Abstract
The role of the endoplasmic reticulum stress-regulated kinase, PERK, in mammary gland function was assessed through generation of a targeted deletion in mammary epithelium. Characterization revealed that PERK is required for functional maturation of milk-secreting mammary epithelial cells. PERK-dependent signaling contributes to lipogenic differentiation in mammary epithelium, and perk deletion inhibits the sustained expression of lipogenic enzymes FAS, ACL, and SCD1. As a result, mammary tissue has reduced lipid content and the milk produced has altered lipid composition, resulting in attenuated pup growth. Consistent with PERK-dependent regulation of the lipogenic pathway, loss of PERK inhibits expression of FAS, ACL, and SCD1 in immortalized murine embryonic fibroblasts when cultured under conditions favoring adipocyte differentiation. These findings implicate PERK as a physiologically relevant regulator of the lipogenic pathway.
Keywords: lipid metabolism, SREBP1, FAS, ACL, fatty acids
Endoplasmic reticulum (ER) homeostasis depends on the balance between protein folding and available chaperones. Perturbations in glucose or oxygen availability can disrupt protein folding; the resultant accumulation of misfolded proteins in the ER triggers the Unfolded Protein Response (UPR). The UPR is a signaling cascade initiated by three ER membrane-bound transducers: the endoribonuclease inositol requiring kinase (Ire)-1 (α/β), the ER transmembrane transcription factor ATF6, and the PKR-like ER kinase (PERK; ref. 1). The UPR either facilitates the restoration of balance between ER load and capacity or promotes cell death. All three transducers reduce protein load through the induction of ER chaperone genes (2), and genes involved in the ER-associated degradation pathway (3, 4). However, PERK functions uniquely to reduce protein synthesis via phosphorylation of eukaryotic translation initiation factor 2 alpha (eIF2α; ref. 5).
PERK is highly expressed in mouse pancreas; perk null mice exhibit neonatal diabetes mellitus and develop exocrine pancreas atrophy (6, 7). Diabetes results from insufficient proliferation of β-cell mass and dysfunctions in insulin secretion (8), whereas atrophy of exocrine pancreas results from cell death of acinar cells (6, 7, 9). PERK deficiency also affects the proliferation and differentiation of osteoblasts (10), collectively implicating PERK as a critical regulator of secretory cell function.
The mammary gland is another secretory tissue that experiences hormone-induced increase in biosynthetic activity and secretory compartment expansion during transition from pregnancy to lactation. Ductal and alveolar mammary epithelial cell proliferation during pregnancy results in the formation of an extensive network of side branches ending in lobular-alveolar clusters that produce milk. At midpregnancy, the expression of biosynthetic enzymes for milk production is induced in a limited number of alveolar cells that begin to secrete select milk components, and by the end of pregnancy, alveolar cells acquire a mature synthetic, secretory phenotype (11).
Although the protein component constitutes ≈12% of mouse milk, the main macromolecular constituent of milk is lipid (30%; ref. 12). The Sterol Regulatory Element Binding Protein (SREBP) family of transcription factors are the major regulators of lipid metabolic genes involved in fatty acid and cholesterol biosynthesis (13, 14). SREBPs localize to the ER as transmembrane precursors and are retained via interaction with SREBP cleavage-activating protein (SCAP) and Insig1 (insulin inducible gene1; ref. 15). When cholesterol levels decline, the Insig1–SCAP complex dissipates and SCAP escorts SREBPs to the Golgi for cleavage by site-1/2 proteases to generate transcription factors that bind to sterol regulatory elements (SRE; refs. 16 and 17).
We have used a conditional gene deletion strategy to generate mice deficient for perk in mammary epithelium. This work demonstrates that PERK contributes to secretory maturation in the mouse mammary gland and serves as a critical regulator of lipid metabolism via regulation of SREBP processing and target gene expression.
Results
Deletion of perk in Mouse Mammary Epithelium Impairs Secretory Activation and Attenuates Pup Growth.
Examination of PERK expression at various stages of mammary gland development revealed PERK expression at all developmental stages (supporting information (SI) Fig. S1A), with activation and accumulation of phosphorylated eIF2α species during lactation (18, 19). We next generated a mammary epithelium-specific knockout of perk. Mammary glands isolated from these mice (cKO) revealed undetectable PERK protein at all developmental stages and a corresponding decrease in levels of p-eIF2α (Fig. S1A). Histological analysis revealed normal development of virgin and pregnant (Fig. S1 B and C) glands in cKO mice. By day 3 of lactation (secretory activation), glands of cKO mice exhibited distinct morphological deficiencies (Fig. 1 A and B). cKO glands were morphologically similar to wild-type pregnancy day 16 displaying reduced alveolar expansion with areas of uncollapsed adipocyte stroma (Fig. 1B).
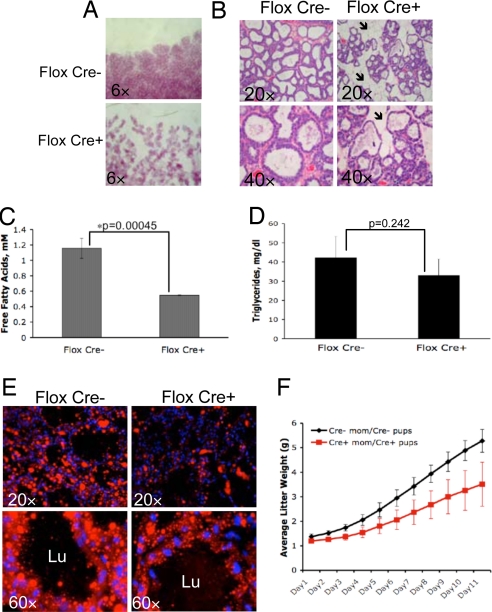
Fig. 1.
Conditional deletion of perk in mouse mammary gland affects mouse milk fat composition, mammary gland lipid content, and pup growth. (A) Whole mounts of the mammary glands from control (Flox Cre−) and cKO (Flox Cre+) mice on lactation day 3. (B) H&E staining of sections from control and cKO glands on lactation day 3. Arrows indicate the areas of uncollapsed adipocyte stroma. (C and D) Free fatty acids or triglyceride content of milk collected from 4-month-old cKO (Flox Cre+, n = 3) and control (Flox Cre−, n = 5) dams on lactation day 12. Error bars represent SD. (E) Nile red staining of mammary glands from 4-month-old control and cKO dams on lactation day 12. Lu, lumen of the alveoli. (F) Average litter weight plotted vs. pups' age in a cohort of 4-month-old control (Cre−, n = 8) dams nursing wild-type pups and cKO (Cre+, n = 9) animals nursing cKO pups. Error bars represent SD.
Because PERK regulates protein translation, we assessed milk protein content in cKO glands. Significantly, the milk protein profile was unchanged (Fig. S2A). Because milk fat is a major milk component and energy source in neonates (12), we analyzed milk lipid content. Qualitative analysis of milk lipids by thin-layer chromatography (Fig. S2B), or quantitative analysis of free fatty acids and triglycerides in mouse milk, revealed a significant reduction in free fatty acid content and a trend toward reduced triglycerides in the milk of cKO dams (Fig. 1 C and D). Nile Red (binds triglycerides and free fatty acids; ref. 20) staining of mammary glands revealed decreased lipid droplet size and formation in cKO glands (Fig. 1E). Consistent with reduced fatty acids and triglyceride in milk, growth of pups nursed by cKO dams was significantly attenuated, and average litter weight was reduced by ≈30% by day 12 of lactation (Fig. 1F). Similar growth retardation was observed in wild-type pups nursed by cKO dams, whereas cKO pups exhibited normal growth when fostered by control dams (Fig. S2C).
Perk Deletion Inhibits the Sustained Induction of Lipogenic Enzymes.
Lactating mice rely on de novo synthesis of glycerol and medium chain fatty acids from glucose and amino acids (12, 21). Thus, the decrease in the amount of free fatty acids in the milk may serve as an indicator of reduced levels of precursors for de novo triacylglycerol synthesis. Because expression of key lipogenic enzymes, ATP citrate lyase (ACL), fatty acid synthase (FAS), stearyl-CoA desaturase-1 and -2 (SCD1/SCD2), is induced during secretory activation (21), we assessed expression of these enzymes in mammary gland samples from control and cKO mice. Increased ACL and SCD1 mRNA levels were apparent by lactation day 3 in control and cKO glands, whereas FAS mRNA levels were slightly increased only in control mice (Fig. 2A). Lipogenic enzyme mRNA levels declined to a much greater extent in the cKO glands than in control mice on lactation day 12 (Fig. 2A). ACL and SCD1 protein levels were lower in cKO mice at all stages examined, whereas FAS protein specifically declined in cKO mice on lactation day 12 (Fig. 2B).
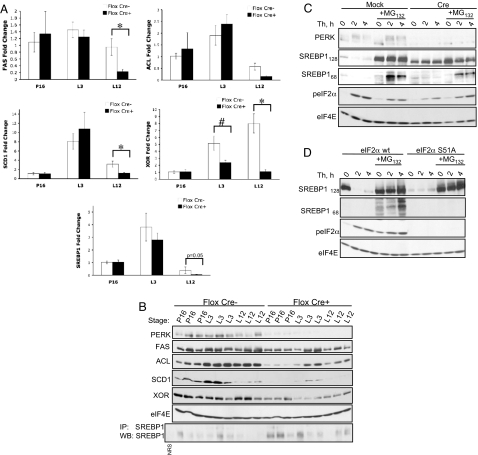
Fig. 2.
Perk deletion in mouse mammary epithelium inhibits SREBP1 activation and the sustained induction of lipogenic enzymes. (A) RT-PCR analysis for SREBP1, FAS, ACL, SCD1, and XOR from mammary extracts of 4-month-old control (Flox Cre−, n = 3) and cKO (Flox Cre+, n = 3) dams at P16, lactation day 3 (L3), and 12 (L12). Error bars represent SEM (*, P < 0.005; #, P < 0.05). (B) Western blot for PERK, FAS, ACL, SCD1, XOR, and eIF4E of mammary gland extracts from the same developmental stages as in A. IP-Western for SREBP1 was performed on the same samples. NRS, nonspecific rabbit serum. (C) Western blot for PERK, Myc-tagged SREBP1 precursor (SREBP1128) or processed isoform (SREBP168), phosho-eIF2α, and eIF4E on samples from perk LoxP pBabe MEFs (Mock) or perk LoxP Cre MEFs (Cre) expressing Myc-SREBP1 and treated as indicated with 50 nM thapsigargin (Th) and 10 μM MG132. (D) Western blot for Myc-SREBP1 precursor and processed isoform, phosho-eIF2α, and eIF4E on samples from eIF2α wild-type (wt) or eIF2α S51A mutant knockin MEFs expressing Myc-SREBP1 and Myc-Insig1. Cells were treated with 50 nM thapsigargin (Th) and 10 μM MG132 where indicated.
Because SREBP transcription factors are major regulators of lipid metabolic genes and can be induced by ER stress (22, 23), we addressed whether PERK deletion inhibited SREBP activation. Indeed, loss of PERK attenuated SREBP1 expression (Fig. 2A), consistent with previous work revealing autoregulation of srebp1 gene expression (24). Additionally, an increased retention of SREBP1 precursor was detected in cKO samples (Fig. 2B), suggesting reduced processing without PERK.
SREBP Maturation Is Induced in a PERK and eIF2α-Dependent Fashion.
The PERK dependence of ER stress-mediated SREBP processing was further tested through utilization of an acute model of PERK deletion in murine embryonic fibroblasts (MEFs); perk LoxP MEFs were transduced with a retrovirus expressing Cre recombinase (PERK Cre) or empty virus (PERK Mock) and treated with thapsigargin (Fig. 2C). To facilitate detection of SREBP1, the cells were engineered to stably express Myc-tagged SREBP1. Acute perk deletion eliminated processing of Myc-SREBP1 (Fig. 2C) and endogenous SREBP1 (Fig. S3A). Although SREBP1 precursor levels rapidly declined upon ER stress, no processed isoform was detected. Because the processed SREBPs are rapidly degraded by the 26S proteasome (25), we inhibited the proteasome with MG132. In the presence of MG132, thapsigargin induced SREBP1 processing with accumulation of the processed SREBP1 in control cells while processing was significantly attenuated in perk−/− cells (Fig. 2C). Because MG132 induces activation of the cytosolic eIF2α kinase, GCN2 (26), incomplete abrogation of SREBP1 processing in PERK Cre MEFs likely reflects compensatory GCN2 action. Indeed, MG132 alone resulted in a small accumulation of processed SREBP1, suggesting basal processing. Importantly, endogenous SREBP1 precursor levels declined independently of MG132 presence, ruling out SREBP1 degradation during ER stress (Fig. S3B). Essentially no processing of SREBP1 was observed in eIF2α S51A MEFs (27) demonstrating that PERK-dependent SREBP1 activation occurs in an eIF2α-dependent manner (Fig. 2D).
SREBP Maturation During ER Stress Is Mediated by Insig1 Depletion.
SREBP1 processing can be regulated via direct depletion of Insig1, an ER-localized protein that anchors the SCAP–SREBP complex in the ER membrane (22). Because Insig1 is a short-lived protein, analogously to cyclin D1 (28), PERK-mediated translation inhibition should result in the rapid depletion of Insig1. We therefore generated a Myc-tagged Insig1 (due to the lack of suitable antibodies for detecting endogenous protein) and transduced wild-type and perk−/− MEFs. Thapsigargin treatment triggered a rapid loss of Insig1 in a PERK- (Fig. 3A) and phospho-eIF2α-dependent manner (Fig. S3C).
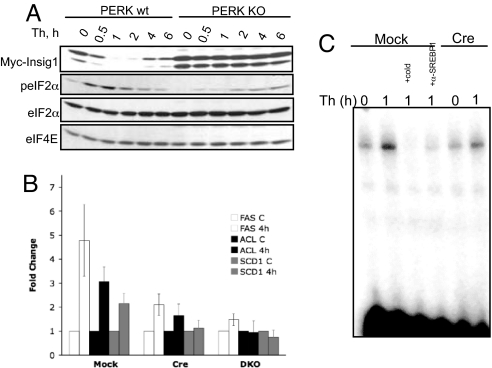
Fig. 3.
PERK-dependent depletion of Insig1 contributes to induction of SREBP1 target genes. (A) Western blot for proteins indicated on samples from wild type (wt) or perk knockout (KO) MEFs treated as indicated. (B) RT-PCR analysis of FAS, ACL, and SCD1 in perk LoxP pBabe MEFs (Mock), perk LoxP Cre MEFs (Cre), or perk/Gcn2 double-knockout (DKO) MEFs. Error bars represent SD for three independent experiments. (C) EMSA analysis of nuclear extracts isolated from perk LoxP pBabe MEFs or perk LoxP Cre MEFs treated with thapsigargin (Th). + Cold indicates the reaction performed in the presence of unlabeled IRS2 duplex; + α-SREBP1 indicates the addition of SREBP1 antibody.
Robust induction of SREBP1 target genes mRNA (FAS, ACL, and SCD1) was observed in wild-type but not perk−/− MEFs (Fig. 3B). Consistent with target induction, a 2-fold increase in SREBP1 DNA binding was noted in thapsigargin-treated wild type but not perk−/− MEFs (Fig. 3C).
PERK Deficiency Inhibits the Lipogenic Pathway During Differentiation of MEFs into Adipocytes.
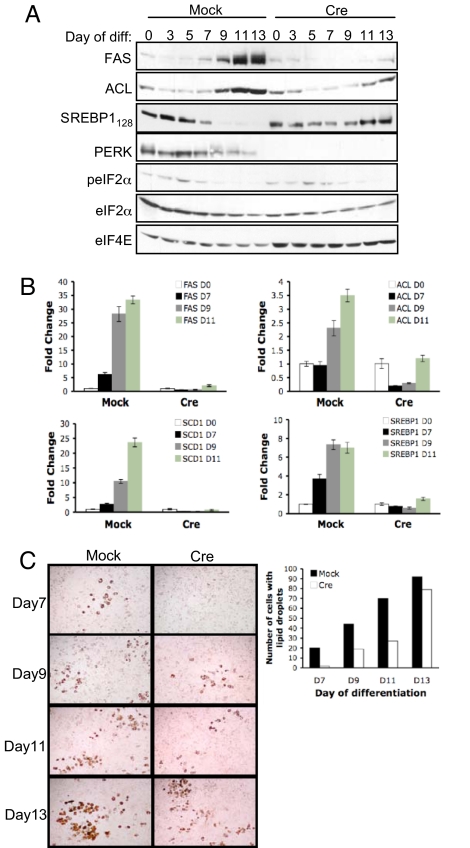
Given the ability of PERK to regulate SREBP1 activation and lipogenic enzymes expression in mouse mammary epithelium, we reasoned that PERK might also contribute to the initiation of a lipogenic program during adipocyte differentiation. To evaluate this possibility, we cultured PERK Mock MEFs and PERK Cre MEFs, engineered to express Myc-tagged SREBP1, in medium favoring adipocytic differentiation and monitored induction of lipogenic enzymes. Increased SREBP1c, FAS, and SCD1 mRNA levels were observed in control cells by day 7, and ACL mRNA levels were induced by day 9 of treatment; this was significantly attenuated in PERK Cre MEFs (Fig. 4B). Increases in FAS and ACL protein levels were also detected by day 9 of treatment in PERK Mock, but not in PERK Cre MEFs (Fig. 4A). Although transient activation of PERK is difficult to detect, and phosphorylation of eIF2α is not completely eliminated by PERK excision (compensation by GCN2), the depletion of Myc-tagged SREBP1 precursor could be clearly observed in PERK Mock, but not in PERK Cre MEFs by day 7 of treatment (Fig. 4A). Consistently, the accumulation of Oil Red O-positive droplets was significantly attenuated in the absence of PERK (Fig. 4C) and was essentially abolished in perk/Gcn2 double knockout (DKO) MEFs (data not shown). Similar results were observed after knockdown of PERK in 3T3-L1 cells (Fig. S4 A and B).
Fig. 4.
PERK affects the lipogenic program during differentiation of immortalized MEFs into adipocytes. (A) Western blot for FAS, ACL, Myc-SREBP1 precursor, PERK, phosho-eIF2α, total eIF2α, and eIF4E in perk LoxP pBabe MEFs (Mock) or perk LoxP Cre MEFs (Cre) infected with virus encoding Myc-SREBP1 and cultured in adipocyte differentiation mixture for 0, 3, 5, 7, 9, 11, or 13 days. (B) RT-PCR quantification of FAS, ACL, SCD1, and SREBP1c from cells treated as in A. (C) Oil Red O staining on perk LoxP pBabe MEFs or perk LoxP Cre MEFs expressing Myc-SREBP1 and cultured in adipocyte differentiation mixture.
Discussion
This work reveals a role for PERK in the regulation of the lipogenic enzyme expression. PERK deletion in mouse mammary epithelium revealed that, although dispensable during the formation of the primitive ductal tree and during pregnancy hormone-induced mammary epithelial cell proliferation, PERK is necessary for the lipogenic maturation of mammary epithelial cells during lactation. Specifically, PERK regulates the expression of key metabolic enzymes, FAS, ACL, and SCD1, necessary for lipid production and storage (XOR, Fig. 2B). PERK deletion significantly reduced mammary gland lipid content and caused a decrease in the amount of free fatty acids present in the milk of conditional knockout mice. In vitro analysis, by using wild-type or perk−/− MEFs, supports a role for PERK in the regulation of SREBP1 activation, which controls expression of key metabolic enzymes necessary for lipid production. Our data reveal a pathway wherein PERK-dependent inhibition of protein translation triggers the depletion of Insig1, an integral ER membrane anchor protein, leading to the Golgi translocation and processing of SREBP1 (Fig. S5). Significantly, loss of PERK-dependent regulation of lipogenic enzymes also leads to the reduced induction of lipid metabolism enzymes in immortalized MEFs cultured under conditions that promote their differentiation into adipocytes, suggesting PERK action may contribute to fat storage in multiple cell types in vivo.
A highlight of perk deletion is the attenuation of mammary gland maturation during lactation and the resultant retarded pup growth, which correlated with reduced milk lipids. PERK is highly active during midlactation, as revealed by the presence of increased levels of phosphorylated eIF2α at lactation day 7 and 12 (Fig. S1A). That Cre-mediated perk excision results in a significant reduction in the levels of phosphorylated eIF2α during lactation suggests that PERK is a major eIF2α kinase in this tissue. Although PERK function does not appear critical during the initial activation of lipogenesis on lactation day 3, PERK-deficient mammary epithelial cells fail to maintain the expression of lipid synthesis genes during midlactation. Akt1 may contribute to SREBP1 activation during transition from pregnancy to lactation via increasing stability of mature SREBP1 through down-regulation of GSK-3β activity (29), or even by promoting the initial SREBP1 export into the Golgi apparatus for processing (30). The importance of PERK in the activation of the lipogenic pathway is further underscored by our observation that deletion of perk reduced kinetics of lipid accumulation during adipocytic differentiation, which coincided with reduced levels of FAS, ACL, and SCD1 in MEFs. Here again, the importance of Insig1 as a regulatory switch was previously noted in that Insig1 overexpression inhibited lipid accumulation and expression of prodifferentiation factors in preadipocytes (31). Our data support a model wherein PERK function maintains lipid synthesis by promoting SREBP1 activation via the phospho-eIF2α-dependent depletion of the Insig1 protein during midlactation (Fig. S5).
Although activation of SREBP by chemical inducers of ER stress has been noted (22, 23), PERK- and phospho-eIF2α dependence was not addressed. Paradoxically, an antagonistic relationship between PERK and SREBP has also been observed (32). A construct containing a drug-inducible dimerization domain fused to the cytosolic kinase domain of PERK molecule (33) was designed resulting in a chimeric PERK that no longer localizes to the ER membrane and may in fact function similarly to another cytosolic eIF2α kinase, GCN2. By using this construct, a decline in the levels of processed SREBP2 protein and in mRNA levels of SREBP2 target genes was observed (32). Interestingly, GCN2 activation by amino acid deprivation results in the suppression of the SREBP pathway in hepatocytes (34). In contrast, overexpression of the targeting subunit of eIF2α phosphatase, Gadd34, in the murine liver decreases hepatosteatosis in animals fed a high-fat diet (35). This is consistent with our findings and argues that eIF2α kinase-dependent signals promote lipogenesis. Importantly, PERK-deficient mice exhibit a reduced ratio of phosphorylated to total eIF2α in the pancreas, lungs, and thymus, but not in liver, suggesting that eIF2α kinases in addition to PERK contribute to translational control in the liver (6). Furthermore, it is likely that physiological signaling by eIF2α kinases, PERK in mammary epithelium and PERK and GCN2 in liver, has a different outcome at the level of the whole organism relative to the acute activation of these kinases via drug-inducible dimerization or amino acid deprivation.
Accumulating data suggest that the UPR has functions beyond monitoring protein folding within the ER. Transgenic expression of Gadd34 can counteract the effects of a high-fat diet on fatty liver (35) and targeted deletion of Xbp1 contributes to reduced lipid accumulation (36), whereas overexpression promotes increased phospholipid production (37). Collectively with our data, these data support a role for the UPR in maintenance of lipid homeostasis.
Methods
Materials.
Tissue culture media and medium supplements were purchased from Invitrogen and HyClone. All chemicals and reagents were purchased from Fisher Scientific, Sigma, and Invitrogen.
Animals and Tissue.
Experiments were conducted in accordance with the Animal Welfare Act and the Department of Health and Human Services Guide. Mice homozygous for the LoxP allele of perk (7) were mated to MMTV-CRE transgenic mice (38). The Cre transgene-bearing offspring were bred to homozygocity for the LoxP allele of perk thus generating mammary gland-specific perk“null.” Littermates not inheriting the Cre transgene were bred to homozygocity for the LoxP allele of perk and used as controls. Mammary glands were harvested from virgin mice, pregnant mice at day 8 and 16, lactating mice at day 0, 3, 7, 12, and involuting glands were harvested at day 3 postweaning. Samples were snap-frozen and stored at −80°C. For analysis of lipid composition, 10 μl of milk was extracted and triglyceride (Cayman Chemical) and free fatty acid (BioVision Research Products) content was measured after lipid extraction by using the method of Bligh and Dyer (39).
Cell Culture.
MEFs were cultured in DMEM supplemented with 4 mM l-glutamine, 10% (vol/vol) FBS, 100 units/ml penicillin/streptomycin, and 55 μM β-mercaptoethanol. Passage immortalized perk LoxP MEFs were derived as previously described (40). Perk LoxP pBabe and perk LoxP Cre MEFs were generated as described in ref. 41 and selected with puromycin. For differentiation, cells were grown to confluence and treated with differentiation mixture (10 μg/ml insulin, 0.5 mM isobutylmethylxanthine, 1 μM dexamethasone, and 5 μM troglitazone) for 7 days. On day 5, 7, and 9 of the differentiation protocol, MEFs were refed fresh differentiation medium (day 5) or 5 μM troglitazone (days 7 and 9).
Plasmids.
The N-terminally Myc-tagged human SREBP1 (BC057388; Open Biosystems) and C-terminally Myc-tagged rat Insig1 (BC078827; Open Biosystems) was cloned into the pBabe retroviral vector. All primer sequences and cloning schemes are available on request. Retroviruses were produced and used as described in ref. 28. The anti-mouse shPERK was from Open Biosystems (clone ID V2MM_51300).
RNA analysis.
RNA was collected with TRIzol (Invitrogen). Five micrograms of total RNA was used for first-strand cDNA synthesis by using SuperScript II Reverse Transcription Kit (Invitrogen). For RT-PCR, input cDNA was analyzed in triplicate. All reactions were performed by using SYBR Green (SuperArray) and 500 nM of the forward/reverse primers (ABI Prizm 7200 Sequence Detector, Applied Biosystems). The following primers were used: mFAS, mAcl, mSCD1, SREBP1c (34), and mXOR, mKeratin18 (42), m18S rRNA (F) 5′-AAATCAGTTATGGTTCCTTTGGTC-3′; (R) 5′-GCTCTAGAATTACCACAGTTATCCAA-3′. In cultured cells, mRNA was normalized to 18S rRNA and each target was determined relative to signal from untreated cells (1-fold). For tissue, target gene levels were normalized to cytokeratin18. The induction of each target expression on day 3 or day 12 of lactation was determined relative to the signal in samples of the corresponding genotype on pregnancy day 16 (1-fold).
EMSA.
An oligonucleotide containing the sterol regulatory element (SRE) from the human insulin receptor substrate 2 (IRS2) promoter (43) was used. Binding assays were performed at room temperature and the DNA–protein complexes were separated by electrophoresis on 4% TBE PAGE gel and visualized by using STORM PhosphorImager/Image Quant software (Molecular Dynamics). Where indicated, 2 μg of SREBP1 (Santa Cruz Biotechnology) antibody was included.
Oil Red O and Nile Red Staining.
Cells were fixed for 2 min in 3.7% formaldehyde, washed with water, and stained with Oil Red O. 10 μM OCT-embedded mammary gland sections were fixed in 4% paraformaldehyde, stained with Hoechst 33258, and coverslipped with Nile red.
Western Analysis.
Cultured cells were lysed in 0.15 M NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS (RIPA buffer). Slices of mammary gland were homogenized in EBC buffer (28). Immunoblot was performed by using following antibodies: phospho-Ser51eIF2α, eIF4E, SCD1 (Cell Signaling); eIF2α (BioSource); PERK (Rockland Immunochemicals); FAS (BD Biosciences); ACL (44); SREBP1 (Thermo Scientific); c-Myc (9E10), XOR (H-110) (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments.
We thank M. Burnbaum for providing 3T3-L1 cells; R. DeBerardinis for assistance with lipid TLC; the AFCRI histology core. This work was supported by National Institutes of Health Grants F32CA1238252 (to E.B.M.) and P01 CA104838 (to J.A.D. and C.B.T.) and a Wyeth Pharmaceutical grant (to J.A.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808517105/DCSupplemental.
References
- 1.Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- 2.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 6.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 7.Zhang P, et al. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, et al. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Iida K, Li Y, McGrath BC, Frank A, Cavener DR. PERK eIF2 alpha kinase is required to regulate the viability of the exocrine pancreas in mice. BMC Cell Biol. 2007;8:38. doi: 10.1186/1471-2121-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei J, Sheng X, Feng D, McGrath B, Cavener DR. PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol. 2008 Aug 6; doi: 10.1002/JCP.21543. [DOI] [PubMed] [Google Scholar]
- 11.McManaman JL, Neville MC. Mammary physiology and milk secretion. Adv Drug Deliv Rev. 2003;55:629–641. doi: 10.1016/s0169-409x(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 12.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res. 2007;9:204. doi: 10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 14.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 15.Yang T, et al. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 16.Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 17.Rawson RB, et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 18.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 19.Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277:18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 20.Fowler SD, Brown WJ, Warfel J, Greenspan P. Use of nile red for the rapid in situ quantitation of lipids on thin-layer chromatograms. J Lipid Res. 1987;28:1225–1232. [PubMed] [Google Scholar]
- 21.Rudolph MC, et al. Metabolic regulation in the lactating mammary gland: A lipid synthesizing machine. Physiol Genomics. 2007;28:323–336. doi: 10.1152/physiolgenomics.00020.2006. [DOI] [PubMed] [Google Scholar]
- 22.Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 23.Colgan SM, Tang D, Werstuck GH, Austin RC. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Miserez AR, Cao G, Probst LC, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2) Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- 25.Sundqvist A, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 27.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 28.Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc Natl Acad Sci USA. 2000;97:12625–12630. doi: 10.1073/pnas.220247197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 30.Du X, Kristiana I, Wong J, Brown AJ. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: A link between a key cell proliferative pathway and membrane synthesis. Mol Biol Cell. 2006;17:2735–2745. doi: 10.1091/mbc.E05-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Takaishi K, Cook W, McCorkle SK, Unger RH. Insig-1 “brakes” lipogenesis in adipocytes and inhibits differentiation of preadipocytes. Proc Natl Acad Sci USA. 2003;100:9476–9481. doi: 10.1073/pnas.1133426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding HP, et al. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2005;2:361–371. doi: 10.1016/j.cmet.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu PD, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sriburi R, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 38.Wagner KU, et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 39.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 40.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, et al. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- 42.Rossiter H, et al. Inactivation of VEGF in mammary gland epithelium severely compromises mammary gland development and function. FASEB J. 2007;21:3994–4004. doi: 10.1096/fj.07-8720com. [DOI] [PubMed] [Google Scholar]
- 43.Ide T, et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol. 2004;6:351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- 44.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.