true, then comparisons between animals, fungi, and plants (which would include organisms derived from each branch of the earliest divergence) would be largely sufficient to diagnose the generalities of the ancestral RNAi machinery. However, because of the detected pattern of lineage specific losses of RNAi components (see above) and the possibility of an alternative eukaryotic rooting in the branch leading to Diplomonadida/Parabasalia, we have included proteins from species belonging to each of five eukaryotic supergroups (whenever possible) in our phylogenetic analyses.
The most likely point of origin of the Argonaute-Piwi, Dicer-like, and RdRP protein families was inferred from the patterns of phyletic distribution and phylogenetic tree topology and on the basis of the parsimony principle (Anantharaman et al. 2002). If a particular protein family is widely represented in all eukaryotic supergroups, the most parsimonious scenario points to its presence in the last common ancestor of eukaryotes. This conclusion is reinforced when the phylogenetic tree for the family in question conforms to the topology of the eukaryotic tree. However, none of the best phylogenetic trees for Ago-Piwi, Dicer-like, or RdRP polypeptides strictly coincided with the consensus tree of Eukaryota (Medina 2005) nor could reconstruct the monophyly of some higher-order eukaryotic groups, as previously reported for individual gene/protein trees (Arisue et al. 2004; Philippe et al. 2004). Besides the usual problems of weakness of phylogenetic signal, lateral gene transfers, hidden paralogy, and tree reconstruction artefacts (Philippe et al. 2004), incorrectly predicted protein models (since some sequences were extracted from draft genomes) may have contributed to greater than usual divergences making the relationship among deep branches difficult to resolve.
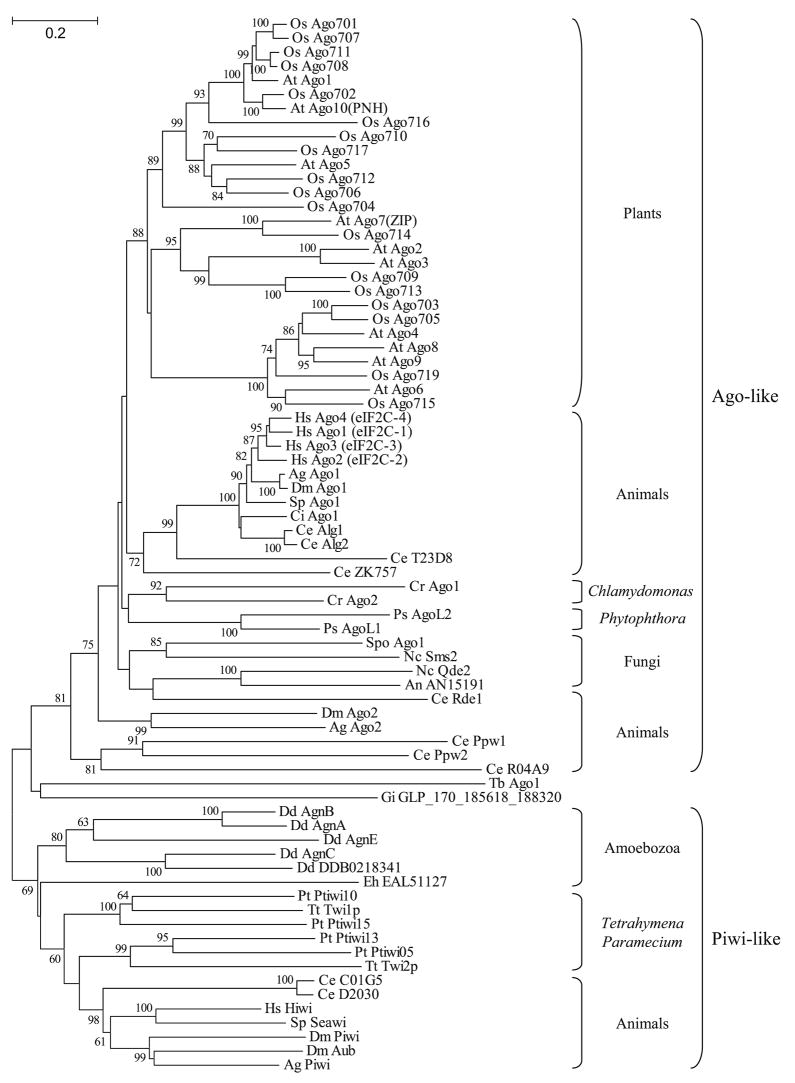
Despite these caveats, the Argonaute-Piwi proteins in present day organisms fell into two relatively well supported, presumably paralogous, groups: the Argonaute-like and the Piwi-like polypeptides (Fig. 2). Fungi (Opisthokonta), green algae and plants (Archaeplastida), and P. sojae (Chromalveolata) appear to encode exclusively Argonaute-like proteins in their genomes. In contrast, Amoebozoa, and T. thermophila and Paramecium tetraurelia (Chromalveolata) seem to encode exclusively Piwi-like proteins. Lastly, animals (Opisthokonta) have representatives of both types of proteins whereas the Excavata sequences (G. intestinalis and T. brucei) could not be reliably resolved in terms of their grouping. A parsimonious interpretation of these data suggests that the last common ancestor of eukaryotes contained both Argonaute-like and Piwi-like proteins and that specific lineages independently lost either one or the other. Only animals appear to have retained both classes of proteins, although this conclusion may need to be reexamined as more sequences from diverse taxonomic groups become available. Interestingly, the Argonaute-Piwi duplication may have preceded the formation of a multidomain, PAZ-containing Dicer protein (see below) since in phylogenetic analyses the PAZ domains of Piwi-like and Dicer-like proteins cluster together whereas the PAZ domains of Argonaute-like proteins behave as an outgroup (data not shown). Thus, domain shuffling from an ancestral Piwi-like gene might have contributed the PAZ motif to Dicer.
Fig. 2.
Neighbor-Joining tree showing the phylogenetic relationship among Argonaute-Piwi proteins. Sequences corresponding to the PAZ and PIWI domains were aligned using the ClustalX program and the tree was drawn using the MEGA 3.1 program. Numbers indicate bootstrap values, as percentage, based on 1,000 pseudoreplicates (only values > 60% are shown). The Ago-like and Piwi-like protein subclasses are shown to the right of the tree. Species are designated by a two-letter abbreviation preceding the name of each protein: Ag A. gambiae; At A. thaliana; An A. nidulans; Ce C. elegans; Cr C. reinhardtii; Ci C. intestinalis; Dd D. discoideum; Dm D. melanogaster; Eh E. histolytica; Gi G. intestinalis; Hs H. sapiens; Nc N. crassa; Os O. sativa; Pt P. tetraurelia; Ps P. sojae; Spo S. pombe; Sp S. purpuratus; Tt T. thermophila; and Tb T. brucei. Accession numbers of proteins used to draw the tree: Ag Ago1 EAA00062; Ag Ago2 EAL41436; Ag Piwi EAA05900; At Ago1 AAC18440; At Ago2 NP_174413; At Ago3 NP_174414; At Ago4 NP_565633; At Ago5 NP_850110; At Ago6 NP_180853; At Ago7(ZIP) NP_177103; At Ago8 NP_197602; At Ago9 NP_197613; At Ago10(PNH) CAA11429; An AN1519 EAA63775; Ce Alg1 NP_510322; Ce Alg2 NP_871992; Ce C01G5 AAB37734; Ce D2030 CAA98113; Ce Ppw1 NP_740835; Ce Ppw2 AAF60414; Ce Rde1 NP_741611; Ce R04A9 NP_508092; Ce T23D8 NP_492643; Ce ZK757 CAB54247; Cr Ago1 v2_C_130206*; Cr Ago2 v2_1200017*; Ci Ago1 C_chr_ 02q000291*; Dd AgnA EAL69296; Dd AgnB EAL62204; Dd AgnC EAL71514; Dd AgnE EAL62770; Dd DDB0218341 EAL66399; Dm Ago1 BAA88078; Dm Ago2 Q9VUQ5; Dm Aub CAA64320; Dm Piwi Q9VKM1; Eh EAL51127 EAL51127; Gi GLP_170_185618_188320 XP_779885; Hs Ago1(eIF2C-1) AAH63275; Hs Ago2(eIF2C-2) AAL76093; Hs Ago3(eIF2C-3) BAB14262; Hs Ago4(eIF2C-4) BAB13393; Hs Hiwi AAC97371; Nc Qde2 AAF43641; Nc Sms2 AAN32951; Os Ago701 XP_468547; Os Ago702 BAB96813; Os Ago703 NP_912975; Os Ago704 XP_478040; Os Ago705 AL606693; Os Ago706 NP_909924; Os Ago707 AP005750; Os Ago708 XP_473529; Os Ago709 XP_473887; Os Ago710 XP_469312; Os Ago711 AP004188; Os Ago712 XP_476934; Os Ago713 XP_473888; Os Ago714 XP_468898; Os Ago715 XP_477327; Os Ago716 XP_464271; Os Ago717 XP_469311; Os Ago719 AP000836; Pt Ptiwi05 CAI44468; Pt Ptiwi10 CAI39070; Pt Ptiwi13 CAI39067; Pt Ptiwi15 CAI39065; Ps AgoL1 v1_C_580101*; Ps AgoL2 v1_C_1000004*; Spo Ago1 O74957; Sp Ago1 XP_782278; Sp Seawi AAG42533; Tt Twi1p AAM77972; Tt Twi2p AAQ74967; Tb Ago1 AAR10810. *Accession numbers according to the annotated draft genomes of C. reinhardtii, C. intestinalis, or P. sojae at http://www.genome.jgi-psf.org/euk_cur1.html
Ago-Piwi proteins have also undergone a marked degree of expansion in certain eukaryotic lineages (Fig. 2), most prominently plants and metazoans, perhaps associated with more extensive diversification of function. In plants, several duplications of Argonaute-like proteins appear to have occurred both before and after the divergence of monocots and dicots, represented by Oryza sativa and A. thaliana, respectively (Fig. 2). Extensive expansion of Argonaute-Piwi proteins has also occurred in the animal lineage. Moreover, in certain species such as C. elegans and D. melanogaster some of these polypeptides are currently so divergent that they do not reconstruct the monophyly of animals (Fig. 2). At least one group of C. elegans Argonaute-like proteins (including PPW1 and PPW2) behaves as paralogous to all other Ago-like polypeptides in animals, fungi, plants, C. reinhardtii, and P. sojae (Fig. 2).
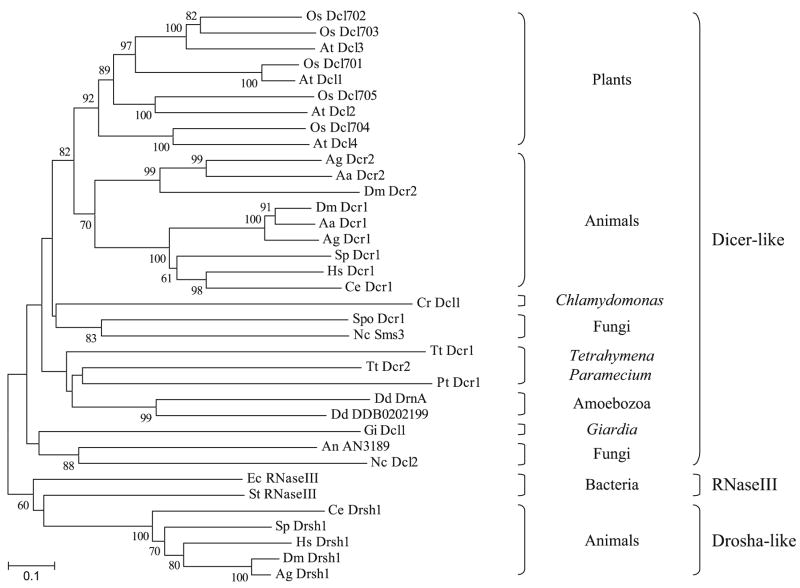
A phylogenetic tree of Dicer-like proteins, constructed based on the alignment of the dual RNaseIII domains, did no allow resolving the relationship among most of these proteins (Fig. 3). This is likely a reflection of the lower sequence (and domain structure) conservation of Dicer-like proteins relative to Ago-Piwi and RdRP polypeptides. However, the animal and plant Dicer-like sequences form a well-supported cluster and appear to be orthologous (Fig. 3). Interestingly, plant Dicer-like sequences underwent significant expansion largely prior to the divergence of monocots and dicots. In contrast, most animals appear to encode a single Dicer sequence, with the exception of insects that contain two. Whereas insect Dcr1 clusters with all other animal Dicers, Dcr2 is much more divergent and forms a paralogous clade (Fig. 3). Intriguingly, insect Ago2 is also much more divergent than Ago1 and does not cluster with most other animal Argonaute-like proteins (the Ago1 clade) (Fig. 2). It remains unclear whether this reflects an ancient RNAi pathway duplication in the animal lineage that was retained only in insects and/or the fast evolution of certain duplicated sequences within the insect lineage. Although a recent report suggests that D. melanogaster Dcr2 and Ago2 are among the fastest evolving genes in this organism, perhaps as a result of a coevolutionary ‘arms race’ with viral pathogens (Obbard et al. 2006).
Fig. 3.
Phylogenetic tree of Dicer-like proteins. Sequences corresponding to the RNaseIIIa and b domains were aligned using the ClustalX program and a Neighbor-Joining tree was constructed and drawn using MEGA 3.1. Numbers show bootstrap values, as percentage, based on 1,000 pseudoreplicates (only values > 60% are shown). The Dicer-like, eubacterial RNaseIII, and Drosha-like protein subclasses are indicated to the right of the tree. Species are designated by a two-letter abbreviation preceding the name of each protein, as shown in the legend to Fig. 2 but also including: Aa Aedes aegypti; Ec Escherichia coli; and St Streptococcus thermophilus. Accession numbers of proteins used to draw the tree: Aa Dcr1 AAW48724; Aa Dcr2 AAW48725; Ag Dcr1 AAO73809; Ag Dcr2 EAA00264; Ag Drsh1 EAL39656; At Dcl1 NP_171612; At Dcl2 NP_566199; At Dcl3 NP_189978; At Dcl4 NP_197532; An AN3189 XP_660793; Ce Dcr1 P34529; Ce Drsh1 NP_492599; Cr Dcl1 v2_C_130110 (at http://www.genome.jgi-psf.org/Chlre3/Chlre3.home.html); Dd DDB0202199 EAL73658; Dd DrnA EAL70472; Dm Dcr1 Q9VCU9; Dm Dcr2 BAB69959; Dm Drsh1 AAD31170; Ec RNaseIII AAC75620; Gi Dcl1 AAO17549; Hs Dcr1 NP_803187; Hs Drsh1 Q9NRR4; Nc Dcl2 CAB91758; Nc Sms3 XP_328976; Os Dcl701 NP_912466; Os Dcl702 XP_463595; Os Dcl703 NP_922059; Os Dcl704 XP_473129; Os Dcl705 XP_463068; Pt Dcr1 CAI44479; Spo Dcr1 Q09884; St RNaseIII AAV62839; Sp Dcr1 XP_790894; Sp Drsh1 XP_790161; Tt Dcr1 BAD34724; Tt Dcr2 BAD34723
A monophyletic origin of animal and plant Dicers is also supported by the comparable domain organization of their sequences (Fig. 1). Moreover, if the eukaryotic tree is truly rooted between unikonts and dikonts, as already discussed, one of the ancestral forms of Dicer may have been similar to the multidomain protein now present in animals and plants. Domain deletion/truncation, domain fusion, as well as sequence divergence could explain the more variable Dicer-like proteins found in other living organisms (which contain various combinations of some of the putative ancestral motifs) (Fig. 1). However, a polyphyletic origin of Dicer-like sequences, and, potentially, the existence of more than one Dicer form in the eukaryotic ancestor, cannot be statistically ruled out (Fig. 3). Interestingly, Drosha, another type of RNaseIII enzyme involved in RNAi via the processing of miRNA precursors in animals (Bartel 2004; Wienholds and Plasterk 2005; Zamore and Haley 2005), is absent from the genome of all other eukaryotes examined (data not shown). Drosha polypeptides form an outgroup with respect to plant and animal Dicers and they seem to be somewhat better related (albeit weakly) to eubacterial RNaseIII enzymes (Fig. 3). Thus, this type of protein may have evolved, independently from Dicer, in the animal lineage.
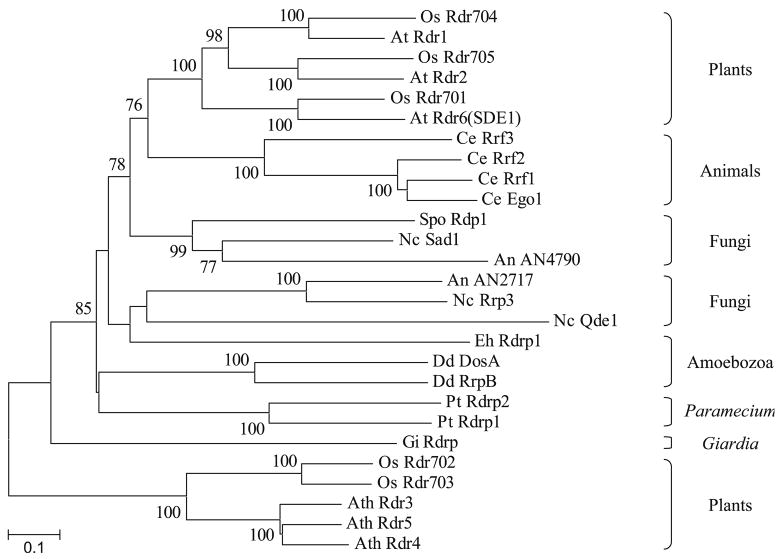
RdRPs are not as widely distributed among eukaryotes as Ago-Piwi and Dicer-like sequences but a phylogenetic tree, constructed by aligning the RdRP domains, supports the monophyletic origin of the proteins found in C. elegans, fungi, Amoebozoa, P. tetraurelia, and a subset of plant RdRPs (Fig. 4). However, the evolutionary relationships among some of these polypeptides as well as the grouping of the G. intestinalis RdRP are not well defined. Besides the already discussed caveats associated with our analyses, the topology of the RdRP tree might also be affected by more prevalent lineage-specific losses of some of these proteins. For instance, there is experimental evidence that A. nidulans has lost, via DNA sequence degeneration, the putative ortholog of the N. crassa RdRP Qde-1 (Hammond and Keller 2005). Intriguingly, plants also contain a subset of RdRPs (including A. thaliana Rdr3, Rdr4, and Rdr5) that behaves as an outgroup to all other RdRPs (Fig. 4). Thus, a parsimonious interpretation of the data is consistent with the existence of at least one RdRP in the common eukaryotic ancestor, but the origin of the subset of more divergent plant RdRPs remains uncertain.
Fig. 4.
Phylogenetic tree of eukaryotic RNA-dependent RNA polymerases. An alignment of sequences corresponding to the RdRP domain was used to construct a Neighbor-Joining tree and its robustness was assessed by a bootstrap test based on 1,000 pseudoreplicates. Bootstrap values higher than 60% are shown on the nodes of the tree. Species are indicated by a two-letter abbreviation preceding the name of each protein, as shown in the legend to Fig. 2. Accession numbers of proteins used to draw the tree: At Rdr1 NP_172932; At Rdr2 NP_192851; At Rdr3 NP_179581; At Rdr4 NP_179582; At Rdr5 NP_179583; At Rdr6(SDE1) NP_190519; An AN2717 XP_660321; An AN4790 XP_662394; Ce Ego1 NP_492132; Ce Rrf1 AAF80368; Ce Rrf2 NP_493057; Ce Rrf3 NP_495713; Dd DosA AAD29638; Dd RrpB CAC41975; Eh Rdrp1 EAL45936; Gi Rdrp AAK97084; Nc Qde1 CAB42634; Nc Rrp3 CAD70515; Nc Sad1 AAK31733; Os Rdr701 NP_918046; Os Rdr702 NP_913147; Os Rdr703 NP_913148; Os Rdr704 AP004880; Os Rdr705 XP_472792; Pt Rdrp1 CAI39057; Pt Rdrp2 CAI39056; Spo Rdp1 O14227
Nature of the ancestral RNAi machinery
RNAi-related phenomena occur in all eukaryotic supergroups (with the possible exception of Rhizaria that remains to be sampled) (Table 1) and phylogenetic analyses suggest that the main components of the RNAi machinery can be traced back to the common ancestor of eukaryotes (Figs. 2, 3, 4). Moreover, several motifs typical of RNAi effectors such as the RdRP (which differs from viral RdRPs), PAZ, and DUF283 domains are restricted to the eukaryotic lineage and have been postulated to be eukaryotic innovations (Anantharaman et al. 2002; Wassenegger and Krczal 2006). Though a somewhat divergent PAZ-like structural fold, lacking primary sequence conservation with the eukaryotic PAZ domain, also occurs in some archaeal and eubacterial proteins (Song et al. 2004; Yuan et al. 2005). The remaining domains in Argonaute-Piwi and Dicer-like proteins such as the PIWI, the SFII RNA helicase, the RNaseIII, and the DSRM motifs appear to have originated in prokaryotic lineages prior to the divergence of eukaryotes (Anantharaman et al. 2002). Thus, the innovative evolution of certain domains as well as the fusion of diverse functional motifs in order to generate Argonaute-Piwi, Dicer-like, and RdRP proteins might have occurred ancestrally in the eukaryotic lineage.
Interestingly, the last common ancestor of living eukaryotes appears to have been a ‘complete’ eukaryotic cell (Richards and Cavalier-Smith 2005; Simpson and Roger 2004). It had a nucleus, endoplasmic reticulum, and Golgi apparatus, and underwent mitosis and meiosis (Ramesh et al. 2005; Simpson and Roger 2004). It also had an endosymbiont-derived mitochondrion, a cilium and centriole, and a complex eukaryotic cytoskeleton (Richards and Cavalier-Smith 2005). The only major eukaryotic features that appear to be of later origin are plastids (Simpson and Roger 2004). Given this complexity it is perhaps not surprising that the ancestral RNAi machinery may have already been well developed. Our analyses (see above) suggest that the last common ancestor of eukaryotes had, at a minimum, one Argonaute-like polypeptide, one Piwi-like protein, one Dicer, and one RdRP. Further, since in the case of Dicer the reconstructed phylogeny did not resolve the relationship among most of these polypeptides (Fig. 3), the existence of Dicer-like paralogs in the last common ancestor of eukaryotes cannot be ruled out.
The function(s) of this ancestral RNAi machinery is unknown. However, several considerations may provide a framework for hypotheses about its possible role(s). First, the ancestral RNAi function(s) very likely was not essential for life since the machinery appears to have been lost from a variety of taxonomically divergent eukaryotes, such as S. cerevisiae, T. cruzi, L. major, C. merolae, and P. falciparum (Table 1). The alternative explanation, that in these species an essential RNAi role(s) was compensated by another mechanism, would require the independent evolution of the latter multiple times. Moreover, in organisms that contain a single Dicer gene such as S. pombe and vertebrates, Dicer-null mutants are RNAi-defective but viable at the cellular level (Giraldez et al. 2005; Kanellopoulou et al. 2005; Martienssen et al. 2005; Murchison et al. 2005; Volpe et al. 2002). Though these mutants commonly show reactivation of transposons and/or repetitive sequences, deficient (hetero)chromatin formation, and/or abnormal chromosome segregation; and vertebrate germ cells fail to differentiate (Fukagawa et al. 2004; Kanellopoulou et al. 2005; Martienssen et al. 2005; Murchison et al. 2005). Second, if a particular function is now widely represented in all examined eukaryotic supergroups, the most parsimonious scenario would suggests that it was already operative in the last common ancestor of eukaryotes. Interestingly, two RNA-mediated processes appear to be widespread among living eukaryotes: the post-transcriptional degradation of cognate RNAs and the transcriptional repression of homologous DNA sequences (Table 2). Third, the existence of duplicated Argonaute-Piwi proteins in the last common ancestor of eukaryotes (Fig. 2) suggests some degree of functional diversification tracing back to the ancestral RNAi machinery (since completely redundant genes are unlikely to be evolutionarily conserved; Ohno 1970; Force et al. 1999; Lynch and Conery 2000; Moore and Purugganan 2005; Presgraves 2004).
In present day eukaryotes both Argonaute-like proteins and Piwi-like proteins have been implicated in a variety of RNAi-related phenomena and several subfamily members appear to have distinct functions (Grishok et al. 2001; Lee et al. 2003; Okamura et al. 2004). For instance, a Piwi-like protein, Twi1, is required for the conjugation-induced accumulation of small RNAs involved in programmed DNA elimination in T. thermophila (Lee and Collins 2006; Liu et al. 2004b). The D. melanogaster Piwi-like genes Aubergine and Piwi have been implicated in both transcriptional, partly associated with histone H3 lysine 9 (H3K9) methylation, and post-transcriptional gene silencing (Aravin et al. 2004; Kavi et al. 2005; Pal-Bhadra et al. 2004). Members of the Argonaute-like class of proteins have been identified as key components of the RISC (Hammond et al. 2001; Rivas et al. 2005; Zamore and Haley 2005). In humans, hAgo2 was shown to be responsible for target RNA cleavage whereas other Argonaute subfamily members (hAgo1, hAgo3, and hAgo4) can associate with both siRNAs and miRNAs but do not mediate cleavage (Liu et al. 2004a; Meister et al. 2004; Rivas et al. 2005). In D. melanogaster, both Ago2 and Ago1 have slicer functions but Ago2 is predominantly involved in siRNA-directed target RNA cleavage whereas Ago1 is necessary for miRNA-directed target RNA cleavage (Miyoshi et al. 2005; Okamura et al. 2004). In plants, specific members of the Argonaute subfamily also show distinct activities. Arabidopsis Ago4 has been implicated in DNA and histone methylation whereas Ago1 is an RNA slicer and much more pleotropic in its functions (Baumberger and Baulcombe 2005; Matzke and Birchler 2005; Qi et al. 2005; Zilberman et al. 2003). Interestingly, in organisms that encode a single Ago-Piwi protein, such as S. pombe and T. brucei, its mutation results in defects in both transcriptional and post-transcriptional silencing (Shi et al. 2004; Sigova et al. 2004).
Given the above constraints, it seems reasonable to hypothesize, as previously suggested, an ancestral (but non-essential) role of the RNAi machinery in defense responses against genomic parasites such as transposable elements and viruses (Baulcombe 2004; Buchon and Vaury 2006; Li and Ding 2005; Matzke and Birchler 2005; Plasterk 2002; Waterhouse et al. 2001). This function has been widely conserved throughout the eukaryotic spectrum (Table 1). Moreover, barring a sampling bias given the relatively small number of taxonomically diverse eukaryotic genomes currently available, all known species that have lost the RNAi machinery are unicellular and possess relatively small genomes. These organisms may be affected by a limited number of genomic parasites and likely have alternative means to control them. Indeed, even in RNAi-positive eukaryotes, partly redundant, RNAi-independent pathways are also involved in the silencing of transposons and other repetitive sequences (Chicas et al. 2004; Jeong et al. 2002; Kuhlmann et al. 2005; Lippman et al. 2003; Lippman and Martienssen 2004; Martienssen et al. 2005; Robert et al. 2005; Tran et al. 2005; van Dijk et al. 2006; Yamada et al. 2005). From a mechanistic standpoint, the ancestral RNAi machinery may have been capable of both siRNA-guided transcript degradation as well as siRNA-guided transcriptional repression of homologous sequences. Again, both roles appear to be widespread among living eukaryotes (Table 2) and this diversification of function could account for the ancestral conservation of duplicated Argonaute-Piwi proteins. It is tempting to speculate that one Ago-Piwi protein might have been predominantly located in the nucleus, as D. melanogaster Piwi (Cox et al. 2000), and perhaps involved in transcriptional silencing whereas another Ago-Piwi protein might have been preferentially located in the cytoplasm, as D. melanogaster Ago2 (Findley et al. 2003; Rehwinkel et al. 2005), and perhaps involved in post-transcriptional silencing. Further roles of the ancestral RNAi machinery are certainly possible but many known RNA-mediated silencing processes show a limited phyletic distribution in present day eukaryotes (Table 2) and may have evolved independently in specific lineages (see below).
Additional (derived?) functions of the RNAi machinery
In Table 2 we have examined the taxonomic distribution of six, experimentally supported, RNA-mediated silencing pathways: dsRNA-induced RNAi, RNAi-mediated (hetero)chromatin formation, RNA-directed DNA methylation, programmed genome rearrangements (DNA elimination), meiotic silencing by unpaired DNA, and miRNA-mediated gene regulation. As discussed above, (degradative) dsRNA-induced RNAi appears to be widespread (Table 2) and likely one of the ancestral functions of the RNAi machinery. However, the sources of long dsRNA are quite variable resulting, in different species, in the silencing of diverse sequences from genomic parasites and repetitive DNA to specific genes (Baulcombe 2004; Chicas et al. 2004; Kavi et al. 2005; Matzke and Birchler 2005; Nakayashiki 2005; Yoshikawa et al. 2005; Zamore and Haley 2005).
The proposed ancestral role of the RNAi machinery in transcriptional gene silencing could have involved siRNA-mediated targeting of chromatin modifications and/or cytosine DNA methylation. RNAi-dependent transcriptional silencing has been demonstrated to entail histone modifications, such as H3K9 methylation, in eukaryotes belonging to at least three different supergroups (Table 2). Moreover, siRNA-triggered transcriptional repression occurs in organisms that lack cytosine DNA methylation such as S. pombe and C. elegans (Grishok et al. 2005; Martienssen et al. 2005; Ponger and Li 2005; Robert et al. 2005; Volpe et al. 2002) and in organisms with very limited DNA methylation such as T. brucei and D. melanogaster (Kavi et al. 2005; Pal-Bhadra et al. 2004; Ponger and Li 2005; Shi et al. 2004; Ullu et al. 2004). In contrast, RNA-directed DNA methylation has, thus far, only been demonstrated in plants and mammals (Kawasaki and Taira 2004; Matzke and Birchler 2005; Morris et al. 2004) and the role of DNA methylation in this type of gene silencing is somewhat debatable in mammals (Ting et al. 2005; Weinberg et al. 2006). Further, several A. thaliana proteins needed for this process, such as the RdRP Rdr2 and subunits of RNA polymerase IV, do not have mammalian counterparts (Chan et al. 2004; Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Xie et al. 2004). Thus, the ancestral RNAi machinery very likely had the capability to target histone modifications, given the widespread phyletic distribution of this function. Conversely, RNA-directed DNA methylation might have arisen independently in specific eukaryotic lineages. Alternatively, if RNA-directed DNA methylation did evolve in the last common ancestor of eukaryotes it appears to have been lost from many lineages and the molecular effectors now differ substantially between plants and mammals.
Interestingly, the mechanism(s) of RNAi-mediated (hetero)chromatin formation also appears to have diverged in present day eukaryotes since, for instance, an RdRP has been implicated in this process in S. pombe (Martienssen et al. 2005; Verdel et al. 2004; Volpe et al. 2002) but it occurs in the absence of RdRPs in D. melanogaster and vertebrates (Fukagawa et al. 2004; Kanellopoulou et al. 2005; Kavi et al. 2005; Pal-Bhadra et al. 2004; Ting et al. 2005; Weinberg et al. 2006). Adding to this complexity, cytosine DNA methylation and histone modifications seem to be interconnected in self-reinforcing feedback loops in higher eukaryotes (Fuks 2005), although the role (if any) of the RNAi machinery in this cycle is not clear. Further, in S. pombe and chicken DT40 cells RNAi-mediated (hetero)chromatin formation may now play a critical role in determining chromosome structure and function during mitosis and/or meiosis (Fukagawa et al. 2004; Martienssen et al. 2005; Wong and Choo 2004). Conversely, in other organisms such as N. crassa the RNAi machinery appears to be dispensable for the methylation of both DNA and H3K9 associated with repetitive sequences (Chicas et al. 2004; Freitag et al. 2004), whereas in mouse there might be cell-type-specific differences in the mechanism(s) of (hetero)chromatin formation (Kanellopoulou et al. 2005; Murchison et al. 2005). Indeed, RNAi-independent pathways for (hetero)chromatin formation and DNA methylation appear to exist in several RNAi-positive eukaryotes (Chicas et al. 2004; Freitag et al. 2004; Goll and Bestor 2005; Jia et al. 2004; Kaller et al. 2006; Laayoun and Smith 1995; Yamada et al. 2005). It remains uncertain to what extent this functional diversity was already present in the unicellular eukaryotic ancestor.
RNAi has also been implicated in the programmed excision of excess DNA in ciliated protozoa such as T. thermophila and P. tetraurelia (Garnier et al. 2004; Mochizuki and Gorovsky 2005; Nowacki et al. 2005; Yao and Chao 2005). This process requires small RNAs, termed scan RNAs, and components of the RNA machinery that direct H3K9 methylation of the chromatin associated with the sequences to be deleted (Garnier et al. 2004; Liu et al. 2004b; Mochizuki and Gorovsky 2005; Yao and Chao 2005). Many of the eliminated sequences appear to be derived from transposons (Lee and Collins 2006; Yao and Chao 2005) and RNA-mediated DNA elimination may have evolved as an extension of the role of the RNAi machinery in the transcriptional silencing of transposon/repetitive sequences. Interestingly, DNA diminution phenomena have also been observed in Ascaris worms and in some species of crustaceans and fish, although it is not known whether these processes are RNAi mediated. Based on this phyletic pattern, Yao and Chao (2005) have recently proposed that programmed genome rearrangements may have arisen by the independent evolution in some eukaryotic lineages of a final (yet uncharacterized) RNAi step, elimination of the (hetero)chromatin induced by small RNAs.
In N. crassa, as a zygotic cell undergoes meiosis (which involves pairing of homologous chromosomes), the presence of an unpaired copy of a gene triggers silencing of all homologous sequences in the genome. This phenomenon has been termed meiotic silencing by unpaired DNA (MSUD) and shown to require an RdRP (Sad1) and an Argonaute-like protein (Lee et al. 2003; Shiu et al. 2001). MSUD also requires RNA production from the unpaired DNA sequence and these transcripts are presumable used as a template by Sad1 to synthesize dsRNA that enters the degradative RNAi pathway (Lee et al. 2004; Matzke and Birchler 2005; Nakayashiki 2005). Thus, even though MSUD originates in the nucleus, it ultimately seems to be a post-transcriptional process that does not involve detectable chromatin alterations at the target locus (Matzke and Birchler 2005; Nakayashiki 2005). MSUD-like phenomena have also been observed in mouse and C. elegans (Maine et al. 2005; Turner et al. 2005). However, both of these processes involve chromatin modifications and transcriptional repression of the unpaired loci. Moreover, (hetero)chromatin formation on unpaired DNA in C. elegans requires the RdRP Ego1 but occurs in the absence of several other RNAi pathway components (Maine et al. 2005). Thus, given its more limited taxonomic distribution and the mechanistic differences in various species, the silencing of unpaired DNA during meiosis is likely a derived, more recently evolved function of RNAi.
The RNAi machinery also plays an important role in gene regulation via microRNAs. However, miRNAs have, thus far, only been identified in multicellular plants and animals (Table 2). They appear to be absent from several unicellular eukaryotes such as S. pombe and T. brucei, where extensive libraries of small RNAs have been sequenced (Djikeng et al. 2001; Reinhart and Bartel 2002), and no miRNA-directed silencing pathway has been documented in fungi (Nakayashiki 2005). In contrast, miRNAs are essential for the development of animals and plants (Bartel 2004; Chen 2005; Kidner and Martienssen 2005; Wienholds and Plasterk 2005). For instance Dicer-deficient vertebrate germ cells are viable but they fail to differentiate (Giraldez et al. 2005; Kanellopoulou et al. 2005; Murchison et al. 2005). Moreover, Dicer is required for morphogenesis (but not for cell fate specification) during zebrafish embryogenesis, and the absence of miRNAs is responsible, at least in part, for this phenotype (Giraldez et al. 2005). Similarly, null alleles of Dicer-like1, necessary for the generation of mature miRNAs, result in embryo lethality in A. thaliana (Chen 2005; Ray et al. 1996; Kidner and Martienssen 2005).
Despite these similarities, plant and animal miRNA pathways vary in multiple aspects (Bartel 2004; Llave et al. 2002; Reinhart et al. 2002). In animals, miRNAs are initially transcribed into long precursor transcripts that are processed to mature miRNAs in a series of steps involving two RNaseIII-like enzymes, Drosha and Dicer (Bartel 2004; Wienholds and Plasterk 2005). Plants lack a Drosha homolog and the production of miRNAs from precursor RNAs appears to be carried out by a single Dicer-like protein (Chen 2005; Kurihara and Watanabe 2004; Millar and Waterhouse 2005). Plant miRNAs are also methylated on the ribose of the last nucleotide, a modification presumably involved in protecting miRNAs from 3′ end uridylation and degradation, whereas animal miRNAs do not appear to be modified (Chen 2005; Li et al. 2005; Yu et al. 2005). Mechanistically, most animal miRNAs are only partly complementary to their targets and mediate silencing primarily by translational repression, although localization to the processing bodies may also affect RNA stability (Humphreys et al. 2005; Liu et al. 2005b; Pillai et al. 2005; Wienholds and Plasterk 2005; Zamore and Haley 2005). In contrast, most plant miRNAs have near-perfect complementarity to their targets and trigger predominantly mRNA cleavage (Carrington and Ambros 2003; Chen 2005; Schwab et al. 2005). These differences, the lack of conservation of particular miRNA genes between plant and animals, and their absence from many eukaryotes suggest that the miRNA pathway may have evolved independently in the lineages leading to multicellular plants and animals (Bartel 2004; Millar and Waterhouse 2005; Wienholds and Plasterk 2005). Conceivably, the appearance of miRNAs may have played a role in the evolution of organisms with complex body patterns (Bartel 2004; Millar and Waterhouse 2005; Wienholds and Plasterk 2005).
Summary and perspective
Double-stranded RNA has been demonstrated to trigger gene silencing in eukaryotes, linking a variety of apparently dissimilar phenomena with RNA interference in animals. Biochemical and genetic studies have led to the identification of three conserved components of the RNAi machinery, namely Dicer, Argonaute-Piwi, and RdRP. We have analyzed the taxonomic distribution and the phylogenetic relationship of these proteins with the goal of inferring the composition and function(s) of the RNAi machinery in the last common ancestor of eukaryotes. This ancestral RNAi machinery likely consisted of, at least, one Argonaute-like polypeptide, one Piwi-like protein, one Dicer, and one RNA-dependent RNA polymerase. The original role of these components may have been non-essential for unicellular life, although important for defense responses against genomic parasites such as transposable elements and viruses. In fact the RNAi machinery in the eukaryotic ancestor may have been able to target both transcript degradation as well as locus-specific histone modifications, resulting in the inactivation of extra-chromosomal and genome integrated parasitic sequences. Other known RNAi-mediated processes show a limited taxonomic distribution in living eukaryotes and may have evolved more recently in specific lineages. RNA-directed DNA methylation, DNA elimination, and meiotic silencing by unpaired DNA possibly arose as an extension of the RNAi machinery role in controlling transposons and retroviruses. In contrast, miRNAs and several kinds of endogenous siRNAs appear to be flexible innovations that allowed gathering the selectivity of RNAi for the regulation of gene expression.
Although the ancestral RNAi machinery seems to have been fairly complex, a considerable degree of functional diversification as well as integration with RNAi-independent pathways for (hetero)chromatin formation and DNA methylation seem to have occurred during eukaryotic evolution. Moreover, the great expansion of RNAi components, particularly Ago-Piwi proteins, in present day plants and animals suggests the possibility of further, still unrecognized, pathway specialization. Much remains to be learned about the extent of subfunctionalization, neofunctionalization, and partial redundancy of gene family members. In addition, the regulation of gene expression by miRNAs may be a relatively easy innovation, particularly when triggering transcript cleavage as in plants. As recently proposed (Allen et al. 2004; Smalheiser and Torvik 2005), at least some miRNA genes may arise from inverted duplications of target gene sequences and the initially produced double stranded foldback transcripts may operate as regulators via the degradative RNAi pathway. Progressive sequence degradation, under selective pressure, may eventually result in the bulged structure typical of miRNA precursors. From this perspective, it seems reasonable to expect that miRNAs will be found in additional eukaryotic lineages, particularly in organisms with complex genomes where new repeats are tolerated and where the regulation conferred by miRNAs provides a selective advantage. Conversely, the RNAi machinery seems to have been entirely lost or extensively simplified in a number of unicellular eukaryotes with small genomes. In the latter, the presence of a recognizable Argonaute-Piwi protein appears to be diagnostic of a functional RNAi pathway, whereas Dicer-like proteins are less conserved and RdRPs may be absent.
Acknowledgments
We are grateful to members of the Cerutti lab for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM62915).
Footnotes
Communicated by R. Bock
References
- Abed M, Ankri S. Molecular characterization of Entamoeba histolytica RNase III and AGO2, two RNA interference hallmark proteins. Exp Parasitol. 2005;110:265–269. doi: 10.1016/j.exppara.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MFJR. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during transacting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- An CI, Sawada A, Fukusaki E, Kobayashi A. A transient RNA interference assay system using Arabidopsis protoplasts. Biosci Biotechnol Biochem. 2003;67:2674–2677. doi: 10.1271/bbb.67.2674. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisue N, Hasegawa M, Hashimoto T. Root of the Eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Mol Biol Evol. 2004;22:409–420. doi: 10.1093/molbev/msi023. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:282–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Buchon N, Vaury C. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity. 2006;96:195–202. doi: 10.1038/sj.hdy.6800789. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chen X. microRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Cogoni C, Macino G. RNAi-dependent and RNAi-independent mechanisms contribute to the silending of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 2004;32:4237–4243. doi: 10.1093/nar/gkh764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Forrest EC, Sepich S, Cogoni C, Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for tandem repeat stabilization in Neurospora crassa. Mol Cell Biol. 2005;25:3793–3801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- DaRocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Bastin P. TbAGO1, an Argonaute protein required for RNA interference is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003;1:2. doi: 10.1186/1741-7007-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Force a, Lynch M, Pickett B, Amores A, Yan Y-L. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Lee DW, Kothe GO, Pratt RJ, Aramayo R, Selker EU. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesellchen V, Boutros M. Managing the genome: microRNAs in Drosophila. Differentiation. 2004;72:74–80. doi: 10.1111/j.1432-0436.2004.07202003.x. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Grishok A, Mello CC. RNAi (Nematodes: Caenorhabditis elegans) Adv Genet. 2002;46:339–360. doi: 10.1016/s0065-2660(02)46012-9. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hammond TM, Keller NP. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics. 2005;169:607–617. doi: 10.1534/genetics.104.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hoa NT, Keene KM, Olson KE, Zheng L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem Mol Biol. 2003;33:949–957. doi: 10.1016/s0965-1748(03)00101-2. [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DIK, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B-R, Wu-Scharf D, Zhang C, Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc Natl Acad Sci USA. 2002;99:1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SIS. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1975. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Kaller M, Euteneuer U, Nellen W. Differential effects of Heterochromatin Protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum. Eukaryot Cell. 2006;5:530–543. doi: 10.1128/EC.5.3.530-543.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kaur G, Lohia A. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem Biophys Res Commun. 2004;320:1118–1122. doi: 10.1016/j.bbrc.2004.06.064. [DOI] [PubMed] [Google Scholar]
- Kavi HH, Fernandez HR, Xie W, Birchler JA. RNA silencing in Drosophila. FEBS Lett. 2005;579:5940–5949. doi: 10.1016/j.febslet.2005.08.069. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. The developmental role of miRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Söderbom F, Hammann C, Nellen W. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005;19:6405–6417. doi: 10.1093/nar/gki952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laayoun A, Smith SS. Methylation of slipped duplexes, snapbacks and cruciforms by human DNA (cytosine-5) methyltransferase. Nucleic Acids Res. 1995;23:1584–1589. doi: 10.1093/nar/23.9.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel gene coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Pratt RJ, McLauhlin M, Aramayo R. An Argonaute-like protein is required for meiotic silencing. Genetics. 2003;164:821–828. doi: 10.1093/genetics/164.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Seong K-Y, Pratt RJ, Baker K, Aramayo R. Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics. 2004;167:131–150. doi: 10.1534/genetics.167.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-W, Ding S-W. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005a;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004a;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005b;7:1161–1166. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004b;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of the guide RNA strand by A. fulgidus PIWI protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Maine EM, Hauth J, Ratliff T, Vought VE, She X, Kelly WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc Natl Acad Sci USA. 2005;102:6630–6635. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Millar AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005;5:129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K, Kim K, Sciulli C, Dowd SR, Minden JS, Carthew RW. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci USA. 2005;102:12023–12028. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Zagorski-Ostoja W, Meyer E. Nowa1p and Nowa2p: Novel putative RNA binding proteins involved in transnuclear crosstalk in Paramecium tetraurelia. Curr Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer; Berlin Heidelberg New York: 1970. p. 150. [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–93. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Lurent J, Moreira D, Muller M, Le Guyader H. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Philos Trans R Soc Lond B Biol Sci. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Piccin A, Salameh A, Benna C, Sandrelli F, Mazzota G, Zordan M, Rosato E, Kyriacou CP, Costa R. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 2001;29:E55. doi: 10.1093/nar/29.12.e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus cG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translation initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- Ponger L, Li W-H. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol. 2005;22:1119–1128. doi: 10.1093/molbev/msi098. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. Evolutionary Genomics: new genes for new jobs. Curr Biol. 2004;15:R52–R53. doi: 10.1016/j.cub.2004.12.053. [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM., Jr A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wand X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Ray A, Lang JD, Golden T, Ray S. SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development. 1996;122:2631–2538. doi: 10.1242/dev.122.9.2631. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- Rohr J, Sarkar N, Balenger S, Jeong B-R, Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- Schroda M. RNA silencing in Chlamydomonas: mechanisms and tools. Curr Genet. 2006;49:69–84. doi: 10.1007/s00294-005-0042-1. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- Shi h, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359–2367. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Stein P, Svoboda P, Anger M, Schultz RM. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA. 2003;9:187–192. doi: 10.1261/rna.2860603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Girke T, Zhu JK. Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res. 2005;33:4443–4454. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Samuels V, Whitley N, DeLaGarza T, Newton RJ. Post-transcriptional gene silencing induced by short interfering RNAs in cultured transgenic plant cells. Genomic Proteomics Bioinformatics. 2004;2:97–108. doi: 10.1016/S1672-0229(04)02015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, Delrow J, Boyle T, Kwong S, Bryson TD, Jacobsen SE, Henikoff S. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 2005;6:R90. doi: 10.1186/gb-2005-6-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Ullu E, Lujan HD, Tschudi C. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell. 2005;4:1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk K, Xu H, Cerutti H. Epigenetic silencing of transposons in the green alga Chlamydomonas reinhardtii. In: Nellen W, Hammann C, editors. Small RNAs—analysis and regulatory functions. Springer; Berlin Heidelberg New York: 2006. pp. 159–178. [Google Scholar]
- Vayssie L, Vargas M, Weber C, Guillen N. Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol. 2004;138:21–28. doi: 10.1016/j.molbiopara.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Watson JM, Fusaro AF, Wang M, Waterhouse PM. RNA silencing platforms in plants. FEBS Lett. 2005;579:5982–5987. doi: 10.1016/j.febslet.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen Z, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Avrova AO, van West P, Jones JT. A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans. Mol Plant Path. 2005;6:153–163. doi: 10.1111/j.1364-3703.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- Wong LH, Choo KHA. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004;20:611–616. doi: 10.1016/j.tig.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RHA. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of transacting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-R, Pei Y, Ma J-B, Kuryavyi V, Zhadina M, Meister G, Chen H-Y, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNaseIII. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]