Abstract
The TATA-binding protein (TBP) is critical for transcription by all three nuclear RNA polymerases. In order to identify factors that interact with TBP, the nonnatural photoreactive amino acid ρ-benzoyl-phenylalanine (BPA) was substituted onto the surface of Saccharomyces cerevisiae TBP in vivo. Cross-linking of these TBP derivatives in isolated transcription preinitiation complexes or in living cells reveals physical interactions of TBP with transcriptional coregulator subunits and with the general transcription factor TFIIA. Importantly, the results show a direct interaction between TBP and the SAGA coactivator subunits Spt3 and Spt8. Mutations on the Spt3-interacting surface of TBP significantly reduce the interaction of TBP with SAGA, show a corresponding decrease in transcription activation, and fail to recruit TBP to a SAGA-dependent promoter, demonstrating that the direct interaction of these factors is important for activated transcription. These results prove a key prediction of the model for stimulation of transcription at SAGA-dependent genes via Spt3. Our cross-linking data also significantly extend the known surfaces of TBP that directly interact with the transcriptional regulator Mot1 and the general transcription factor TFIIA.
Keywords: SAGA, TATA-binding protein, transcription activation
Transcription of mRNA in eukaryotes is a highly regulated process that requires the sequential and coordinated assembly of general transcription factors, coregulators, and RNA Polymerase II at promoters to form a preinitiation complex (PIC). Central to the formation of the PIC at TATA-containing promoters is the binding of TATA-binding protein (TBP) to the TATA element (Hahn 2004; Thomas and Chiang 2006). Since TBP plays a critical role in transcription, considerable effort has been made to determine which proteins interact with and control the entry of TBP into the PIC. Studies have identified a broad range of candidates including general transcription factors, activators, and coactivator proteins (Lee and Young 1998).
The best-characterized TBP interactions are those between TBP and the general transcription factors TFIIA and TFIIB, which form a complex with TBP–DNA. High-resolution crystallographic studies as well as biochemical experiments with purified proteins have mapped these interactions on TBP (Kim et al. 1993; Nikolov et al. 1995; Bryant et al. 1996; Geiger et al. 1996; Tan et al. 1996; Tang et al. 1996). The negative regulatory complex NC2 is thought to act in part by physically precluding TFIIA and TFIIB binding to TBP, thereby interfering with PIC formation (Inostroza et al. 1992; Goppelt and Meisterernst 1996; Goppelt et al. 1996). The structure of NC2–TBP–DNA has been solved by X-ray crystallography, showing how NC2 binds TBP–DNA and blocks the binding of TFIIB (Kamada et al. 2001). Mot1, another well-characterized negative regulator of TBP, uses energy from ATP hydrolysis to dissociate TBP from promoter DNA (Auble et al. 1994; Sprouse et al. 2006). The interaction of Mot1 with TBP has been mapped to the C-terminal helices on the TBP saddle surface. Both NC2 and Mot1 also have positive roles at many yeast promoters, although the mechanism by which they positively control transcription is still under study (Willy et al. 2000; Creton et al. 2002; Dasgupta et al. 2002; Geisberg and Struhl 2004a, b; Schluesche et al. 2007).
In Saccharomyces cerevisiae, the coactivator complexes TFIID and SAGA are critical for TBP recruitment (Dudley et al. 1999b; Kuras et al. 2000; Li et al. 2000; Bhaumik and Green 2002). While yeast TFIID tends to regulate promoters of “housekeeping” genes, yeast SAGA typically acts at highly regulated genes that are modulated by stress (Lee et al. 2000; Huisinga and Pugh 2004). SAGA is a multisubunit complex that is directly recruited to promoters by activators (Utley et al. 1998; Bhaumik and Green 2001; Brown et al. 2001; Larschan and Winston 2001; Fishburn et al. 2005) and was originally identified as a histone acetyltransferase (HAT) complex containing the HAT subunit Gcn5 (Grant et al. 1997). Importantly, many genes that require SAGA for transcriptional activation do not require Gcn5 activity, demonstrating that SAGA has a HAT-independent coactivator function (Dudley et al. 1999b; Sterner et al. 1999; Lee et al. 2000). Chromatin immunoprecipitation (ChIP) studies have revealed that the Spt3 and Spt8 subunits of SAGA are necessary for TBP binding at several SAGA-dependent promoters (Bhaumik and Green 2002). Spt3 and Spt8 have been shown to interact genetically with TBP (Eisenmann et al. 1992; Laprade et al. 2007), but Spt8 and Ada1 are the only known SAGA subunits that physically interact and/or cross-link with recombinant TBP in vitro (Madison and Winston 1997; Sterner et al. 1999; Warfield et al. 2004; Sermwittayawong and Tan 2006). However, it has also been proposed that Spt8 interacts with the DNA-binding surface of TBP and that this interaction does not occur when TBP is bound to DNA (Sermwittayawong and Tan 2006), leaving unresolved the mechanism of SAGA–TBP interaction in functional transcription complexes. Biochemical studies have so far failed to show a direct interaction between Spt3 and TBP (Madison and Winston 1997; Sterner et al. 1999; Sermwittayawong and Tan 2006), a key feature of previously proposed models linking SAGA and TBP.
TBP is also associated with TAFs (TBP-associated factors) that are required for TBP recruitment to TFIID-dependent promoters (Dynlacht et al. 1991; Reese et al. 1994; Poon et al. 1995), although it is not precisely known how many of the TAFs contact TBP in this complex. Taf1 can assemble with TBP on DNA in a purified system (Chen et al. 1994). Taf1 also contains an N-terminal domain (TAND) that binds the TBP DNA-binding surface in vitro, and this interaction is blocked upon TBP binding to DNA (Kokubo et al. 1998). It is likely that both the activator and the promoter sequence dictate whether SAGA and/or TFIID are utilized for TBP recruitment to specific promoters (Cheng et al. 2002; Li et al. 2002; Mencia et al. 2002).
In this work, we inserted a nonnatural photoreactive amino acid at specific positions on the surface of yeast TBP to identify and map protein–protein interactions within the transcription machinery. This approach allows the mapping of interactions both in vivo and in vitro, in the context of isolated PICs. Importantly, our results revealed a direct interaction between TBP and the SAGA subunit Spt3. Mutations constructed on the Spt3-interacting surface of TBP strongly reduce the physical interaction of TBP and SAGA, the level of transcription activation observed in vivo and in vitro, and TBP occupancy at the GAL1 promoter in vivo, showing a direct functional interaction between these two factors. We also found multiple additional factors cross-linking to TBP at a SAGA-dependent promoter, and we further extend the known TBP surfaces that interact with the transcriptional repressor Mot1 and with the general transcription factor TFIIA.
Results
Incorporation of the photo-cross-linker ρ-benzoyl-phenylalanine (BPA) into TBP
The photoreactive, nonnatural amino acid BPA was incorporated on the surface of S. cerevisiae TBP using the using an Escherichia coli nonsense suppressor tRNA/tRNA synthetase system (Supplemental Fig. 1; Chin et al. 2003; Chen et al. 2007). The TBP gene with 13 Myc epitope tags fused at the N terminus and under the control of the strong ADH1 promoter was cloned to a high-copy 2μ vector. TBP was mutagenized to individually substitute the codons of 61 different exposed surface residues with the amber codon TAG (Supplemental Table 1). Each plasmid bearing a TAG codon in the TBP ORF was transformed into a yeast TBP shuffle strain along with a plasmid expressing both an E. coli tRNATyr amber suppressor tRNA and the E. coli tyrosyl-tRNA synthetase variant selected to charge the suppressor tRNA with BPA (Chen et al. 2007). TBP residues that could be substituted by BPA to generate a functional protein were identified by a plasmid shuffle assay on media containing BPA and the drug 5-FOA to select against the plasmid-borne wild-type copy of TBP. About one-third of these substitutions gave rise to viable yeast, and the growth of nearly all of these viable strains required the addition of BPA to the media (Supplemental Table 1). The TBP mutant strains that were viable in the absence of BPA showed very slow growth, and Western blot analysis showed about 20-fold lower levels of TBP when BPA was omitted. Interestingly, insertion of tyrosine at each of the 61 positions in TBP gave viable cells, suggesting that it is the incorporation of BPA rather than the loss of the wild-type residue that results in lethality. Together, our results show that BPA can be incorporated at 24 surface-exposed positions on TBP to give rise to viable cells.
TBP cross-linking in the PIC
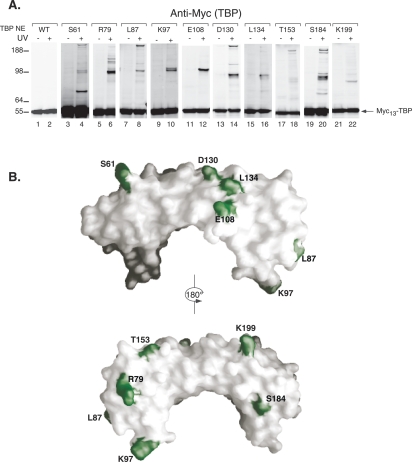
Nuclear extracts were prepared from each of the viable strains containing the BPA-substituted TBP, and all supported levels of Pol II transcription in vitro that were comparable to those observed with wild-type nuclear extracts (data not shown). Next, the nuclear extracts and the activator Gal4-VP16 were used to form PICs on immobilized DNA templates bearing the HIS4 promoter and a single Gal4-binding site. The PICs were washed to remove nonspecifically bound proteins, and one-half of the reaction was irradiated with UV light for 5 min while the other half received no UV treatment. Samples were analyzed by SDS-PAGE and Western blot (Fig. 1A). Myc-tagged TBP migrates at ∼55 kDa, and no cross-linking was observed using extracts with wild-type TBP (Fig. 1A, lanes 1,2). In contrast, BPA substituted at 10 different positions on TBP gave numerous cross-links that were apparent as UV-dependent shifts in the mobility of TBP (Fig. 1A, lanes 3–22). The remaining 14 viable TBP variants showed no reproducible cross-linking (Supplemental Table 1). The altered mobility of cross-linked TBP was used to estimate the size of the cross-linked polypeptides, which ranged from 17 to >190 kDa (Supplemental Table 2). As expected, positions that are close to each other in the TBP structure most often cross-link to proteins of the same mobility (for example, compare cross-linked proteins in variants K97 and L87) (Fig. 1A,B). Although several substituted positions of TBP were observed to cross-link to multiple polypeptides, from Figure 1 we cannot determine if TBP is contacting multiple proteins simultaneously or if the PICs represent a mixture of complexes in which the TBP makes alternative interactions (see below). Polypeptides that have been implicated previously in PIC formation and that have molecular weights similar to the predicted sizes of the cross-linking candidates were individually tested for cross-linking to TBP by Flag epitope-tagging the potential candidate gene and testing for a change in mobility of the cross-linked protein. Supplemental Table 3 lists the proteins that were successfully identified by this method as well as other factors that were tested but did not appear to be cross-linked based on the mobility shift assay.
Figure 1.
Photo-cross-linking of TBP derivatives on the immobilized HIS4 promoter. (A) Western analysis of TPB-BPA cross-linking reactions. The positions of the BPA-substituted amino acids within yeast TBP are indicated above each blot. TBP NE refers to the nuclear extract made from each TBP derivative. Molecular weight markers are indicated on the left. (B) Model of TBP structure showing positions of residues (green) where BPA substitution results in cross-links at HIS4.
TBP position S184 cross-links to Ncb2
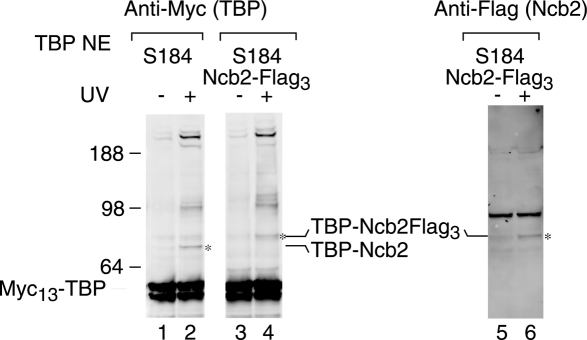
TBP with BPA substituted at position S184 cross-links to a polypeptide with an observed mobility of 17 kDa. The NC2 complex that is known to interact with TBP has two subunits, Ncb1 and Ncb2, and the latter has a predicted size of 16.6 kDa. To test whether Ncb2 cross-links to TBP at position 184, the NCB2 gene was triple Flag tagged in this strain, and the cross-linking experiment was repeated. Figure 2 (lanes 2,4) shows that the predicted 17-kDa cross-linked polypeptide shifts mobility in the Flag-tagged strain to a new estimated size of 22 kDa, the size difference predicted to be conferred by the addition of the triple Flag tag (Chen et al. 2007). This shifted cross-linking product is also detected by an antibody to the Flag epitope tag on Ncb2, confirming that this cross-linked complex contains both TBP and Ncb2 (Fig. 2, lanes 5,6). Using similar criteria, we showed that BPA at TBP position S61 also cross-links to Ncb2 (Supplemental Table 3; data not shown). The cross-linking of TBP to Ncb2 at these positions is in agreement with the X-ray structure of hTBP and hNC2, which shows that these residues in human TBP are close to the NC2β subunit of the NC2 complex (Kamada et al. 2001). These findings demonstrate that our cross-linking method detects specific and biologically relevant interactions.
Figure 2.
Cross-linking identifies an interaction between TBP and Ncb2. Western analysis of TBP cross-linking at HIS4 using extracts with the S184 → BPA substitution (lanes 1,2) and extracts with the substitution in addition to a Flag tag on Ncb2 (lanes 3,4). TBP was visualized using anti-Myc antibodies (lanes 1–4), and Ncb2-Flag was visualized using the anti-Flag antibody (lanes 5,6). The cross-linking product corresponding to TBP–Ncb2 is indicated by an asterisk (*). The uncross-linked Ncb2 protein is of a very small size (17 kDa) and is not visible on this blot.
TBP cross-links to Mot1 at positions S184, D130, and S61
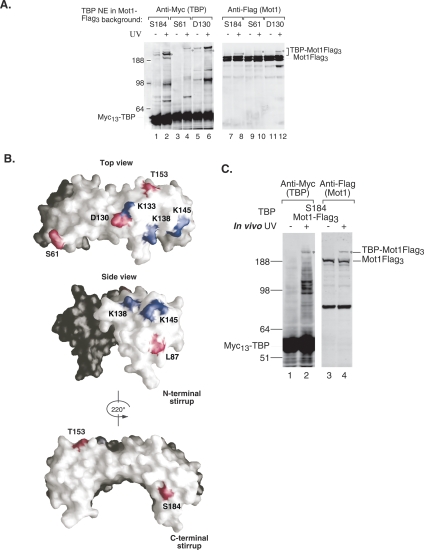
Several TBP derivatives exhibit very slow mobility upon cross-linking, suggesting that they may be cross-linked to Mot1, the largest known TBP-interacting factor at 210 kDa (Auble et al. 1994). To determine whether Mot1 was the cross-linked factor, Mot1 was Flag-tagged in these TBP–BPA strains, and the cross-linking was repeated. Due to the large size of Mot1, a triple Flag tag on Mot1 does not produce a visible mobility shift. However, probing the Western blot with anti-Flag showed a UV-dependent shift in the mobility of Mot1-Flag to a mobility identical to that of the cross-linked TBP derivatives (Fig. 3A, cf. lanes 1–6 and 7–12). This confirms that Mot1 is one of the large polypeptides cross-linked at TBP positions 61, 130, and 184. These three positions of TBP also cross-link to at least one other large polypeptide that has not yet been identified (Fig. 3A). The cross-linked Mot1 at TBP position S184 displays a faster mobility than those at TBP positions 61 and 130 (Fig. 3A), and this is likely due to cross-linking at a different position within Mot1 giving rise to a branched polypeptide with altered mobility compared with cross-linking at other positions. TBP S184 has the strongest relative cross-linking to Mot1 compared with cross-linking at positions D130 and S61. We also found that Mot1 cross-links to TBP at positions T153 and L87 (Fig. 1A; data not shown).
Figure 3.
TBP cross-links to Mot1 in vitro and in vivo. (A) Western analysis of TBP cross-linking at HIS4 assembled using extracts with the S184 → BPA substitution and a Flag tag on Mot1. TBP cross-linking was visualized using anti-Myc antibodies (lanes 1–6), and Mot1 cross-linking was visualized using anti-Flag antibodies. The cross-linked products corresponding to the Mot1–TBP fusion protein are indicated by an asterisk (*). (B) Model of TBP showing the positions where TBP cross-links to Mot1 (red residues). In addition to S184, S61, and D130, Mot1 cross-linking is also observed at L87 and T153 (data not shown). Blue residues indicate positions where radical mutations disrupt TBP–Mot1 interaction (Auble and Hahn 1993). (C) Mot1 cross-links to TBP in vivo. Western analysis of yeast whole-cell extracts prepared from S184 → BPA-substituted cells that were (+) or were not (−) irradiated with UV in vivo. The TBP–Mot1 fusion protein is indicated by an asterisk (*).
Figure 3B summarizes the positions on the TBP surface that cross-link to Mot1 on immobilized templates (red residues). Previously, it was shown that mutation of three positions on the TBP surface disrupts interaction with Mot1 in an in vitro binding assay (Fig. 3B, blue residues) (Auble and Hahn 1993). Our cross-linking results significantly extend the known surface of TBP interacting with Mot1 across the top and both legs of TBP.
TBP cross-links to Mot1 in vivo
Since the cross-linker BPA is incorporated in vivo, we tested whether this can be used as a probe for protein–protein interactions in living yeast cells. For this experiment, we used the TBP strain substituted with BPA at position S184 and a Flag tag on Mot1, as TBP S184 gives rise to strong, reproducible in vitro cross-links with several polypeptides, including Mot1. The strain was grown in minimal media and the culture was irradiated with UV light to activate the cross-linker. Cells were then collected by centrifugation and disrupted to generate whole-cell extracts. Extracts from irradiated and nonirradiated cells were separated by SDS-PAGE and were probed by Western blotting. Upon irradiation, TBP position S184 cross-links to several polypeptides, including a high-molecular-weight protein (Fig. 3C, lane 2). This high-molecular-weight polypeptide is also recognized by the anti-Flag antibody (Fig. 3C, lane 4), confirming that Mot1-Flag cross-links to TBP position S184 in vivo. While this cross-linking is seen on the immobilized promoter as well as in vivo, other cross-links are also observed when the cells are irradiated, potentially reflecting the interactions made by TBP at other types of Pol II promoters or in association with Pol I or Pol III factors. Ncb2, which cross-links to TBP at position 184 on the immobilized promoter, does not visibly cross-link in vivo, suggesting that that a greater fraction of TBP in growing cells is interacting with Mot1 compared with NC2 (Fig. 3C; data not shown).
TBP helix 2 contacts TFIIA
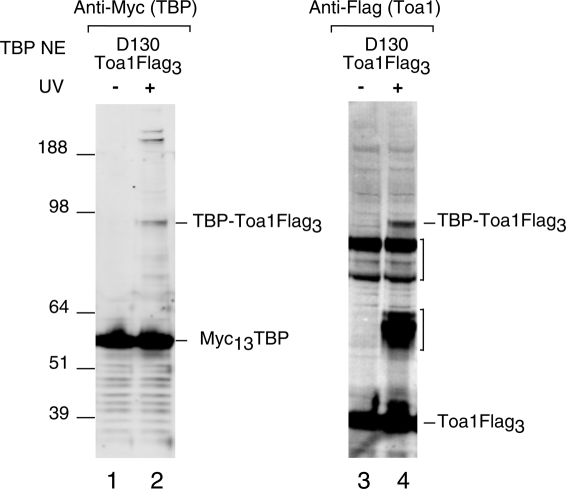
The structure of the yeast TBP–TFIIA–DNA complex reveals that the two protein factors interact primarily through the N-terminal leg and stirrup of TBP (Geiger et al. 1996; Tan et al. 1996). However, the TFIIA used in these structures lacked a 160-amino-acid internal segment (residues 60–220) of Toa1, the large TFIIA subunit. Mutations on TBP helix H2, located on the saddle surface, result in loss of TFIIA interaction and suggested that TFIIA makes additional interactions with TBP that were not observed in the X-ray structure (Buratowski and Zhou 1992; Bryant et al. 1996; Geiger et al. 1996; Tang et al. 1996). We found that position D130, located in TBP helix H2, cross-links to a protein with an estimated size of 49 kDa, similar to that of Toa1. To determine whether this cross-linked polypeptide was the large TFIIA subunit, Toa1 was Flag tagged in this strain and the cross-linking experiment was repeated. In this new strain, Toa1 shows a UV-dependent mobility shift at the exact mobility observed with cross-linked TBP (Fig. 4, lanes 2,4). This result directly demonstrates that wild-type TFIIA contacts TBP through interactions with TBP helix 2.
Figure 4.
TBP cross-links to TFIIA. Western analysis of TBP cross-linking in PICs that were assembled using extracts with the D130 → BPA substitution and a Flag tag on Toa1. Brackets on the right indicate background bands, one of which appears to be UV-specific and may be a degradation product of the cross-linked product.
TBP interacts directly with Spt3 and Spt8 in the PIC
The Spt3 and Spt8 subunits of SAGA interact genetically with TBP (Eisenmann et al. 1992, 1994; Laprade et al. 2007), and Spt8 has been shown to interact biochemically with TBP in purified systems (Sterner et al. 1999; Warfield et al. 2004; Sermwittayawong and Tan 2006). However, to date there has been no biochemical evidence to show a direct interaction between Spt3 and TBP. The HIS4 promoter used for our cross-linking studies is SAGA-dependent and TFIID-independent (Fishburn et al. 2005) and therefore provides an ideal system to test TBP–SAGA interactions within the PIC.
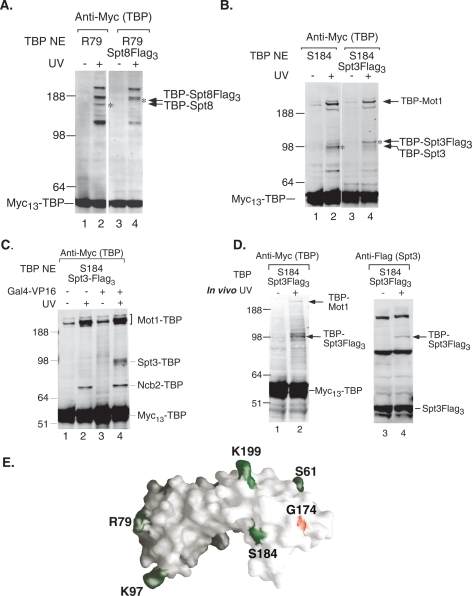
TBP position R79 cross-links to several proteins ranging in size from 54 kDa to >191 kDa (Fig. 5A, lane 2). This position is near the surface of the TFIIA four-helix bundle in the TBP–TFIIA–DNA complex, which has been shown to cross-link to Spt8 in PICs (Warfield et al. 2004). To determine if one of these cross-linked polypeptides was Spt8, SPT8 was Flag tagged in this strain and the cross-linking was repeated. This resulted in a mobility shift of the p70 cross-linked polypeptide by ∼5 kDa, conclusively identifying p70 as Spt8 (Fig. 5A, lane 4). BPA positioned at S184 cross-links to a protein with an estimated size of 43 kDa, the size of Spt3 (Fig. 5B, lane 2). Residue S184 is also close to the TBP residue G174, which, when mutated, genetically interacts with Spt3 (Fig. 5E; Eisenmann et al. 1992). SPT3 was Flag tagged in this strain and the cross-linking was repeated. A mobility shift of the p43 cross-linked polypeptide was observed upon Flag fusion, identifying this polypeptide as Spt3 (Fig. 5B, lanes 2,4). Cross-linking experiments were also performed with extracts from a strain bearing a deletion of SPT3 and with BPA at TBP position S184. As predicted, TBP no longer cross-links to the p43 polypeptide, again confirming the identity of Spt3 as the cross-linked p43 factor (data not shown). Together, our analysis provides direct evidence for specific interactions between key subunits of SAGA and TBP in the PIC.
Figure 5.
TBP cross-links to Spt8 and Spt3. (A) Several cross-linking products are revealed when the unnatural amino acid is positioned at R79. When Spt8 is Flag-tagged in the R79 mutant strain, the p70 (*) band shifts by 5 kDa, identifying this product as TBP–Spt8. (B) Cross-linking experiments were performed with extracts containing Flag-tagged (right panel) or untagged (left panel) Spt3. In the presence of UV light (+), the p43 band shifts upward by 5 kDa when Spt3 bears a Flag tag (*), identifying this product as TBP–Spt3. (C) TBP cross-linking to Spt3 is dependent on activator and independent of Mot1 and Ncb2. Cross-linking was performed with HIS4 templates that were (lanes 3,4) or were not (lanes 1,2) preincubated with Gal4-VP16. (D) TBP cross-links to Spt3 in vivo. Western analysis of yeast whole-cell extracts prepared from S184 → BPA-substituted cells that were (+) or were not (−) irradiated with UV. The TBP–Spt3 fusion protein is indicated by arrows. (E) Model of TBP showing the location of TBP → BPA-substituted residues (green), and indicating the position of S184 relative to residue G174 (red).
Our results show that TBP contacts at least three different proteins at position S184: Ncb2, Mot1, and Spt3. Since previous work has shown that SAGA is recruited to promoters by activators (Bhaumik and Green 2001; Larschan and Winston 2001), we investigated whether any of these cross-links were activator-dependent. The cross-linking experiment was repeated as before except that the activator Gal4-VP16 was omitted. Our results show that cross-linking of TBP to Mot1 and Ncb2 is not dependent on the presence of the activator (Fig. 5C, lane 2) while cross-linking to Spt3 is detected only when the activator is present (Fig. 5C, lane 4). Western analysis of these reactions, probing for the SAGA subunits Ada1 and Spt3, showed that SAGA is recruited to the immobilized promoter only in the presence of activator (data not shown). Our results suggest that Mot1 and Ncb2 contact TBP in complexes that are separate from PICs that are formed in the presence or absence of activator. This is consistent with the observation that TBP is present in approximately fivefold excess over other general transcription factors, and Pol II is bound to the immobilized promoter (Ranish 1999). In order to confirm that the cross-links between TBP and Mot1, Ncb2, and Spt3 are not dependent on each other, we repeated the cross-linking experiment using extracts that were immunodepleted of SAGA via a Flag tag on Spt3. As shown in Supplemental Figure 2B, this results in a 75% depletion of Spt3 (lane 4) as compared with mock-depleted extract (lanes 1–3), and the TBP cross-link to Spt3 is correspondingly reduced without reduction in the cross-linking to Mot1 or Ncb2 (Supplemental Fig. 2A). Together, our results show that the cross-links between TBP, Mot1, and Ncb2 are not dependent on Spt3, and suggest that Mot1, NC2, and SAGA are present in separate complexes with TBP.
TBP cross-links to Spt3 in vivo
To determine whether the contact between TBP and Spt3 occurs efficiently in vivo, cells containing BPA at TBP S184 and a Flag tag on Spt3 were grown in galactose media. The cells were irradiated, whole-cell extracts were prepared, and cross-linked proteins were assayed by Western blot. As seen in Figure 5D (left panel), probing with the anti-Myc antibody reveals a TBP-cross-linked polypeptide with an estimated size of 43 kDa. This cross-linking product is also visible when the Western blot is probed with anti-Flag antibody, which detects Spt3-Flag (Fig. 5D, right panel), indicating that the cross-linked product contains both TBP and Spt3. This experiment demonstrates that the TBP–Spt3 interaction occurs efficiently in vivo, confirming the interaction observed in PICs.
TBP cross-linking is not DNA-dependent
Our studies above showed that TBP contacts several factors on immobilized promoter templates. To determine whether these proteins interact with TBP only at the promoter, we performed cross-linking in extracts in the absence of any exogenously added promoter DNA. We tested for TBP cross-linking to Mot1 and Toa1 at TBP position D130 and to Spt3 at TBP position S184 by irradiating yeast nuclear extracts and analysis by Western blot. Supplemental Figure 3 shows that TBP cross-links to Flag-tagged Mot1, Toa1, and Spt3 in extracts. This indicates that a fraction of TBP is associated with these factors in extracts before binding to promoter DNA.
Functional interactions of TBP with Spt3
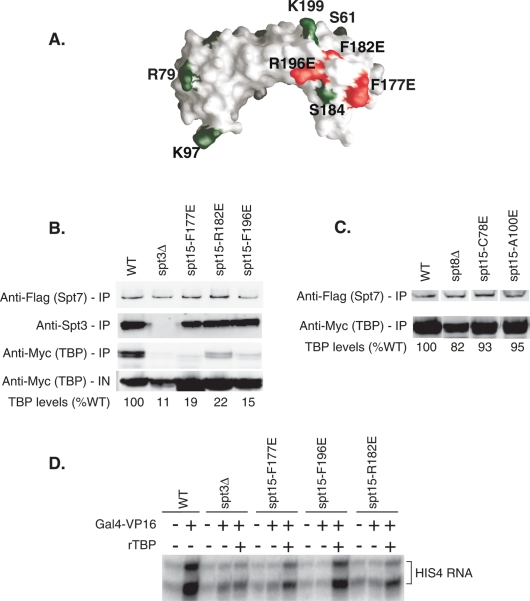
To investigate the significance of the Spt3–TBP interaction, mutations were generated in surface-exposed residues of TBP located near the site of Spt3 cross-linking and near residue G174, previously shown to genetically interact with TBP (Fig. 5E; Eisenmann et al. 1992). Several mutations near TBP residue 184 at positions Y185, E186, L189, and F190 were lethal. In contrast, other nearby residues at TBP positions A176, F177, G180, T181, F182, S183, I194, and R196 were viable and were tested for growth phenotypes consistent with a SAGA defect (Fig. 6A; Supplemental Table 4). Mutations at positions F177, F182, and R196 all displayed a slow growth phenotype in liquid culture and a cold-sensitive phenotype at 18°C, similar to that of an spt3Δ mutant. These three mutations also showed slow growth on YP GAL (+anti-mycin) with mutations F177E and R196E showing the strongest phenotypes, which were similar to, but not as severe as, a complete SPT3 deletion. Mutations F177E, R196E, and spt3Δ also showed a strong growth phenotype on plates containing the drug sulfometuron methyl (SM), which mimics amino acid starvation and requires induction of Gcn4-regulated genes for growth. In contrast, mutations near the site of Spt8 cross-linking at residue R79 (G77, C78, L80, D81, L82, A100, A117, S118, K120, and F148) were all viable and had no detectable growth phenotype in our assays, comparable to the lack of growth phenotype observed in an spt8Δ strain. Mutation of residues L80 and K120 was lethal.
Figure 6.
Disruption of TBP–Spt3 contacts results in a loss in activation in vitro. (A) Model of TBP showing the positions of radical surface mutations (red) relative to TBP → BPA substitutions (green). (B) Western analysis of SAGA immunoprecipitations from wild-type (WT), Δspt3, and TBP radical mutant strains near S184. (IN) Input. TBP levels were visualized using the Myc antibody, Spt3 levels with a rabbit polyclonal anti-Spt3 antibody, and Spt7 levels with an anti-Flag antibody. Westerns were scanned and quantitated using the LiCor Odyssey. TBP levels are expressed relative to the wild-type TBP immunoprecipitation (set to 100%). (C) Western analysis of SAGA immunoprecipitations from wild-type (WT), spt8Δ, and TBP radical mutant strains near R79. (D) In vitro transcription using nuclear extracts made from wild-type, spt3Δ, and TBP radical mutant strains. Basal and activated single-round transcription was measured from the HIS4 promoter in the absence or presence of the activator Gal4-VP16 and recombinant TBP.
A likely reason for the growth phenotypes of mutations F177, F182, and R196 is that these TBP mutations disrupt the physical interaction between TBP and Spt3. We were unable to test for disruption of this protein–protein interaction using the cross-linking assay since the combination of BPA insertion at S184 and the individual TBP mutations is lethal. As an alternative assay, we measured the association of TBP and SAGA in extracts made from strains containing either of the TBP mutations or a deletion of the SPT3 gene. Previous analysis found that SAGA purified using the TAP-tag method from whole-cell extracts contains very low levels of TBP (Laprade et al. 2007). However, we found that a single-step affinity purification of SAGA via a triple Flag tag on the SAGA subunit Spt7 retains associated TBP (Fig. 6B). In contrast, an spt3 deletion eliminates ∼90% of the SAGA-associated TBP. Similarly, the three SPT15 mutations (SPT15 is the gene encoding TBP) F177E, F182E, and R196E all had lower levels of SAGA-associated TBP with levels ranging from 15%–22% of wild type (Fig. 6B). This demonstrates that these mutations in TBP reduce the TBP–Spt3 interaction, resulting in a phenotype similar to an SPT3 deletion. In contrast to these results, an SPT8 deletion mutation did not significantly disrupt SAGA–TBP coprecipitation, consistent with the lack of growth phenotype in this strain (Fig. 6C). Radical mutant substitutions around the TBP–Spt8 interaction site, including C78E and A100E, also lacked a TBP interaction defect (Fig. 6C).
To confirm that the defect in Spt3–TBP interactions could lead to a transcription activation defect, in vitro transcription assays were performed using nuclear extracts from cells with wild-type TBP or TBP bearing the radical mutations F177, F182, or R196 or from cells containing an SPT3 deletion. Single-round transcription reactions were performed using the SAGA-dependent HIS4 core promoter. Activation is nearly lost in the spt3Δ mutant extract and was severely reduced in extracts from the three TBP mutations (Fig. 6D). The activation levels in these three strains were ∼10% of wild-type levels. As expected, adding recombinant TBP to the transcription reactions suppresses the activation defect caused by the TBP mutations, but not in the spt3Δ extract.
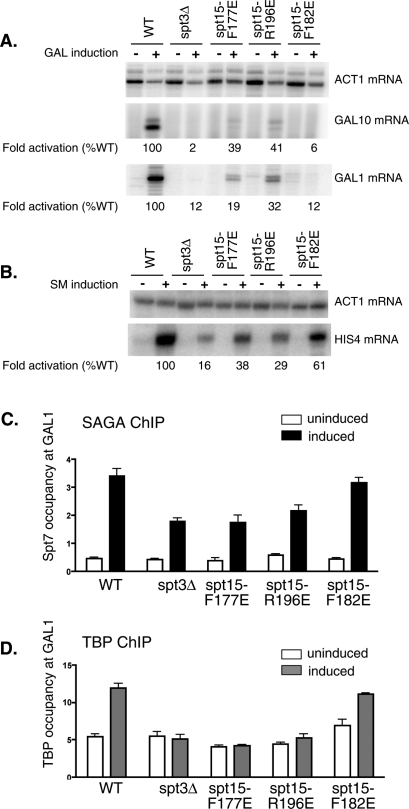
To check for activation defects in vivo, Gal4 and Gcn4-dependent transcription was measured in the mutant strains. Wild-type yeast, spt3Δ cells, or the strains bearing the TBP mutations were induced with galactose or with SM. RNA was harvested and primer extension was used to quantitate transcripts from the SAGA-independent gene ACT1, and the SAGA-dependent genes GAL1, GAL10, and HIS4. Activated transcription from the GAL10, GAL1, or the HIS4 genes is impaired 50-fold, eightfold, and sixfold, respectively, in the Δspt3 strain (Fig. 7A,B). Similarly, activated transcription of all three genes is also reduced by the TBP mutations. The reduction in expression ranges from 16-fold to 2.5-fold for GAL1,10 genes and sixfold to 1.6-fold for HIS4, while the expression of the SAGA-independent gene ACT1 is unaffected.
Figure 7.
Mutations in the TBP–Spt3 contact surface result in reduced activation and TBP recruitment in vivo. (A,B) RNA was extracted from yeast cells grown in glucose or galactose as indicated, and the indicated transcripts were monitored by primer extension. Transcript levels were quantitated using Image Quant software and normalized to ACT1 transcripts. Levels of activation are indicated relative to activation in a strain with wild-type TBP (set to 100%). (C) ChIP to detect Spt7 occupancy at the GAL1 promoter in cells grown in either glucose (uninduced) or galactose (induced). (D) ChIP to detect myc-TBP occupancy at the GAL1 promoter in cells grown in either glucose (uninduced) or galactose (induced).
In order to determine whether the reduction in activated transcription caused by the TBP mutations reflects a defect in TBP recruitment to promoters in vivo, we performed ChIP assays to measure the level of TBP occupancy at the GAL1 promoter. Upon galactose induction, SAGA is recruited to the GAL1 UAS, and recruitment is reduced about twofold in the Δspt3 strain (Fig. 7C). It is well established that the SAGA complex remains intact in the absence of the Spt3 subunit (Wu and Winston 2002). The strains containing the three TBP mutants also show SAGA recruitment to the GAL UAS, at a level equal to or greater than that observed in the Δspt3 strain. In contrast, upon galactose induction, TBP recruitment to the GAL1 promoter was abolished in the Δspt3 strain as well as in the TBP mutants F177E and R196E. Reduced TBP recruitment was observed in the TBP mutant F182E. Together, our results confirm the functional significance of the direct Spt3–TBP interaction in SAGA-dependent transcription observed in our cross-linking assay.
Discussion
We substituted the photoreactive amino acid BPA at specific residues on the surface of TBP to map protein–protein contacts of TBP within the PIC. This cross-linker has a very short length (∼6.5 Å from the Cα carbon) and thereby probes for proteins in very close contact with TBP. Cross-linking of TBP bound to an immobilized promoter template assembled from a yeast nuclear extract shows that TBP makes numerous site-specific contacts with other polypeptides. Interestingly, many TBP residues cross-link to more than one factor, but it is likely that not all of these protein–protein interactions occur simultaneously. For example, stoichiometry measurements have shown that there is approximately fivefold excess of TBP bound to the immobilized templates compared with other PIC components (Ranish 1999). The model that TBP exists in complexes outside of PICs is also consistent with the observed cross-linking of TBP to the repressors Mot1 and NC2, which, under our conditions, likely inhibit PIC formation.
The most significant finding of our study is the identification of a direct contact between Spt3 and TBP. Based on genetic evidence and the fact that Spt3 is required for recruitment of PICs at many SAGA-dependent promoters, it was proposed that direct Spt3–TBP interaction is responsible for recruitment of TBP (Eisenmann et al. 1992; Dudley et al. 1999a, b; Bhaumik and Green 2001, 2002; Larschan and Winston 2001). Despite genetic evidence and much effort, no convincing biochemical evidence for a direct interaction between these two factors was observed in previous studies. Coprecipitation between TBP and Spt3 was observed previously in yeast extracts, but only when both these factors were overexpressed, and no biochemical interactions were observed with purified factors in vitro (Eisenmann et al. 1992; Madison and Winston 1997; Sermwittayawong and Tan 2006). Since the TBP–Spt3 interaction is readily observed by the cross-linking assay in vivo and in vitro, but not with purified proteins, it is likely that this interaction is influenced by other subunits of SAGA and possibly either activators and/or other PIC components. Interestingly, previous in vitro experiments using recombinant TBP and SAGA showed that TBP binding involved the Spt8 subunit rather than Spt3. Our experiments show the opposite result, where TBP–SAGA binding in extracts without added recombinant TBP is dependent on Spt3 rather than Spt8. Together these results raise the possibility that recombinant TBP interacts with SAGA differently compared with assembly of the SAGA–TBP in cells. Exploring this difference will be an important area for future study.
Our ability to detect the Spt3–TBP interaction in vivo suggests that this is a relatively efficient association and confirms the previous genetic data suggesting a protein–protein interaction between these factors (Eisenmann et al. 1992; Laprade et al. 2007). TBP mutations based on the Spt3 cross-linking results defined a surface of TBP involved in Spt3 interaction. Disruption of the TBP–Spt3 contacts by these mutations near the TBP C-terminal stirrup resulted in a significant reduction in the physical interaction between TBP and SAGA measured in extracts and a corresponding reduction in activated transcription and recruitment of TBP to the GAL1 promoter. Our results, taken together with previous findings, demonstrate that the HAT-independent coactivator activity of SAGA is dependent upon the direct contact between Spt3 and TBP, leading to stable binding of TBP at promoters.
Further questions regarding this critical interaction remain to be determined. For example, the details of the protein–protein recognition between TBP and Spt3 are not yet clear. Previous genetic studies have shown that mutations in Spt3 residues L178, K179, D182, M188, Y193, W196, Q201, and A202 give a dominant-negative Spt− phenotype (Laprade et al. 2007). The phenotype for Y193C can be suppressed by mutation in the C-terminal region of TBP, which, according to our results, interacts directly with Spt3. Together, this suggests that at least some of these residues in Spt3 are important for directly contacting TBP. This C-terminal Spt3 segment is also thought to interact with an N-terminal segment of Spt3 via an intramolecular interaction to form an atypical histone fold domain, similar to that formed by human Taf11–Taf13 (Birck et al. 1998). As discussed above, the structure of this domain and therefore the affinity of the interaction with TBP may be indirectly influenced by other SAGA subunits, PIC components, or possibly by interaction of SAGA with activation domains. Also not resolved is whether the Spt3–TBP interaction results only in recruitment of TBP, or if there is an additional step involved after TBP binding. Finally, it remains to be determined whether a preassembled SAGA–TBP complex is the form of SAGA recruited by activator in which TBP binds to the TATA element in a slower rate-limiting step.
Our work has also shown that TBP contacts the Spt8 subunit of the SAGA complex at position R79. However, compared with the strong requirement for Spt3 during activation, in vitro activation from the HIS4 promoter was unaffected in extracts from an spt8Δ mutant. In addition, mutations on the putative TBP–Spt8 interaction surface did not impair transcription from HIS4 or from the previously characterized Spt8-dependent genes PHO84 and VTC3 (Bhaumik and Green 2002; data not shown). Since Spt8 has been shown to negatively regulate basal transcription under certain conditions (Belotserkovskaya et al. 2000; Warfield et al. 2004), it is possible that the TBP–Spt8 interaction is primarily a negative interaction that prevents coactivator function of SAGA in the absence of activator. Further experiments will be required to investigate the mechanism and functional significance of the Spt8–TBP interaction.
Previous cross-linking studies performed with purified SAGA and TBP identified an interaction between TBP and the Ada1 protein (Sermwittayawong and Tan 2006); however, our studies failed to uncover this interaction in the PIC. Reasons for this include (1) the interaction between TBP and SAGA subunits may be different in the context of cellular assembly or in the presence of other PIC components, or (2) it is possible that the Ada1–TBP interaction takes place through a region of TBP that is not close to a substituted residue. Two-thirds of the TBP residues substituted with BPA were lethal, and we avoided all surface residues that are close to the DNA-binding surface of TBP to minimize effects on DNA binding. Since the cross-linker is very short, BPA must be positioned very close to the protein–protein interaction surface for detectable cross-linking.
Our results show that the Toa1 subunit of TFIIA has a broader interaction with TBP than was determined in X-ray studies. TBP interacts with TFIIA through Helix 2 on its N-terminal saddle. It is likely that this interaction takes place through the large portion of Toa1 that was omitted from the structural studies of TFIIA, as this segment inhibits crystal formation (Geiger et al. 1996; Tan et al. 1996). Although this region of TFIIA is not well conserved in other species, mutations on TBP Helix2 were shown to decrease the binding of TFIIA to human and yeast TBP, underscoring the significance of this interaction (Buratowski and Zhou 1992; Bryant et al. 1996).
Genetic studies have shown an interaction between TBP and TFIIB through residue K239 (Buratowski et al. 2002). While we did not substitute this residue with BPA, we did substitute other residues such as R171 and E173, which are very close to K239 on the TBP stirrup. However, BPA substitution at these residues gave a lethal phenotype, perhaps due to the disruption of a critical interaction between TBP and TFIIB. In fact, many other residues that are close to the TBP–TFIIB-binding surface as revealed by the TBP–TFIIB–DNA cocrystal (Nikolov et al. 1995) cannot be substituted by BPA, pointing to the critical role of this TBP surface. As a result, we were not able to detect TFIIB binding to TBP in our cross-linking studies.
Future studies that are currently underway are aimed at using alternative methods to identify protein cross-linking products, allowing more rapid identification of protein partners. Since we were able to detect several protein–protein interactions in vivo using the site-specific cross-linkers, the methods used here should be generally applicable to the study of interactions in other systems.
Materials and methods
Construction of plasmids
TBP was cloned into the LEU2 2μ plasmid pRS425 (Christianson et al. 1992). The ADH1 promoter was cloned upstream of the TBP ORF. A 13× Myc tag was inserted at the TBP N terminus after the first codon using the QuikChange method (Stratagene) (Geiser et al. 2001), yielding the plasmid pNM10. QuikChange was used to substitute 61 different codons in the ORF with the amber codon TAG in order to create the site-specific mutations that were subsequently confirmed by sequencing. pLH157 (TRP1) containing the E. coli nonsense suppressor tRNA/tRNA synthetase system was a gift from Linda Warfield (Chen et al. 2007).
Yeast strains and media
All strains are derived from BY4705 (Brachmann et al. 1998). In the TBP shuffle strain, the entire gene encoding TBP (SPT15) was deleted using the KanMX marker. The amber-substituted TBP plasmids and pLH157 were cotransformed into this shuffle strain. To prepare solid media containing BPA, 10 mL of 50 mM BPA (BACHEM) dissolved in 1N HCl was added to 1 L of media along with 10 mL of 1N NaOH to a final BPA concentration of 0.5 mM. The transformants were streaked onto −Trp −Leu +BPA medium. TBP residues that could be substituted by BPA to generate a functional protein were identified by a plasmid shuffle assay on media containing BPA and the drug 5-FOA to select against the wild-type URA3 plasmid-borne copy of SPT15. Strains were Flag-tagged at genomic loci using standard methods, and SPT3 and SPT8 were deleted using the hygromycin marker. Anti-mycin A (1 μg/mL; Sigma) was used where indicated.
Photo-cross-linking reactions, immunoprecipitations, and transcription
Nuclear extracts were made from the TBP amber mutants strains as described previously (Ranish et al. 1999), except that 0.5 mM BPA was added to the media during cell growth. The extracts were used to form PICs as described previously using immobilized HIS4 promoter DNA (Ranish et al. 1999). The complexes were washed with and resuspended in 100 μL of transcription buffer. UV-treated samples were then placed in a Spectrolinker XL-1500a UV cross-linker (Spectronics Corp.) and irradiated twice at energy setting 7500. Samples were then resuspended in LDS Sample Buffer and run on a 4%–12% BIS-Tris gel (Invitrogen) in MOPS buffer for 80–120 min at 200 V. To detect photo-cross-linking, TBP was visualized by Western analysis using commercially available mouse monoclonal anti-Myc antibodies (1:2000; Covance), and Flag-tagged proteins were visualized using M2-Flag antibody (1:2000; Sigma) using the Odyssey infrared imaging system (LiCor Biosciences). For immunodepletion of SAGA from nuclear extracts that were used in cross-linking, we prepared Protein G Dynabeads (Invitrogen) conjugated to the M2-anti-Flag antibody. Five milligrams of nuclear extract were depleted using 20 μL of Protein G Dynabeads that had been conjugated to 1 μg of M2 antibody. Depletions were carried out for 2 h at room temperature with flicking to resuspend the beads every 15 min. Mock-depleted extracts were incubated with Protein G Dynabeads that were not conjugated to antibody. For in vivo cross-linking, cells were grown in minimal media with glucose or galactose to OD600 ∼1.0, harvested and resuspended in 10 mL of minimal media, and poured into an 8-cm square Petri dish. Cells were irradiated as described above for PICs, washed in extraction buffer containing in 100 mM Tris (pH 8.0), 250 mM ammonium sulfate, 10% glycerol, 1 mM EDTA, 0.1% Triton X-100, 1 mM DTT with protease inhibitors, and resuspended in 1 mL of extraction buffer in a 2-mL screwcap tube with ∼400 μL of glass beads. Cells were disrupted using a Mini Bead Beater (Biospec Products) for 5 min to prepare whole-cell extracts. Extracts were spun at 4°C, 20,000g in an Eppendorf 5417C microfuge for 20 min, and frozen on dry ice. To demonstrate DNA-independent cross-linking to TBP, nuclear extracts were prepared and 50 μg of extract were irradiated with UV light. Ten micrograms of the irradiated as well as nonirradiated extract was separated by SDS-PAGE and analyzed by Western blotting. For SAGA immunoprecipitations, 0.5 mg of extract was prepared as above without UV irradiation and incubated with 20 μL of M2-Flag beads (Sigma) for 1–2 h at 4°C. The beads were washed two to three times as above in extraction buffer, and SAGA was eluted with 200 ng/μL Flag peptide (Sigma) in 60 μL of TBS. In vitro transcription assays were carried out as described previously (Ranish et al. 1999), and used plasmid pSH515 with the HIS4 promoter and an upstream Gal4-binding site along with yeast nuclear extract and recombinant activator Gal4-VP16.
RNA extraction and primer extension
For induction of the GAL genes, 100 mL of cells was grown at 30°C to OD600 ∼0.8–1.0 in YPD and shifted to 2% galactose for 5 h. For induction of Gcn4-dependent genes, cells were grown in −isoleucine, −valine minimal media, and sulfometuron methyl dissolved in DMSO (Sigma) was added to a final concentration of 5 μg/mL for 30 min. RNA was extracted using hot acid phenol and chloroform. Twenty micrograms to 40 μg of RNA were used per 10-μL primer extension reaction and incubated with radioactive end-labeled primers to recognize transcripts from ACT1, GAL1, GAL10, and HIS4. MMLV-RT (Invitrogen) was used for the reverse transcription, which was performed as described previously for in vitro transcription (Ranish et al. 1999). Reactions were run on an 8% sequencing gel and visualized by phosphorimager. Transcript levels were quantitated using ImageQuant software.
ChIP
ChIP assays were performed as described previously (McConnell et al. 2004), with a few modifications. For GAL1, 10 induction, strains were cultured in rich media with 2% glucose and then shifted to galactose for 5 h. Chromatin was solubilized by sonication using a Bioruptor (Diagenode, Inc.) for 30 min total (30 sec ON/30 sec OFF) with samples cooled on ice after 15 min. Chromatin containing ∼500 mg of protein per reaction was incubated overnight at 4°C with 6 μg of the following antibodies: for TBP immunoprecipitationss, anti-Myc 9E10 (Covance); for Spt7 immunoprecipitationss, anti-M2-Flag (Sigma). Twenty microliters of Dynabeads Protein G (Dynal) were washed with FA buffer + 5 mg/mL IgG-free BSA, then added to each overnight reaction and allowed to bind for 90 min at 4°C. Beads were washed as described (McConnell et al. 2004) and eluted with 50 μL of 20 mM Tris (pH 8.0), 100 mM NaCl, 20 mM EDTA, 1% SDS for 15 min at 65°C. Elutions were repeated and combined. One-hundred microliters of TE were added and cross-linking was reversed as described, then DNA was purified with a PCR purification kit (Invitrogen) into a 50-μL final volume. Ten percent of the chromatin input was processed to reverse cross-links along with immunoprecipitation samples. qPCR was carried out in 15-μL reaction volumes using Power SYBR PCR master mix (Applied Biosystems), 300 nM each forward and reverse primers, and 1.5 μL of 1:4 diluted DNA. PCR program: 2 min at 50°C, 10 min at 95°C, 40× (15 sec at 95°C, 1 min at 60°C). Samples were run in duplicate, and relative amounts of DNA were calculated using a standard curve generated from serial dilutions of yeast genomic DNA using POL1 primers. Experiments were performed in biological duplicate. Spt7 occupancy was determined by amplifying a region surrounding the GAL1 UAS, while TBP occupancy was determined by amplifying a region surrounding the GAL1 TATA. Primers in a Pol1 ORF were used as a nontranscribed region control.
The following primers were used to amplify the indicated sequences: GAL1-UAS-FP, AGTAATACGCTTAACTGCTC ATTGCT; GAL1-UAS-RP, ACGCACGGAGGAGAGTCTTC; GAL1-TATA-FP, GCGATTAGTTTTTTAGCCTTATTTCTG; GAL1-TATA-RP, GGTTATGCAGCTTTTCCATTTATATAT CT; POL1-ORF-FP, TTTCTGCTGAGGTGTCTTATAGAAT TCA; POL1-ORF-RP, GCTTTGGGCCCATGCAT.
The ratios of GAL1 UAS or TATA to control signals (Pol I) in immunoprecipitation samples were normalized for the corresponding ratios for Input (IN) samples, and the resulting values were plotted in histograms as “occupancy.” Three independent experiments were performed.
Acknowledgments
We thank members of the Hahn laboratory, I. Whitehouse, and E.T. Young for their comments and suggestions throughout the course of this work; L. Warfield for advice on the use of nonnatural amino acids; and I. Whitehouse and B. Moorefield for comments on the manuscript. This work was supported by NIH Grants GM075114 and GM054351.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1724408.
References
- Auble D.T., Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes & Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- Auble D.T., Hansen K.E., Mueller C.G., Lane W.S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes & Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Sterner D.E., Deng M., Sayre M.H., Lieberman P.M., Berger S.L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R., Green M.R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R., Green M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck C., Poch O., Romier C., Ruff M., Mengus G., Lavigne A.C., Davidson I., Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- Brachmann R.K., Yu K., Eby Y., Pavletich N.P., Boeke J.D. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17:1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Howe L., Sousa K., Alley S.C., Carrozza M.J., Tan S., Workman J.L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Bryant G.O., Martel L.S., Burley S.K., Berk A.J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes & Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992;255:1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- Buratowski R.M., Downs J., Buratowski S. Interdependent interactions between TFIIB, TATA binding protein, and DNA. Mol. Cell. Biol. 2002;22:8735–8743. doi: 10.1128/MCB.22.24.8735-8743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Attardi L.D., Verrijzer C.P., Yokomori K., Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Chen H.T., Warfield L., Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.X., Floer M., Ononaji P., Bryant G., Ptashne M. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 2002;12:1828–1832. doi: 10.1016/s0960-9822(02)01257-5. [DOI] [PubMed] [Google Scholar]
- Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., Schultz P.G. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Creton S., Svejstrup J.Q., Collart M.A. The NC2 α and β subunits play different roles in vivo. Genes & Dev. 2002;16:3265–3276. doi: 10.1101/gad.234002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Darst R.P., Martin K.J., Afshari C.A., Auble D.T. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A.M., Gansheroff L.J., Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912Δ promoter in Saccharomyces cerevisiae. Genetics. 1999a;151:1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle C., Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999b;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B.D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eisenmann D.M., Arndt K.M., Ricupero S.L., Rooney J.W., Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann D.M., Chapon C., Roberts S.M., Dollard C., Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J., Mohibullah N., Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Geiger J.H., Hahn S., Lee S., Sigler P.B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K. Cellular stress alters the transcriptional properties of promoter-bound Mot1–TBP complexes. Mol. Cell. 2004a;14:479–489. doi: 10.1016/j.molcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004b;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M., Cebe R., Drewello D., Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–90. 92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- Goppelt A., Meisterernst M. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goppelt A., Stelzer G., Lottspeich F., Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Duggan L., Cote J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K.L., Pugh B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Inostroza J.A., Mermelstein F.H., Ha I., Lane W.S., Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R.G., Meisterernst M., Burley S.K. Crystal structure of negative cofactor 2 recognizing the TBP–DNA transcription complex. Cell. 2001;106:71–81. doi: 10.1016/s0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J.H., Hahn S., Sigler P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kokubo T., Swanson M.J., Nishikawa J.I., Hinnebusch A.G., Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Kosa P., Mencia M., Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Laprade L., Rose D., Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Young R.A. Regulation of gene expression by TBP-associated proteins. Genes & Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton H.C., Holstege F.C., Shen W.C., Hannett N., Jennings E.G., Winston F., Green M.R., Young R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Bhaumik S.R., Green M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- Li X., Bhaumik S., Zhu X., Li L., Shen W., Dixit B., Green M. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- Madison J.M., Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell A.D., Gelbart M.E., Tsukiyama T. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 2004;24:2605–2613. doi: 10.1128/MCB.24.7.2605-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia M., Moqtaderi Z., Geisberg J.V., Kuras L., Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- Nikolov D.B., Chen H., Halay E.D., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K. Crystal structure of a TFIIB–TBP–TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Poon D., Bai Y., Campbell A.M., Bjorklund S., Kim Y.J., Zhou S., Kornberg R.D., Weil P.A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish J.A. “Mechanisms of transcription by RNA polymerase II.” Ph.D. thesis. University of Washington; 1999. [Google Scholar]
- Ranish J.A., Yudkovsky N., Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: Holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes & Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J.C., Apone L., Walker S.S., Griffin L.A., Green M.R. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- Schluesche P., Stelzer G., Piaia E., Lamb D.C., Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D., Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: Implications for TBP recruitment. EMBO J. 2006;25:3791–3800. doi: 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse R.O., Brenowitz M., Auble D.T. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant P.A., Roberts S.M., Duggan L.J., Belotserkovskaya R., Pacella L.A., Winston F., Workman J.L., Berger S.L. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Hunziker Y., Sargent D.F., Richmond T.J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–151. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- Tang H., Sun X., Reinberg D., Ebright R.H. Protein-protein interactions in eukaryotic transcription initiation: Structure of the preinitiation complex. Proc. Natl. Acad. Sci. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda K., Grant P.A., Cote J., Steger D.J., Eberharter A., John S., Workman J.L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Warfield L., Ranish J.A., Hahn S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes & Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy P.J., Kobayashi R., Kadonaga J.T. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- Wu P.Y., Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]