Abstract
Calcium-dependent protein kinases (CDPKs) of Apicomplexan parasites are crucial for the survival of the parasite throughout its life cycle. CDPK1 is expressed in the asexual blood stages of the parasite, particularly late stage schizonts. We have identified two substrates of Plasmodium falciparum CDPK1: myosin A tail domain-interacting protein (MTIP) and glideosome-associated protein 45 (GAP45), both of which are components of the motor complex that generates the force required by the parasite to actively invade host cells. Indirect immunofluorescence shows that CDPK1 localizes to the periphery of P. falciparum merozoites and is therefore suitably located to act on MTIP and GAP45 at the inner membrane complex. A proportion of both GAP45 and MTIP is phosphorylated in schizonts, and we demonstrate that both proteins can be efficiently phosphorylated by CDPK1 in vitro. A primary phosphorylation of MTIP occurs at serine 47, whereas GAP45 is phosphorylated at two sites, one of which could also be detected in phosphopeptides purified from parasite lysates. Both CDPK1 activity and host cell invasion can be inhibited by the kinase inhibitor K252a, suggesting that CDPK1 is a suitable target for antimalarial drug development.
Calcium dependent protein kinases (CDPKs)3 are enzymes found in plants and organisms in the Alveolate lineage (1). These include Apicomplexan parasites such as Toxoplasma gondii and Plasmodium falciparum, the causative agent of the most severe form of malaria. CDPKs are characterized by their unique regulatory structure. They possess a catalytic kinase domain that is tethered to a calmodulin-like regulatory domain by a junction domain. Activation of the kinase occurs upon binding of Ca2+ to the calmodulin-like domain with associated binding of the junction domain by the N-lobe of the calmodulin-like domain. Inhibition of the kinase active site is relieved when this binding occurs, allowing phosphorylation of substrates (2).
CDPKs in Plasmodium are present as a multigene family comprising five members (3). Each gene is expressed predominantly at a distinct phase of the parasite life cycle. For example, CDPK3 is expressed in ookinetes. Plasmodium berghei parasites without a functional CDPK3 gene (ΔCDPK3) have a reduced ability to invade the midgut of the mosquito host and also show a reduction in the gliding motility phenotype characteristic of this parasite stage. However, sporozoites from ΔCDPK3 parasites are indistinguishable from wild-type parasites, with normal gliding motility, and the asexual blood stages develop and invade red blood cells normally (4, 5). PbCDPK4 is up-regulated in sexual stages of P. berghei and has been shown to be crucial in the development of parasites at a very specific stage of the life cycle; it regulates cell cycle progression in the male gameto-cyte and is therefore important in sexual reproduction and transmission to the mosquito host (6). These data suggest that different CDPKs are functional at different stages of the parasite life cycle.
Zhao et al. (7) first identified PfCDPK1, and it has been proposed that the protein is associated with the parasitophorous vacuole membrane (PVM) (8). During the asexual cycle of P. falciparum, expression of CDPK1 is restricted to late blood stages of the parasite and is at its highest in mature schizonts (9). The protein is membrane-associated by virtue of dual acylation of its N terminus, with myristate added at Gly2 and palmitate at Cys3, followed by a stretch of amino acids enriched in basic residues. Mutagenesis studies have shown the importance of each of these three features in conferring membrane localization to the protein (8). Disruption of the cdpk1 gene in P. berghei has not been possible, suggesting that it is indispensable for parasite viability in the asexual blood stages.4 Although CDPK1 has been shown to phosphorylate the exogenous substrates casein and histone, only one Plasmodium protein, Raf kinase inhibitor protein (RKIP), has been shown to be phosphorylated by PfCDPK1 in vitro; it was proposed to be a regulator of PfCDPK1, perhaps by modulating the activity of the kinase to other substrates (10).
It is possible that the substrates of CDPKs are directly involved in motility and cell invasion; CDPK3 knock-out parasites or parasites treated with inhibitors of CDPKs show reduced gliding and invasion in both Plasmodium and the related parasite T. gondii (4, 5, 11). We have therefore reinvestigated the subcellular location of CDPK1 in asexual blood stages and examined phosphorylation by CDPK1 of components of the motor complex. Myosin A (MyoA), the myosin light chain known as myosin tail domain interacting protein (MTIP), and the 50- and 45-kDa glideosome-associated proteins (GAP50 and GAP45) are present in a heterotetrameric complex at the inner membrane complex (IMC), under the plasma membrane of the invasive stages of Plasmodium (12). GAP45 is a membrane-anchored protein of unknown function. Like CDPK1, it undergoes dual acylation of its N terminus, resulting in the addition of myristate and palmitate groups that, together, are capable of providing a stable membrane anchorage (13). MTIP binds strongly to the short tail of MyoA and shares homology with calmodulin and related proteins such as myosin light chains (14, 15). The force required for the parasite to invade its host cell is generated by translocation of actin filaments by MyoA. The actin filaments themselves are linked, indirectly, to adhesins that traverse the parasite plasma membrane and interact with receptors on the host erythrocyte surface (16). The net result of the myosin power stroke is forward propulsion of the parasite into the forming parasitophorous vacuole within the host cell.
Here we reinvestigate the subcellular localization of CDPK1 and identify substrates. We demonstrate that CDPK1 is located at the periphery of merozoites and can phosphorylate both GAP45 and MTIP. We identify phosphorylated peptides of GAP45 and MTIP and show that both proteins are phosphorylated in the parasite. The potential role of these phosphorylation events in regulating the actin-myosin motor is discussed.
EXPERIMENTAL PROCEDURES
Parasite Culture—P. falciparum clone 3D7 was cultured, and merozoites were purified according to previously described methods (17). Invasion inhibition assays were carried out by the addition of K252a (Biomol) to late stage schizonts at a starting parasitaemia of 0.5%, with 2% hematocrit. The solvent used was 10% Me2SO, and inhibitor was added to the appropriate final concentration with a final Me2SO concentration of 0.5%. Invasion rates were measured using a FACSCalibur machine (Becton Dickinson) 48 h after the addition of inhibitor according to previously described methods (18). These data were confirmed by microscopic examination of Giemsa-stained smears of parasite cultures.
Total parasite lysates were prepared by the addition of reducing SDS-PAGE sample buffer to cell pellets, followed by heating the sample to 95 °C for 5 min. Carbonate extracts of merozoites were prepared by incubating merozoites in 0.1 m Na2CO3, pH 11.0, on ice for 20 min. Supernatants were collected by ultra-centrifugation at 100,000 × g in a Beckman TL-100 ultracentrifuge. The carbonate-insoluble pellet was resuspended in reducing SDS-PAGE sample buffer. For immunoblots, carbonate-soluble and insoluble fractions were separated by SDS-PAGE on a 12% BisTris NuPAGE gel (Invitrogen), transferred to Protran membrane (Whatman), and incubated with the appropriate antibodies.
Recombinant Proteins and Antibodies—Open reading frames for PfCDPK1, PfMTIP, PfGAP50, and PfGAP45 were amplified from late schizont cDNA and cloned into the expression vectors pET30XaLIC, (PfCDPK1 and PfMTIP; for Fig. 3 only), pET46EkLIC (PfMTIP, in all experiments barring those in Fig. 3), pET30XaLIC (PfGAP50), and pQE60 (PfGAP45). PfCDPK1 was expressed in BL21(DE3) pLysS cells, PfMTIP, and PfGAP50 in BL21(DE3) cells and PfGAP45 in BL21 cells (all Stratagene). After induction with 1 mm isopropyl β-d-thiogalactopyranoside, the cells were lysed in Bugbuster (Novagen) and His-tagged proteins purified on nickel-nitrilotriacetic acid-agarose (Qiagen) in the cases of CDPK1, GAP50, and MTIP and TALON resin (Clontech) in the case of GAP45. Recombinant MSP7-22 was a gift from Dr. Madhusudan Kadekoppala (National Institute for Medical Research).
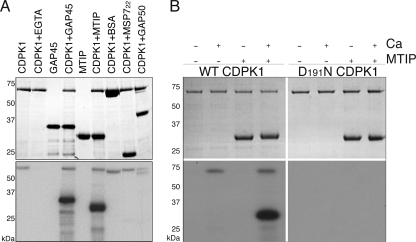
FIGURE 3.
GAP45 and MTIP are substrates of CDPK1. A, the upper panel shows a Coomassie Blue-stained gel of an in vitro kinase assay performed with recombinant CDPK1 and MTIP, GAP45, bovine serum albumin, MSP722, or GAP50 substrates. The lower panel is an autoradiograph of duplicate reactions incubated in the presence of [γ-32P]ATP. Only MTIP and GAP45 are phosphorylated by CDPK1. In all cases we see autophosphorylation of CDPK1, except when EGTA is included in the reaction mixture. Recombinant GAP50 was treated with factor Xa to remove the 12-kDa thioredoxin tag encoded by the pET32 vector. Residual factor Xa has cleaved a proportion of the tag present on CDPK1 (∼5 kDa, encoded in the pET30 vector), resulting in CDPK1 running as a doublet in this lane. B, phosphorylation of MTIP is absolutely dependent on the presence of calcium, and a substitution of Asn for Asp191 in the catalytic triad of CDPK1 results in an inactive protein. The two upper panels show Coomassie-stained gels of kinase reactions; the left panel shows reactions with wild-type CDPK1, and the right panel shows reactions with the Asp191 to Asn substitution. The lower panels show autoradiographs of the same reactions incubated in the presence of [γ-32P]ATP.
A kinase-dead mutant of CDPK1 was produced in which Asn substituted Asp191 of the catalytic triad. This was achieved using a QuikChange II mutagenesis kit (Stratagene) according to the manufacturer's instructions with the primers: 5′-CATAAACATAATATTGTACATCGAAATATTAAACCAG-3′ and 5′-CTGGTTTAATATTTCGATGTACAATATTATGTTTATG-3′. Similarly, recombinant MTIP with single amino acid substitutions (Ser47, Ser51, Ser55, Ser58, and Ser61 substituted with alanine) or double S47A/S51A substitutions were generated using a QuikChange II mutagenesis kit (Stratagene).
A fragment encoding the calmodulin-like domain of CDPK1 was also amplified using the primers 5′-GACGACGACAAGATGGCAACAATATTAATAAAAGTGATCAA-3′ and 5′-GAGGAGAAGCCCGGTTTATGAAGATTTATTATCACAAATTTTGTG-3′, where the bases in italics represent vector-specific sequence to generate overhangs for ligation-independent cloning into pET46EkLIC. Purified recombinant protein was used as an antigen. The antisera were raised in Balb/c mice and New Zealand White rabbits for each of the purified recombinant proteins, using standard procedures. Other antibodies used were rabbit anti-PTRAMP (19), mouse monoclonal 89.1 against MSP1 (20), rabbit anti-MSP7 (21), and rabbit anti-PfCRT (from the Malaria Reagent and Research Resource Centre (MR4)).
Immunofluorescence—Thin smears of parasites were air-dried and then fixed using 3% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. The cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min followed by blocking in 3% bovine serum albumin in PBS overnight at 4 °C. Antibodies were diluted in blocking buffer and incubated on the slide for 1 h followed by washing in PBS. Antigen visualization was achieved using Alexa-Fluor-488- or 594-conjugated anti-species antibodies (Molecular Probes). The images were processed using a DeltaVision cooled charge-coupled device imaging system (Applied Precision Inc.). Images from the fluorescence microscope were collected and analyzed with Softworx and prepared for publication with Adobe Photoshop.
Cell Lysis and Shrimp Alkaline Phosphatase Treatment—Synchronized schizonts (∼40-45 h) were lysed in 0.15% saponin in Tris-buffered saline. Parasites were lysed in a buffer containing 7 m urea, 2 m thiourea, 4% CHAPS, 25 mm Tris (all PlusOne reagents; GE Healthcare), using 250 μl/1 × 108 cells. To ensure complete lysis, the cells were frozen in a dry ice/ethanol bath and thawed at 29 °C five times. Insoluble material was pelleted by centrifugation. An alkaline phosphatase reaction was set up using 300 μg of cell lysate, 3 μl of protease inhibitor mix (GE Healthcare), 20 units of shrimp alkaline phosphatase (SAP; Promega), in a total of 300 μl. A control reaction was set up in parallel as above but with the addition of 6 μl of phosphatase inhibitor mixture I (Sigma) and 20 mm imidazole. The lysates were incubated at 37 °C for 30 min and then heated to 65 °C for 15 min to inactivate the SAP. The proteins were precipitated using methanol/chloroform, followed by an acetone precipitation, and resuspended in rehydration buffer containing 6 m urea, 2 m thiourea, and 4% CHAPS.
Two-dimensional Electrophoresis and Western Blotting—Rehydration buffer containing 0.01% dithiothreitol, 0.002% bromphenol blue and 0.02% immobilized pH gradient buffer was added to 50 μg of control treated or SAP treated schizont lysate to give a total volume of 125 μl/sample. Isoelectric focusing was carried out on IPGPhor (GE Healthcare) using 7-cm immobilized pH gradient strips (pH 3-10NL; GE Healthcare) to a maximum voltage of 5000 V, collecting a total of 22000 Vh. After isoelectric focusing, immobilized pH gradient strips were incubated for 15 min at room temperature in equilibration buffer (2% SDS, 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% (v/v) glycerol, 0.002% bromphenol blue) with 10 mg/ml dithiothreitol, followed by a second incubation in equilibration buffer containing 25 mg/ml iodoacetamide. The strips were run in parallel in the second dimension using NuPAGE 4-12% BisTris Zoom gels (Invitrogen). The gels were blotted onto polyvinylidene difluoride membrane and probed with affinity-purified rabbit anti-MTIP (4 μg/ml) or polyclonal rabbit anti-GAP45 (1/500), using an horseradish peroxidase-conjugated anti-rabbit IgG (1/15,000; Sigma) as a secondary antibody. The signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Samples from a parasite time course were lysed in the same buffer but separated by one-dimensional SDS-PAGE. Scanning densitometry of autoradiographs was performed using Image J software (22).
Kinase Assays and Kinetics—Radiolabeled phosphate incorporation kinase assays were carried out with 15 pmol of CDPK1 and 200 pmol of substrate protein in 20 mm Tris-HCl, pH 8.0, 20 mm MgCl2, 100 μm ATP, 0.1 MBq of [γ-32P]ATP, 1 mm CaCl2. The reactions were incubated for 15 min at 30 °C, stopped by the addition of reducing sample buffer, and analyzed by SDS-PAGE. After fixation in a solution of 30% methanol plus 5% glycerol, the gels were dried and exposed to BiomaxMR film (Kodak). To prepare samples for mass spectrometry and fluorimetric analysis, 3.5 nmol of substrate proteins were incubated with 150 pmol of CDPK1 under the same conditions (without radiolabeled ATP) for 16 h at room temperature.
Kinase activity measurements were made using a continuous spectrophotometric assay based on one described by Cook et al. (23). A typical assay contained the following components in a total volume of 0.5 ml in a 10-mm-pathlength cuvette: 50 mm Hepes (pH 7.5), 1 mm CaCl2, 10 mm MgCl2, 6 units of pig heart l-lactic dehydrogenase, 4 units of rabbit muscle pyruvate kinase, 1 mm phosphoenolpyruvate, 250 mm NADH. Concentrations of ATP and MTIP (or GAP45) were varied as appropriate. In experiments for determining the Km for MTIP and GAP45, the concentration of ATP was fixed at 700 μm. The reactions were initiated by the addition of CDPK1 (at 10-15 μg/ml), and the rates (at 30 °C) were obtained by recording absorbance changes at 340 nm. Kinetic parameters were determined by direct nonlinear least squares fits to the Michaelis-Menten equation using CURFIT software (Dr. S. R. Martin). This coupled assay was validated by comparison with a radio-metric assay using [γ-32P]ATP (24).
GAP45 Purification from Merozoites—Merozoites were lysed in 1% n-octyl-β-d-glucopyranoside, 50 mm Tris-HCl, 5 mm EDTA, 150 mm NaCl, pH 8.0, on ice for 20 min. The supernatants were collected by ultracentrifugation at 100,000 × g in a Beckman TL-100 ultracentrifuge. GAP45 was purified from these supernatants using an immunoaffinity column of rabbit anti-GAP45 antibodies cross-linked to cyanogen bromide-activated Sepharose (GE Healthcare). After extensive washing with lysis buffer and PBS, GAP45 was eluted using 0.1 m glycine, pH 2.7, and separated by SDS-PAGE, and the relevant band was excised from the gel. The protein bands were reduced with dithiothreitol and alkylated using iodoacetamide. The gel was dried and reswollen in a sufficient volume to cover of 2 ng/μl modified sequencing grade trypsin or 1 ng/μl AspN (both Promega) in 5 mm ammonium bicarbonate. After overnight digestion at 32 °C, the supernatant was acidified by the addition of a 1/10th volume of 4% trifluoroacetic acid.
Purification of Phosphopeptides—Recombinant proteins were phosphorylated by CDPK1, subjected to digestion with trypsin (MTIP) or AspN (GAP45) and phosphopeptides enriched by purification on either gallium spin columns (Pierce) for GAP45 or TALON magnetic beads (Clontech) for MTIP according to the manufacturer's instructions.
Peptide Mass Fingerprinting by MALDI-TOF Mass Spectrometry—Peptides from protease digestion were analyzed by MALDI-TOF, and the spectra were examined to identify phosphorylated peptides. Peptide mass fingerprinting was performed using a Reflex III MALDI time-of-flight mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany), equipped with a nitrogen laser and a Scout-384 probe, to obtain positive ion mass spectra of digested protein with pulsed ion extraction in reflectron mode. An accelerating voltage of 26 kV was used with detector bias gating set to 2 kV and a mass cut-off of m/z 650. The matrix surfaces were prepared using recrystallized α-cyano-4-hydroxycinammic acid and nitrocellulose using the fast evaporation method (25). 0.4 μl of acidified digestion supernatant was deposited on the matrix surface and allowed to dry prior to desalting with water.
RESULTS
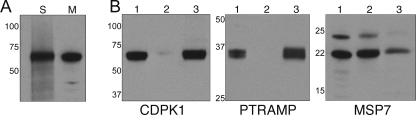
Expression of PfCDPK1 in Merozoites and Localization to the Parasite Plasma Membrane—We used anti-CDPK1 antibodies in a Western blot with schizont and merozoite cell lysates. It is evident that PfCDPK1 is present in free merozoites, as well as schizonts (Fig. 1A). In previously published work it was suggested that PfCDPK1 is located in the PVM of developing parasites (8), but because free merozoites do not contain a PVM, it is clear that CDPK1 must be present in another location in the parasite. The protein from merozoites is still associated with membranes, as demonstrated by partitioning of CDPK1 in the insoluble fraction of merozoites extracted with a high pH carbonate buffer (Fig. 1B). The same fraction contains membrane-associated proteins such as PTRAMP (19). By contrast, peripheral membrane proteins such as MSP7 are solubilized by this buffer (Fig. 1B).
FIGURE 1.
CDPK1 is an integral membrane protein in merozoites. A, Western blot analysis using rabbit anti-CDPK1 antibodies. S, schizont lysate; M, merozoite lysate. B, Western blots of merozoites extracts using anti-CDPK1, anti-PTRAMP, and anti-MSP7 antibodies. Lanes 1, total merozoite lysate; lanes 2, carbonate buffer soluble fraction; lanes 3, carbonate buffer insoluble fraction. The numbers on the left of each panel indicate the positions of molecular mass markers in kilodaltons.
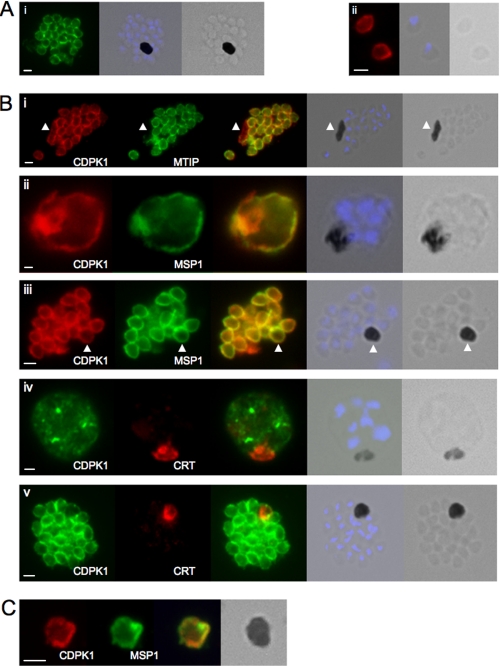
We examined the subcellular location of CDPK1 in free merozoites and schizonts by immunofluorescence. CDPK1 is present at the periphery of merozoites, both within schizonts, and released (Fig. 2A, panels i and ii, respectively). From this image we cannot distinguish between the plasma membrane and membranes of the IMC. However, dual staining of schizonts with anti-CDPK1 antibodies and antibodies raised against MTIP (a marker for the IMC) and MSP1 (a marker for the parasite plasma membrane) revealed interesting differences. CDPK1 colocalizes with MTIP around developing merozoites in segmented schizonts but is also present in membranes around the mature food vacuole/residual body of the schizonts, whereas MTIP is absent (Fig. 2B, panel i). CDPK1 and MSP1 colocalize entirely. In young schizonts (∼39 h post-invasion) where segmentation has not begun, both CDPK1 and MSP1 are present in the parasite plasma membrane (Fig. 2B, panel ii). In more mature schizonts where segmentation is complete, both CDPK1 and MSP1 are seen in the plasma membrane around the food vacuole (Fig. 2B, panel iii). To exclude the possibility that CDPK1 is present within the food vacuole membrane, we stained parasites with antibodies against PfCRT, a known marker of the food vacuole membrane. There was no coincidence of staining in early schizonts and a small amount of colocalization in segmented schizonts (Fig. 2B, panel iv and v). We also see CDPK1and MSP1 in the membranes surrounding residual bodies, structures that consist of food vacuoles containing hemozoin that are discarded when a schizonts ruptures. Residual bodies are surrounded by parasite plasma membrane, a consequence of the mechanism by which merozoites are pinched off from the syncytium within the erythrocyte (26, 27) and therefore stain with known markers of the plasma membrane such as MSP1 (Fig. 2C).
FIGURE 2.
CDPK1 localizes to the plasma membrane of merozoites. A, an anti-CDPK1 antibody detects CDPK1 at the parasite periphery in schizonts (panel i) and free merozoites (panel ii). The second panel shows a merged image of nuclear staining with 4′,6′-diamino-2-phenylindole (blue) and the bright field image, and the third panel shows the bright field image. B, panel i, CDPK1 colocalizes with MTIP around the periphery of merozoites but is also found around the residual body, marked with an arrowhead, from which MTIP is absent. In young (panel ii) and segmented (panel iii) schizonts, CDPK1 colocalizes with the plasma membrane marker MSP1. Both proteins are also detected around the residual body, marked with an arrowhead. CDPK1 does not colocalize with the food vacuole marker PfCRT in either immature (panel iv) or mature (panel v) schizonts. The third panels in all of the images show the merged images of the antibody staining, beside which is a merged image of the bright field image and the nuclei of the parasites stained with 4′,6′-diamino-2-phenylindole (blue), followed by the bright field image alone. C, both CDPK1 (red) and MSP1 (green) can be detected on residual bodies that are released upon schizont rupture. The merged images of the antibody staining are shown in the third panels, followed by the bright field image. In all cases the white scale bar in the first panel of each set of images represents 1 μm.
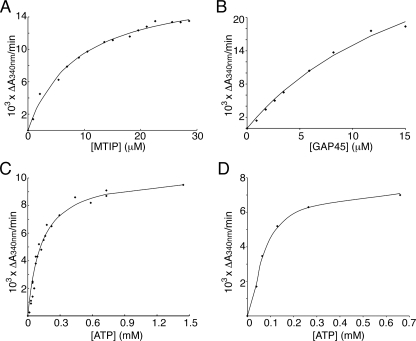
GAP45 and MTIP Are Substrates for PfCDPK1 in Vitro—Because of its close proximity to the IMC and its similar membrane anchorage to GAP45, a known component of the motor complex in the IMC, we tested the ability of recombinant CDPK1 to phosphorylate GAP45, GAP50, and MTIP in vitro. Initially, kinase assays were performed in which the incorporation of 32P from radiolabeled ATP into substrate proteins was measured. The proteins were separated by SDS-PAGE, and phosphorylation was detected by autoradiography. Both MTIP and GAP45 were efficiently phosphorylated by CDPK1, whereas another component of the motor complex, GAP50, was not. Further control proteins, MSP722, and bovine serum albumin were also not phosphorylated by CDPK1 (Fig. 3A). The absolute calcium dependence of this phosphorylation and the inability of a “kinase-dead” mutant of CDPK1 to autophosphorylate or modify MTIP is shown in Fig. 3B. The kinase-dead mutant was made by modifying Asp191 in the HRD catalytic triad of CDPK1 to Asn. We saw no effect of MTIP or GAP45 on autophosphorylation of CDPK1, in contrast to the increased autophosphorylation of CDPK1 in the presence of RKIP (10). Further coupled-enzyme kinase assays were used to determine the kinetic parameters for the enzyme substrate pairs. We determined the Km for MTIP to be 9 ± 0.9 μm (Fig. 4A). The Km calculation for GAP45 was carried out with a somewhat restricted data set because of limiting amounts of GAP45; however, a Km of 18 ± 5 μm was calculated (Fig. 4B). The Km of CDPK1 for ATP was calculated to be 125 ± 13 μm for MTIP and 96 ± 28 μm for GAP45 (Fig. 4, C and D). The activity of CDPK1 was calculated to be 0.2 μmol min-1 mg-1 enzyme with MTIP as a substrate and 0.5 μmol min-1 mg-1 enzyme with GAP45 as a substrate. Previous calculations of CDPK1 activity toward different substrates have been made. When phosphorylating the exogenous substrate casein, CDPK1 had an activity of 0.28 μmol min-1 mg-1 enzyme, whereas with the parasite protein RKIP as a substrate, the activity was 0.04 μmol min-1 mg-1 enzyme (10).
FIGURE 4.
Kinetic studies of CDPK1 and its substrates. Enzyme-coupled kinase assays measuring the reduction in NADH (ΔA340/min) are shown for CDPK1 while varying the substrate concentration. A, MTIP; B, GAP45. The ATP concentration in both cases was 700 μm. From these the Km for MTIP is calculated to be 9 ± 0.9 μm, and that for GAP45 is 18 ± 5 μm. ΔA340/min plotted against ATP concentration, to give the Km for ATP of CDPK1 with MTIP (C) or GAP45 (D) as 125 ± 13 and 96 ± 28 μm, respectively. These experiments were carried out at constant substrate concentration (8.2 μm MTIP or 3.5 μm GAP45). In all cases data points (♦) are plotted, with the solid line representing the best fit curve calculated by direct nonlinear least squares fit to the Michaelis-Menten equation using in-house software.
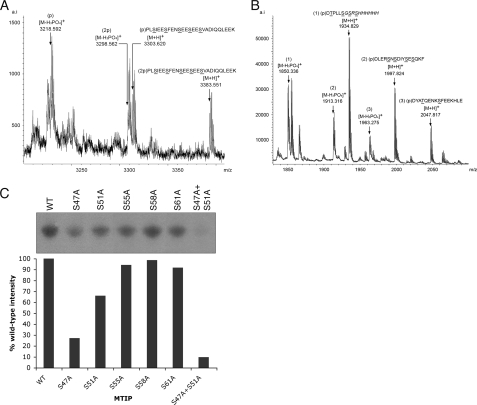
Phosphopeptides from MTIP and GAP45 were identified by MALDI-TOF mass spectrometry. Metastable assignments were confirmed by post-source decay analysis (data not shown). Recombinant proteins were phosphorylated by CDPK1 and subjected to digestion with trypsin (MTIP) or AspN (GAP45), and phosphopeptides were enriched by purification on either gallium spin columns or TALON magnetic beads. A single peptide from MTIP was detected, KPLSIEESFENSEESEES-VADIQQLEEK, which spans amino acids 44-71 in the MTIP sequence. The peptides were isolated in which either one or two of the five serines (Ser47, Ser51, Ser55, Ser58, and Ser61) were phosphorylated, along with their metastable products, because of phosphoric acid loss (Fig. 5A). For GAP45, two phosphopeptides and their metastable products were identified, each containing a single phosphorylated residue. The GAP45 phosphopeptides are DYATQENKSFEEKHLE and DLERSNSDIYSESQKF, spanning amino acids 81-96 and 97-112, respectively (Fig. 5B). The GAP45 spectrum also shows a phosphorylated peptide (m/z 1934.829) that is derived from the vector tag of pQE60.
FIGURE 5.
Analysis of phosphopeptides from recombinant MTIP and GAP45. A, MTIP tryptic peptides eluted from TALON magnetic beads. A single peptide is seen, with either one or two phosphates, along with metastable post-source decay products. B, phosphopeptides of GAP45 eluted from a gallium spin column. Three phosphopeptides are detected. Peptide 1 is derived from the vector-encoded protein tag, whereas peptides 2 and 3 are from GAP45. C, incorporation of 32P into MTIP mutants was measured by scanning densitometry of autoradiographs of in vitro kinase assays and compared with wild-type protein (WT). The S47A mutant showed 32P incorporation of 27%, the and S51A mutant showed 66% compared with wild-type MTIP. Mutation of Ser55, Ser58, and Ser61 did not significantly affect incorporation of 32P. A double mutant (S47A/S51A) gave a signal of 10% compared with WT MTIP.
To identify the specific serine residues in MTIP that are phosphorylated by CDPK1, recombinant MTIP proteins were made where serine residues 47, 51, 55, 58, and 61 were individually mutated to alanine by site-directed mutagenesis. Radio-metric kinase assays were performed, and the incorporation of 32P into the proteins was compared with that of the wild-type MTIP protein (Fig. 5C). Substitution of serines 55, 58, and 61 did not result in a significant reduction in phosphorylation of MTIP by CDPK1. In contrast, mutation of Ser47 or Ser51 resulted in a 73 and 34% reduction in the incorporation of 32P, respectively. When a double S47A/S51A mutant of MTIP was used, a reduction of 32P incorporation of 90% was observed. These data suggest that the primary phosphorylation of MTIP by CDPK1 occurs on Ser47, with a secondary phosphorylation occurring nonstoichiometrically on Ser51. Because the signal in the assay did not drop to zero when the S47A/S51A MTIP mutant was used, it would seem that there is a third site in MTIP that is phosphorylated at a much lower efficiency that remains unidentified.
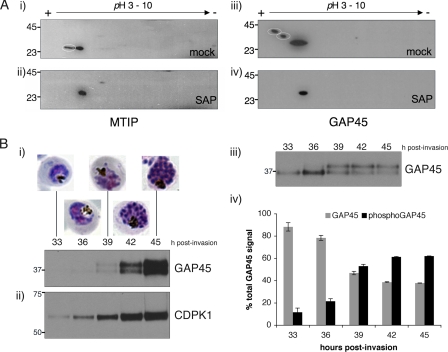
MTIP and GAP45 Are Phosphoproteins in the Parasite—It is crucial to establish that both GAP45 and MTIP are phosphorylated in parasites. To this end, schizont extracts were separated by two-dimensional SDS-PAGE and then immunoblotted with antibodies to either MTIP or GAP45. MTIP migrates as two spots that differ in charge (Fig. 6A, panel i). When phosphate groups were removed by treating the lysate with SAP, MTIP migrates as a single spot (Fig. 6A, panel ii). When the same extracts were probed using an anti-GAP45 antibody, GAP45 was identified as three spots that differ in both mass and charge in the mock treated lysate (Fig. 6A, panel iii). In SAP-treated samples GAP45 also migrates as a single species (Fig. 6A, panel iv). We further examined the phosphorylation of GAP45 during parasite development. Cell lysates from parasites ranging from late trophozoite (33 h post-invasion) to late schizont stages (45 h post-invasion) were separated by one-dimensional SDS-PAGE and blotted with an anti-GAP45 antibody. Expression of GAP45 is just detectable at early time points, and its expression peaks at 45 h post-invasion, where it migrates as a doublet. The upper band (containing phosphorylated GAP45) increases in intensity in relation to the lower band (nonphosphorylated GAP45) as development of the parasite progresses (Fig. 6B, panel i). If the amount of protein loaded is adjusted to normalize GAP45 levels, the relative proportion of phosphorylated versus nonphosphorylated GAP45 at a given time point can be calculated (Fig. 6B, panel iii and iv). The proportion of phosphorylated GAP45 increases from 12% at 33 h to 62% at 45 h. This increase mirrors the expression profile of CDPK1 (Fig. 6B, panel ii).
FIGURE 6.
GAP45 and MTIP are phosphoproteins in the parasite. A, protein extracted from late-stage schizonts was subjected to treatment with SAP, separated by two-dimensional SDS-PAGE and Western blotted with anti-MTIP or anti-GAP45 antibodies. Both MTIP (panel ii) and GAP45 (panel iv) antibodies detect only a single spot. In mock-treated samples, two spots are visible in the anti-MTIP blot (panel i) and three spots in the anti-GAP45 blot (panel iii). The phosphorylated proteins that disappear upon SAP treatment are circled in panels i and iii. B, lysates from a developmental time-course of parasites were probed with anti-GAP45 (panel i) and anti-CDPK1 antibodies (panel ii). Panel iii, loading of lysates normalized according to GAP45 levels shows that the relative proportion of phosphorylated GAP45 (upper band) increases compared with unphosphorylated GAP45 as parasite development progresses. Panel iv, scanning densitometry of anti-GAP45 immunoblots showing the proportion of phosphorylated and unphosphorylated GAP45 at each developmental time point. Each value represents the average of two experiments, with error bars showing one standard deviation from the mean.
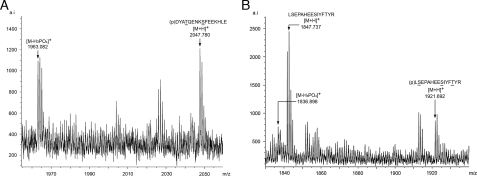
GAP45 Phosphorylation in Parasites: Identification of Phosphopeptides—GAP45 was purified from merozoites using an anti-GAP45 antibody affinity column. Purified protein was excised from an SDS-PAGE gel and digested with either AspN or trypsin, and the resultant peptides were analyzed by MALDI-TOF to identify phosphorylated peptides. Two phosphopeptides were identified: DYATQENKSFEEKHLE in the AspN digested sample (Fig. 7A) and an additional tryptic peptide LSEPAHEESIYFTYR corresponding to residues 141-155 (Fig. 7B). The AspN peptide is identical to one of the peptides identified in in vitro phosphorylation of GAP45 by CDPK1. The detection of a phosphopeptide in parasite-derived GAP45 that was absent from the tryptic digest of recombinant GAP45 phosphorylated by CDPK1 (data not shown) is strong evidence that an as yet unidentified parasite protein kinase phosphorylates this site.
FIGURE 7.
MALDI-TOF analysis of phosphopeptides from affinity-purified parasite-derived GAP45. Phosphopeptides (p) and their post-source decay products ([M-H3PO4]+) from AspN digestion (A) and trypsin digestion of merozoite-derived GAP45 (B).
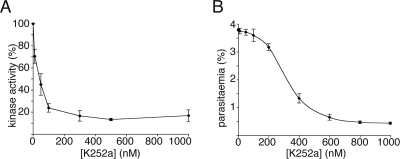
Inhibition of CDPK1 by the Bisindolocarbazole K252a—The effect of the kinase inhibitor KT5926 has been tested in T. gondii. The inhibitor blocked attachment of parasites to host cells and reduced the activity of TgCDPK1 with similar IC50 values of ∼100 nm (11). We tested the effect of a related inhibitor, K252a, on P. falciparum cultures and on the ability of CDPK1 to phosphorylate recombinant MTIP in vitro. K252a potently inhibited CDPK1 activity in vitro, with an IC50 of 45 nm. P. falciparum invasion of erythrocytes was inhibited in the presence of K252a. At very high concentrations (0.8-5 μm) invasion of erythrocytes was completely blocked. On examination of Giemsa-stained smears of the culture, it was clear that the egress of merozoites from schizonts was blocked rather than invasion of erythrocytes by merozoites. At intermediate concentrations of K252a (0.4 μm), invasion was inhibited by 65%. At these intermediate concentrations of K252a, egress of merozoites from schizonts was not prevented; however, invasion of erythrocytes by merozoites was significantly inhibited, with half-maximal inhibition occurring at 348 nm (Fig. 8).
FIGURE 8.
Inhibition of CDPK1 and parasite invasion by K252a. A, CDPK1 activity on recombinant MTIP in the presence of K252a was assayed using radiolabeled ATP. The IC50 for the enzyme was calculated to be 45 nm. B, the rate of invasion of parasites treated with K252a. Half-maximal inhibition of invasion occurred at 348 nm K252a. In both graphs, the values plotted are the averages of triplicate experiments, with the error bars showing one standard deviation from the mean.
DISCUSSION
We have shown that CDPK1 is present in both schizonts and free merozoites in a membrane-bound fraction, which is consistent with the dual acylation of the protein. The protein is located at the plasma membrane of the developing intracellular schizont both before and after segmentation as indicated by colocalization with MSP1. In segmenters it can be detected on the plasma membrane that surrounds the food vacuole, which is identified by the presence of the hemozoin pigment and reactivity with antibodies to CRT (28, 29). Notably, MTIP, a marker for the IMC, is absent from this membrane. Following schizont rupture and release of merozoites, the protein is still detected at the periphery of the discarded residual body derived from the food vacuole but most importantly is located at the periphery of the free merozoite. In free merozoites we cannot exclude that it is also present on other peripheral membranes, particularly the IMC. It has previously been proposed that CDPK1 is exported from the schizont to the PVM of the intracellular parasite, which would require transfer of this protein across the parasite plasma membrane followed by insertion into the PVM (8). Although we cannot exclude that this transfer occurs, our results indicate that CDPK1 is largely present in a membrane bound form at the periphery of both schizonts and free merozoites. We suggest therefore that in free merozoites the protein is located in the space between the plasma membrane (PM) and the inner membrane complex (IMC), which is the location of the actomyosin motor that drives merozoite invasion of red blood cells (30). The dual acylation of CDPK1 is sufficient to retain it at the membrane, and this post-translational modification is identical to that of GAP45, which is also myristoylated and palmitoylated at the N terminus (13). It is noteworthy that GAP45, which is a component of the motor complex, is thought to be located in the space between the IMC and the PM but attached to the IMC membrane (12, 31); it may be that CDPK1 can be associated with both the inner face of the PM and the outer face of the IMC.
We have expressed and purified active CDPK1 from recombinant bacteria. The activity has an absolute requirement for calcium, and the inactivity of the kinase-dead form of the protein resulting from replacement of Asp by Asn in the active site is consistent with the detected kinase activity being attributable to CDPK1. Of particular interest is that the enzyme can phosphorylate two proteins of the motor complex, MTIP and GAP45, whereas a third component of the complex, GAP50 and other control proteins, are not modified by CDPK1. Both MTIP and GAP45 are good substrates for CDPK1 with Km values in the low micromolar range that is well within the suggested threshold of a Km ≤ 50 μm for potential substrates (1). Similarly the activity of CDPK1 with both MTIP and GAP45 is in the same range as its activity with the exogenous substrate casein (0.2-0.5 μmol min-1 mg-1 enzyme) and well above the reported activity with the parasite protein RKIP as a substrate (0.04 μmol min-1 mg-1 enzyme) (10). These data confirm that P. falciparum CDPK1 is a calcium-dependent protein kinase and show that two components of the merozoite motor complex, MTIP and GAP45, are good substrates for phosphorylation. Both MTIP and GAP45 are located in the same cytoplasmic location as CDPK1, between the PM and the IMC.
To examine the phosphorylation of MTIP and GAP45 further, we studied the sites of modification by CDPK1 of the recombinant expressed proteins in vitro by peptide digestion and analysis by mass spectrometry. Bioinformatic predictions suggest that many residues in both MTIP and GAP45 are capable of being phosphorylated by serine-threonine protein kinases. We identified experimentally one peptide from MTIP containing five serines that was phosphorylated once or twice and by mutagenesis have shown that the primary phosphorylation occurs at Ser47, with a minor phosphorylation at Ser51. The two phosphopeptides from GAP45 contain either one threonine and one serine or four serines, respectively, which were both phosphorylated only in one position. It is unlikely that the threonine residue in GAP45 peptide 81-96 is phosphorylated because the phosphorylated protein did not react with phospho-threonine specific antibodies (data not shown), and therefore the phosphorylated residue is Ser89. The presence of multiple serines in peptides 97-112 and 141-155 of GAP45 and poor fragmentation of the peptides in the mass spectrometer have not allowed a definitive identification of the specific residues that are modified. This information will require further studies with peptides or modified recombinant proteins.
Although we showed that both MTIP and GAP45 could be phosphorylated in vitro, it was important to show that this also occurred in vivo. The phosphorylation of both proteins in the parasite was confirmed by two-dimensional gel analysis of the proteins with or without phosphatase treatment. In both cases multiple forms of the protein in the absence of phosphatase treatment were replaced by a single spot, consistent with the removal of phosphate residues by the enzyme. At least one of the forms of each of the proteins in the parasite prior to phosphatase treatment migrated in the same location as the enzyme-treated proteins, suggesting that the phosphorylation is only partial and substoichiometric in schizonts. We have also demonstrated that phosphorylation of GAP45 increases as parasite development progresses and that this mirrors CDPK1 expression. To confirm the phosphorylation of the proteins in vivo, we wished to purify them from parasite extracts and subject them to protease digestion and analysis by mass spectrometry of phosphorylated peptides. We were unable to purify sufficient MTIP for analysis, but sufficient GAP45 was obtained to carry out some analyses. One of the two phosphopeptides observed in recombinant GAP45 was identified, confirming that this sequence is phosphorylated both in vitro and in vivo and implicating CDPK1 as the kinase responsible. In addition, a second novel phosphorylated peptide was identified, suggesting that GAP45 is also the substrate for a second kinase. A recent study has shown that GAP45 is a substrate of PfPKB in vitro (41); it is possible that PfPKB is responsible for the phosphorylation of GAP45 at the “non-CDPK1” site identified here.
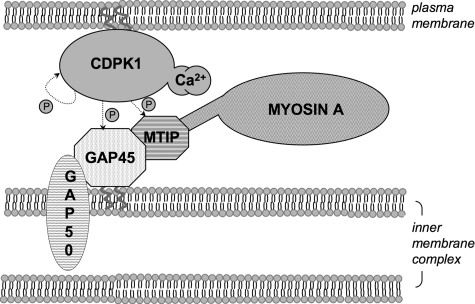
A diagram summarizing our findings is shown in Fig. 9. Activation of plasma membrane-situated CDPK1 by a calcium signal results in phosphorylation of GAP45 and MTIP associated with the IMC. These two membranes must be sufficiently close for this phosphorylation to occur. Our data also suggest that a calcium signal of sufficient magnitude to activate CDPK1 exists in merozoites prior to erythrocyte attachment and invasion.
FIGURE 9.
CDPK1 and the motor complex. After activation by calcium, plasma membrane associated CDPK1 phosphorylates GAP45 and MTIP. GAP45 is directly associated with the outer membrane of the IMC, in a complex with MTIP, GAP50, and myosin A. GAP45 is further phosphorylated by an unidentified kinase in merozoites.
It is of interest to note that the phosphorylated residues in MTIP are in the N-terminal region that was not present in the recently described structure (32). The N-terminal 70 residues extend beyond the region of MTIP that is similar to calmodulin and other myosin light chains and binds the C-terminal tail of myosin A. Furthermore, phosphorylation of MTIP does not affect its binding to a MyoA tail peptide.5 The role of this N-terminal region of MTIP is not known, but it is possible that it may be important for binding to GAP45 because GAP45 is in the motor complex but does not bind directly to the myosin tail.5 Thus it is possible that phosphorylation of either or both MTIP and GAP45 may modulate the interaction between them.
Another possibility is that phosphorylation of MTIP, which is a myosin light chain, is important in control of the motor. Phosphorylation of the regulatory light chains of myosin (RLCs) has been shown to be important in modulating the myosin motor in several systems. In skeletal muscle, phosphorylation of RLC by Ca2+/calmodulin-dependent skeletal muscle myosin light chain kinase leads to enhanced skeletal muscle contraction (33). In cardiac muscle, phosphorylation of RLC results in increased cross-bridge formation and enhanced force from contraction (34). In smooth muscle, phosphorylation of the RLC of myosin II by the Ca2+/calmodulin-dependent myosin light-chain kinase and dephosphorylation by a type 1 phosphatase are the principal mechanisms of regulation of smooth muscle tone (35). Given this evidence, it is possible that phosphorylation of MTIP and/or GAP45 will have consequences on the force generated by MyoA.
Apart from the potential role of GAP45 phosphorylation in an interaction with MTIP or in motor control as suggested above, are there further possibilities? GAP45 has only been detected in Apicomplexan parasites, and apart from the N-terminal acylation sites and a 50-amino acid C-terminal domain, the protein is poorly conserved and of low complexity composition. It is possible that phosphorylation confers structure by adding additional negative charge to this low complexity region or facilitates interaction with other molecules important in motor function, such as GAP50 or others that have yet to be identified.
We have shown that K252a can inhibit CDPK1 in vitro, with an IC50 of 45 nm. Treatment of late stage P. falciparum cultures with K252a also inhibited invasion of erythrocytes by merozoites, but with a much higher IC50 of 348 nm. At high concentrations of K252a the egress of merozoites from schizonts was inhibited, whereas at intermediate concentrations egress was not affected, but merozoites failed to invade erythrocytes at the same rate as solvent-treated control parasites. The two distinct effects of K252a on parasite development could be due to non-specific inhibition of parasite kinases at high concentrations of inhibitor and a more specific inhibition of CDPK1 at intermediate concentrations. However, we cannot exclude that there are other CDPK1 substrates that play a crucial role in merozoite egress from schizonts or that GAP45 and MTIP are important in this process, as well as their role in invasion. While this paper was under review Kato et al. (36) published an article in which ontology-based pattern identification was used to establish potential targets of CDPK1. In this analysis proteins of the motor complex, including MTIP and GAP45, were identified as possible substrates. The authors identified a 2,4,6-trisubstituted purine compound, purfalcamine, that inhibits CDPK1 activity in vitro and inhibits merozoite egress from schizonts in culture. It is interesting that, like treatment with high concentrations of K252a in our study, treatment of parasites with purfalcamine also results in an inhibition of merozoite egress from the schizont. Previous investigations have demonstrated that parasite kinase activity is important for invasion of erythrocytes by merozoites. Treatment of parasites with the broad spectrum kinase inhibitor staurosporine resulted in merozoites arrested on the surface of erythrocytes, after the formation of a junction between the two cells (37). In addition, the importance of calcium in invasion and parasite motility has been shown in several studies (38-40). Phosphorylation of motor complex components by a calcium-dependent kinase may therefore be a crucial regulator of motility and host cell invasion by Plasmodium.
Acknowledgments
We thank Dr. Madhusudan Kadekoppala (National Institute for Medical Research) for the gift of recombinant MSP722. Thanks also to Solabami Ogun for technical assistance with antibody generation.
This work was supported, in whole or in part, by National Institutes of Health Grant 0600370P7601176. This work was also supported by the United Kingdom Medical Research Council and Wellcome Trust Grant 066742. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: CDPK, calcium-dependent protein kinase; IMC, inner membrane complex; MTIP, myosin A tail domain interacting protein; GAP, glideosome-associated protein; PVM, parasitophorous vacuole membrane; MSP, merozoite surface protein; RKIP, Raf kinase inhibitor protein; MyoA, myosin A; BisTris, bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane HCl; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; SAP, shrimp alkaline phosphatase; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; PM, plasma membrane; RLC, regulatory light chain of myosin.
R. Tewari, personal communication.
J. L. Green and S. R. Martin, unpublished observation.
References
- 1.Harper, J. F., and Harmon, A. (2005) Nat. Rev. Mol. Cell. Biol. 6 555-566 [DOI] [PubMed] [Google Scholar]
- 2.Chandran, V., Stollar, E. J., Lindorff-Larsen, K., Harper, J. F., Chazin, W. J., Dobson, C. M., Luisi, B. F., and Christodoulou, J. (2006) J. Mol. Biol. 357 400-410 [DOI] [PubMed] [Google Scholar]
- 3.Ward, P., Equinet, L., Packer, J., and Doerig, C. (2004) BMC Genomics 5 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishino, T., Orito, Y., Chinzei, Y., and Yuda, M. (2006) Mol. Microbiol. 59 1175-1184 [DOI] [PubMed] [Google Scholar]
- 5.Siden-Kiamos, I., Ecker, A., Nybäck, S., Louis, C., Sinden, R. E., and Billker, O. (2006) Mol. Microbiol. 60 1355-1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billker, O., Dechamps, S., Tewari, R., Wenig, G., Franke-Fayard, B., and Brinkmann, V. (2004) Cell 117 503-514 [DOI] [PubMed] [Google Scholar]
- 7.Zhao, Y., Kappes, B., and Franklin, R. M. (1993) J. Biol. Chem. 268 4347-4354 [PubMed] [Google Scholar]
- 8.Möskes, C., Burghaus, P. A., Wernli, B., Sauder, U., Dürrenberger, M., and Kappes, B. (2004) Mol. Microbiol. 54 676-691 [DOI] [PubMed] [Google Scholar]
- 9.Bozdech, Z., Llinas, M., Pulliam, B. L., Wong, E. D., Zhu, J., and DeRisi, J. L. (2003) PLoS Biol. 1 85-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kugelstadt, D., Winter, D., Plückhahn, K., Lehmann, W. D., and Kappes, B. (2007) Mol. Biochem. Parasitol. 151 111-117 [DOI] [PubMed] [Google Scholar]
- 11.Kieschnick, H., Wakefield, T., Narducci, C. A., and Beckers, C. (2001) J. Biol. Chem. 276 12369-12377 [DOI] [PubMed] [Google Scholar]
- 12.Baum, J., Richard, D., Healer, J., Rug, M., Krnajski, Z., Gilberger, T. W., Green, J. L., Holder, A. A., and Couchman, A. F. (2006) J. Biol. Chem. 281 5197-5208 [DOI] [PubMed] [Google Scholar]
- 13.Rees-Channer, R. R., Martin, S. R., Green, J. L., Bowyer, P. W., Grainger, M., Molloy, J. E., and Holder, A. A. (2006) Mol. Biochem. Parasitol. 149 113-116 [DOI] [PubMed] [Google Scholar]
- 14.Green, J. L., Martin, S. R., Fielden, J., Ksagoni, A., Grainger, M., Yim Lim, B. Y., Molloy, J. E., and Holder, A. A. (2006) J. Mol. Biol. 355 933-941 [DOI] [PubMed] [Google Scholar]
- 15.Bergman, L. W., Kaiser, K., Fujioka, H., Coppens, I., Daly, T. M., Fox, S., Matuschewski, K., Nussenzweig, V., and Kappe, S. H. (2003) J. Cell Sci. 116 39-49 [DOI] [PubMed] [Google Scholar]
- 16.Jewett, T. J., and Sibley, L. D. (2003) Mol. Cell 11 885-894 [DOI] [PubMed] [Google Scholar]
- 17.Blackman, M. J., and Holder, A. A. (1992) Mol. Biochem. Parasitol. 50 307-315 [DOI] [PubMed] [Google Scholar]
- 18.Bergmann-Leitner, E. S., Duncan, E. H., Mullen, G. E., Burge, J. R., Khan, F., Long, C. A., Angov, E., and Lyon, J. A. (2006) Am. J. Trop. Med. Hyg. 75 437-442 [PubMed] [Google Scholar]
- 19.Green, J. L., Hinds, L., Grainger, M., Knuepfer, E., and Holder, A. A. (2006) Mol. Biochem. Parasitol. 150 114-117 [DOI] [PubMed] [Google Scholar]
- 20.Holder, A. A., and Freeman, R. R. (1982) J. Exp. Med. 156 1528-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachebat, J. A., Kadekoppala, M., Grainger, M., Dluzewski, A. R., Gunaratne, R. S., Scott-Finnigan, T. J., Ogun, S. A., Ling, I. T., Bannister, L. H., Taylor, H. M., Mitchell, G. H., and Holder, A. A. (2007) Mol. Biochem. Parasitol. 151 59-69 [DOI] [PubMed] [Google Scholar]
- 22.Abramoff, M. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 23.Cook, P. F., Neville, M. E., Vrana, K. E., Hartl, F. T., and Roskoski, R. (1982) Biochemistry 21 5794-5799 [DOI] [PubMed] [Google Scholar]
- 24.Hastie, C. J., McLauchlan, H. J., and Cohen, P. (2006) Nat. Protocols 1 968-971 [DOI] [PubMed] [Google Scholar]
- 25.Vorm, O., and Roepstorff, P. (1994) Biol. Mass Spectrom. 23 734-740 [DOI] [PubMed] [Google Scholar]
- 26.Bannister, L. H., Hopkins, J. M., Dluzewski, A. R., Margos, G., Williams, I. T., Blackman, M. J., Kocken, C. H., Thomas, A. W., and Mitchell, G. H. (2003) J. Cell Sci. 116 3825-3834 [DOI] [PubMed] [Google Scholar]
- 27.Huff, C. G., Pipkin, A. C., Weathersby, A. B., and Jensen, D. V. (1960) J. Biophys. Biochem. Cytol. 7 93-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fidock, D. A., Nomura, T., Cooper, R. A., Su, X., Talley, A. K., and Wellems, T. E. (2000) Mol. Biochem. Parasitol. 110 1-10 [DOI] [PubMed] [Google Scholar]
- 29.Fidock, D. A., Nomura, T., Talley, A. K., Cooper, R. A., Dzekunov, S. M., Ferdig, M. T., Ursos, L. M., Sidhu, A. B., Naudé, B., Deitsch, K. W., Su, X. Z., Wootton, J. C., Roepe, P. D., and Wellems, T. E. (2000) Mol. Cell 6 861-871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinder, J. C., Fowler, R. E., Dluzewski, A. R., Bannister, L. H., Lavin, F. M., Mitchell, G. H., Wilson, R. J., and Gratzer, W. B. (1998) J. Cell Sci. 111 1831-1839 [DOI] [PubMed] [Google Scholar]
- 31.Gaskins, E., Gilk, S., DeVore, N., Mann, T., Ward, G., and Beckers, C. (2004) J. Cell Biol. 165 283-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosch, J., Turley, S., Roach, C. M., Daly, T. M., Bergman, L. W., and Hol, W. G. (2007) J. Mol. Biol. 372 77-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryder, J. W., Lau, K. S., Kamm, K. E., and Stull, J. T. (2007) J. Biol. Chem. 282 20447-20454 [DOI] [PubMed] [Google Scholar]
- 34.Sweeney, H. L., and Stull, J. T. (1986) Am. J. Physiol. 250 C657-C660 [DOI] [PubMed] [Google Scholar]
- 35.Hirano, K., Derkach, D. N., Hirano, M., Nishimura, J., and Kanaide, H. (2003) Mol. Cell. Biochem. 248 105-114 [DOI] [PubMed] [Google Scholar]
- 36.Kato, N., Sakata, T., Breton, G., Le Roch, K. G., Nagle, A., Andersen, C., Bursulaya, B., Henson, K., Johnson, J., Kumar, K. A., Marr, F., Mason, D., McNamara, C., Plouffe, D., Ramachandran, V., Spooner, M., Tuntland, T., Zhou, Y., Peters, E. C., Chatterjee, A., Schultz, P. G., Ward, G. E., Gray, N., Harper, J., and Winzeler, E. A. (2008) Nat. Chem. Biol. 4 347-356 [DOI] [PubMed] [Google Scholar]
- 37.Ward, G. E., Fujioka, H., Aikawa, M., and Miller, L. H. (1994) Exp. Parasitol. 79 480-487 [DOI] [PubMed] [Google Scholar]
- 38.Lovett, J. L., and Sibley, L. D. (2003) J. Cell Sci. 116 3009-3016 [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto, Y., Perry, G., Scheibel, L. W., and Aikawa, M. (1987) Eur J. Cell Biol. 45 36-43 [PubMed] [Google Scholar]
- 40.Wasserman, M., and Chaparro, J. (1996) Parasitol. Res. 82 102-107 [DOI] [PubMed] [Google Scholar]
- 41.Vaid, A., Thomas, D. C., and Sharma, P. (2008) J. Biol. Chem. 283 5589-5597 [DOI] [PubMed] [Google Scholar]