Abstract
Lipins are the founding members of a novel family of Mg2+-dependent phosphatidate phosphatases (PAP1 enzymes) that play key roles in fat metabolism and lipid biosynthesis. Despite their importance, there is still little information on how their activity is regulated. Here we demonstrate that the functions of lipin 1 and 2 are evolutionarily conserved from unicellular eukaryotes to mammals. The two lipins display distinct intracellular localization in HeLa M cells, with a pool of lipin 2 exhibiting a tight membrane association. Small interfering RNA-mediated silencing of lipin 1 leads to a dramatic decrease of the cellular PAP1 activity in HeLa M cells, whereas silencing of lipin 2 leads to an increase of lipin 1 levels and PAP1 activity. Consistent with their distinct functions in HeLa M cells, lipin 1 and 2 exhibit reciprocal patterns of protein expression in differentiating 3T3-L1 adipocytes. Lipin 2 levels increase in lipin 1-depleted 3T3-L1 cells without rescuing the adipogenic defects, whereas depletion of lipin 2 does not inhibit adipogenesis. Finally, we show that the PAP1 activity of both lipins is inhibited by phosphorylation during mitosis, leading to a decrease in the cellular PAP1 activity during cell division. We propose that distinct and non-redundant functions of lipin 1 and 2 regulate lipid production during the cell cycle and adipocyte differentiation.
The phospholipid composition of biological membranes is crucial for many aspects of cell physiology, including growth, differentiation, and transport (1, 2). Phospholipids are also active participants of signaling cascades that control diverse cellular functions (3). Two key precursors of phospholipid biosynthesis are PA3 and DAG, both of which have essential functions in signaling cascades, energy storage, and lipid biosynthetic pathways. Early biochemical studies identified a soluble Mg2+-dependent PA phosphatase (PAP1) activity that is key to catalyzing PA conversion to DAG (4–6). DAG has multiple functions. First, DAG is used for the synthesis of the most abundant phospholipids found in biological membranes, phosphatidylcholine, and phosphatidylethanolamine (6). Second, DAG is also used for the synthesis of the neutral lipid triacylglycerol, an essential storage form of energy and fatty acids, which accumulates as lipid droplets in adipocytes (7, 8). Despite the key role of the PAP1 reaction, the identity of the enzyme(s) responsible for this activity remained unknown until recently. Han et al. (9) have demonstrated that Pah1p/Smp2p is the PAP1 enzyme controlling biosynthetic production of phospholipids and triacylglycerol in yeast. A separate study found that Pah1p regulates transcription of many phospholipid biosynthetic enzymes, nuclear/ER membrane biogenesis, and nuclear structure (10).
Mammals express three Pah1p-related proteins called lipins 1, 2, and 3 (11). Two closely related lipin 1 isoforms have been described, theα form and theβ form, which contains a short insertion close to its N-terminal end (12). All members of the Pah1p/lipin family exhibit the same primary organization; they are soluble proteins containing a very conserved N-terminal domain of ∼100 amino acid residues of unknown function and a C-terminal haloacid dehalogenase-like domain that contains the catalytic site. Recent studies have demonstrated that mammalian lipins possess PAP1 activity (9, 13, 14). Lipin 1 was originally identified through positional cloning as the mutated gene in the fatty liver dystrophy (fld) mouse (11). These mice are characterized by significantly reduced adipose tissue mass throughout the body, insulin resistance, and a progressive peripheral neuropathy (11). Consistent with its metabolic functions in adipogenesis, high levels of lipin 1 in transgenic mice promotes obesity (15). Recent studies have also implicated lipin 1 in the regulation of gene expression of hepatic lipid metabolic enzymes (16).
Despite this progress, there is still relatively little information about the function of lipin 1 in cell physiology, especially outside adipocytes, and essentially no functional information about lipins 2 and 3. If lipins, like Pah1p, are responsible for the production of biosynthetic pools of phospholipids, then temporal and spatial regulation of their activity would be expected to play an important role in membrane biogenesis. Interestingly, the three lipins exhibit distinct but overlapping expression patterns in many tissues (14), raising the question of the functional differences of the various paralogues expressed within the same cell. We report here that the function of lipins is evolutionarily conserved from unicellular eukaryotes to mammals. We provide the first evidence that lipin 1 and 2 exhibit distinct intracellular localization in HeLa M cells and show that lipin 1 is the major PAP1 enzyme in HeLa M cells. siRNA-mediated depletion of the two lipins shows that their expression is reciprocally regulated both in HeLa M cells and differentiating adipocytes. Finally, we demonstrate that the activity and solubility of both lipins is regulated in a cell cycle-dependent manner by mitotic phosphorylation.
EXPERIMENTAL PROCEDURES
Reagents, Plasmids, and Yeast Methods—Unless otherwise stated, all of the reagents were supplied by Sigma-Aldrich. MGC clones (Invitrogen) containing the full-length cDNA for human lipin 1 (3906281) and mouse lipin 2 (5101211) were subcloned into high copy (YEplac181-LEU2) yeast expression vectors. The original lipin 2 clone contained a nonconserved 5′ 115-nucleotide sequence that was removed from all subsequent constructs. Single HA tag or GFP tags were added to the C termini of lipin 1 and 2 using standard PCR cloning techniques, and expression in HeLa M cells was driven by cytomegalovirus promoter-based vectors. PAH1-, lipin 1-, and lipin 2-GFP C-terminal fusions were cloned into a high copy vector (YEplac181) and expressed in the pah1Δ strain (10). Transformations, protein extracts, and microscopy of yeast cells were performed as previously described (10).
Lipin 1 and Lipin 2 Antibody Production—The nucleotide sequences of human lipin1 (corresponding to amino acid residues 190–526) and mouse lipin2 (331–548) were cloned into pGEX4T1, and glutathione S-transferase-tagged fusion proteins were purified from bacterial lysate supernatants using glutathione-Sepharose 4B (Amersham Biosciences). Antibodies against the glutathione S-transferase fusion proteins (500 μg of protein/injection) were raised in rabbits by three subcutaneous injections over 66 days. Affinity purification of the antibody was carried out by coupling the glutathione S-transferase fusion proteins to CNBr-activated Sepharose (Amersham Biosciences) following the product specifications. Rabbit sera from the final bleeds were then incubated with the CNBr-activated Sepharose fusion protein complex for 2 h at 4 °C before washing with PBS. Subsequent elution of the bound antibodies was carried out using 200 mm glycine-HCl (pH 2.3), which was then neutralized immediately with 1 m Tris-HCl (pH 7.4).
Cell Culture, Transfections, and siRNA Knockdowns—HeLa M cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2 mm l-glutamine in a humidified 37 °C incubator with 5% CO2. Transfections of HeLa M cells with plasmids and siRNA oligonucleotides were carried out using the standard protocols supplied with TransIT-HeLaMONSTER transfection kit (Mirus, MIR2900) and oligofectamine reagent (Invitrogen), respectively, with the exception that the final concentration of siRNA oligonucleotides was reduced to 10 nm for all experiments. The siRNA oligonucleotides were purchased from Dharmacon Inc. as SMART pool siGENOME duplexes for human lipin 1 (NM_145693) and human lipin 2 (NM_014646). The siCONTROL nontargeting siRNA#1 was used as the control for each siRNA knockdown experiment. Unless otherwise stated, HeLa M cells were seeded 24 h prior to transfection. The medium was changed 24 h after transfection, and the cells were collected for analysis on day 4, 72 h post-transfection. One single siGENOME duplex was chosen to represent each SMART pool, for lipin1 siGENOME duplex number 2 (D-017427-02) was used and for lipin2 siGENOME duplex number 2 (D-013458-02). 3T3-L1 preadipocytes were cultured and differentiated as previously described (17).
Flow Cytometry—HeLa M cells were collected by trypsin-EDTA release. The cell suspension was centrifuged at ∼800 × g, and the cell pellet was washed once with chilled PBS containing 1 mg/ml BSA. The cells were then fixed with 70% ice-cold ethanol and stored at -20 °C for at least 4 h. DNA was labeled with propidium iodide (15 μg/ml) in the presence of RNase (0.2 mg/ml), 1.1% trisodium citrate and 0.1% Triton X-100 for 30 min. The samples were then passed through a 70-μm Nylon cell strainer (BD Falcon) into a tube prior to flow cytometric determination of DNA content using a Becton Dickinson FACSCalibur Flow cytometer. Propidium iodide levels were measured by using the FLA-3 laser, and the cell cycle distributions were determined through analysis on FlowJo software.
Retrovirus-mediated shRNA Knockdown—Two independent short sequences specifically targeting lipin 1 and lipin 2 were designed and cloned into the RNAi-Ready pSIREN-RetroQ vector (Clontech) according to the manufacturer's instructions. Briefly, the primer pairs L101Bam/L101Eco, L103Bam/L103Eco, L209Bam/L209Eco, and L210Bam/L210Eco were annealed and ligated directly into the BamHI-EcoRI open RNAi-ready pSIREN-RetroQ vector to yield the constructs shRNA-L1A and shRNA-L1C for lipin 1 or shRNA-L2A or shRNA-L2B for lipin 2, respectively. The pSIREN-Luc vector containing shRNA targetting luciferase (Clontech) was used as a control. Retroviral packaging BOSC-HEK293 cells were transfected with these constructs to generate retroviruses to infect 3T3-L1 cells. Virus collection, preparation, and infection of cells were essentially as previously described in Ref. 17.
Cell Cycle Synchronization—For the double thymidine block, HeLa M cells were arrested with 2.5 mm thymidine for 19 h and released by washing two times in medium and allowing them to rest for 9 h, followed by a second 16 h arrest with 2.5 mm thymidine. The cells were then released from the block and allowed to progress through the cell cycle. Mitotic cells were collected by shake-off every 30 min, between 8 and 11 h after release from the second cell cycle arrest. The nocodazole block was achieved through an initial cell cycle arrest with 2.5 mm thymidine for 19 h. The cells were then released from the block by washing two times in medium and allowing them to recover for 4 h. This was followed by a second 12-h arrest using 50 ng/ml of nocodazole. Mitotic cells were collected by shake-off.
Reverse Transcription-PCR—Total RNA was isolated using the RNeasy kit (Qiagen) following the manufacturer's protocol. Reverse transcription and quantitative real time PCR was performed as previously described (18). The primers used are described in Table 1. All reverse transcription-PCRs were carried out using 18 S rRNA as an internal control.
TABLE 1.
Primer Primer pairs F18S/R18S were used for the quantification of 18 S rRNA. As an internal control, hL1F/hL1R and hL2F/hL2R were used for the quantification of lipin 1 and lipin 2, respectively. The remaining primers were used for the construction of the shRNAs targeting lipin 1 and 2, as described under “Experimental Procedures.”
| Primer name | Primer sequence |
|---|---|
| F18S | CGGCTACCACATCCAAGGAA |
| R18S | GTCGGAATTACCGCGGCT |
| hL1F | CCGACCAGCTGATGTGTATTCA |
| hL1R | CTATCCTTTAATGGGTGACAACCA |
| hL2F | CCCTGGACTTATAGACAATCCTAACC |
| hL2R | GAGCTGGATGGCAGGTCACT |
| L1O1Bam | GATCCGGGAACTCTGTAGACAGAATTTCAAGAGAATTCTGTCTACAGAGTTCCCTTTTTTCTCGAGG |
| L1O1Eco | AATTCCTCGAGAAAAAAGGGAACTCTGTAGACAGAATTCTCTTGAAATTCTGTCTACAGAGTTCCCG |
| L1O3Bam | GATCCGGTGGAGAGCACCTCCGACATTCAAGAGATGTCGGAGGTGCTCTCCACCTTTTTTCTCGAGG |
| L1O3Eco | AATTCCTCGAGAAAAAAGGTGGAGAGCACCTCCGACATTCAAGAGATGTCGGAGGTGCTCTCCACCG |
| L2O9Bam | GATCCGCCTCAGATTTCATCGCTATTTTCAAGAGAAATAGCGATGAAATCTGAGGCTTTTTTCTCGAGG |
| L2O9Eco | AATTCCTCGAGAAAAAACCTCAGATTTCATCGCTATTTCTCTTGAAAATAGCGATGAAATCTGAGGCG |
| L2O10Bam | GATCCGGGTAAAGACTGGACGCATCTTTCAAGAGAAGATGCGTCCAGTCTTTACCCTTTTTTCTCGAGG |
| L2O10Eco | AATTCCTCGAGAAAAAAGGTAAAGACTGGACGCATCTTCTCTTGAAAGATGCGTCCAGTCTTTACCCG |
Total Cell Lysates and Cell Fractionation—All of the steps were performed at 4 °C. HeLa M cells were washed with PBS, lysed, and centrifuged as described above. Supernatants were diluted to yield 50 μg of total protein/25 μl of loading volume, heated to 95 °C for 5 min, and resolved by SDS-PAGE followed by Western blotting.
For cell fractionation, HeLa M cells were lysed as above in 50 mm Tris-HCl (pH 7.4), 100 μm 4-(2-aminoethyl) benzenesulfonyl fluoride, protease inhibitor complex tablets, phosphatase inhibitor mixture I, 1 μg/ml DNase, 4 mm MgCl2, with or without 1 m NaCl, 1% Triton-X100, or both. Lysed cells were homogenized by passing through a 25GA needle 10 times before centrifuging at 100,000 × g for 1 h at 4 °C. Equivalent volumes of the supernatants and the pellets were mixed with sample buffer, heated to 95 °C for 5 min, and resolved by SDS-PAGE followed by Western blotting. 3T3-L1-derived adipocyte extracts were prepared in the same way, except that instead of syringing, the cells were lysed by sonication.
Cell Extract PAP Assays—HeLa M cell pellets were disrupted by homogenization in 200 μl of 50 mm Tris-HCl (pH 7.5) buffer containing 0.25 m sucrose, 10 mm 2-mercaptoethanol, protease inhibitors (1 mm benzamidine, 0.5 mm phenylmethylsulfonyl fluoride, 5 μg/ml of aprotinin, leupeptin, and pepstatin). The cell lysates were centrifuged at 1,000 × g for 10 min at 4 °C, and the supernatant was used for the PAP assays as described in Ref. 9. Briefly, total PA phosphatase activity (Mg2+-dependent and Mg2+-independent) was measured for 20 min at 30 °C in a total volume of 100 μl containing 50 mm Tris-HCl (pH 7.5), 1 mm MgCl2, 10 mm 2-mercaptoethanol, 0.2 mm [32P]PA (10,000 cpm/nmol), 2 mm Triton X-100, and enzyme protein. The Mg2+-independent PA phosphatase activity was measured separately under the same reaction condition except for the substitution of 1 mm EDTA for 1 mm MgCl2. The Mg2+-dependent PA phosphatase activity was calculated by subtracting the Mg2+-independent enzyme activity from total PA phosphatase activity. The assay was performed in triplicate for each sample. A unit of PA phosphatase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min.
Indirect Immunofluorescence and Confocal Microscopy—The cells were grown on sterile glass coverslips until they were ∼50% confluent and then washed three times with PBS before being fixed in 3% (v/v) formaldehyde in PBS for 20 min. The coverslips were then washed three times in PBS with the second wash containing 50 mm glycine. The cells were permeabilized for 4 min with 0.01% Triton X-100 and then washed two times with PBS. Each coverslip was blocked with PBS containing 1 mg/ml BSA for 20 min prior to incubation with primary antibodies for 30 min. This was followed by three washes of 5 min in PBS containing 1 mg/ml BSA. The coverslips were then incubated with the secondary antibodies for 30 min in the dark. The coverslips were then incubated with PBS containing 1 mg/ml BSA and 200 nm 4′,6-diamidino-2-phenylindole for 5 min, followed by two washes in PBS containing 1 mg/ml BSA. The coverslips were mounted onto glass slides using ProLong Gold antifade reagent (Invitrogen). The cells were visualized with a 63× or 100× Plan Apochromat objective (numerical aperture, 1.4) on a Zeiss Axiovert 200M inverted microscope with an LSM 510 confocal laser scanning attachment. For live imaging, the cells were grown and imaged into CO2-independent medium.
Antibodies—The following antibodies were used in this study: anti-glyceraldehyde-3-phosphate dehydrogenase (Abcam; ab9485), anti-HA[12CA5] (Santa Cruz; sc-805), anti-LaminB (Santa cruz; sc-6217), anti-MPM2 (Upstate; 05-368), anti-transferrin receptor [clone H678.4] (Invitrogen, Zymed Laboratories Inc.; 13-6800), anti-cytochrome c (BD Pharmingen; 556432), anti-giantin (Abcam; ab24586), HRP-conjugated anti-rabbit IgG light chain specific (Jackson Immunoresearch; 211-032-171), HRP-conjugated anti-rabbit IgG (BD Pharmingen; 554021), HRP-conjugated anti-mouse (BD Pharmingen; 554002), HRP-conjugated anti-goat IgG (Novus Biologicals; NB 710-H), anti-goat IgG (H & L) (fluorescein isothiocyanate) preadsorbed (Abcam; ab7121), anti-mouse IgG (H & L) highly cross-adsorbed (Molecular probes; A11029), and anti-rabbit IgG (H & L) highly cross-adsorbed (Molecular probes; A11037). Crude rabbit sera from the final rabbit bleeds were used for all lipin Western blots. Affinity-purified antibodies were used for all lipin immunoprecipitations.
Immunoprecipitations and Lipin PAP1 Assays—HeLa M cells were lysed in 50 mm Hepes (pH 7.4), 1% Triton X-100, 150 mm NaCl, 100 μm 4-(2-aminoethyl) benzenesulfonyl fluoride, protease inhibitor complex tablets (Roche Applied Science), phosphatase inhibitor mixture I (Sigma), 1 μg/ml DNase, 4 mm MgCl2 at 4 °C. Lysed cells were homogenized by passing through a 25GA needle 10 times before centrifuging the homogenates at 16,000 × g for 30 min. Supernatants were diluted to the same final concentration and mixed with the antibody-protein G-Sepharose complex. After incubation at 4 °C for 2 h, the Sepharose beads were washed three times with a 40-fold excess of lysis buffer followed by three washes with a 40-fold excess of PBS.
To dephosphorylate lipins, immune pellets were washed three times with a 40-fold excess of wash buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl and 1 mm EDTA). One twentieth of the immune pellets was removed for Western blot analysis, and the remainder was assayed for PAP1 activity, essentially as previously described (9). Briefly, immune pellets were centrifuged at 3000 × g for 1 min at 4 °C. The pellets were resuspended and incubated in 100 μl of the reaction buffer (50 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 10 mm 2-mercaptoethanol, 2 mm Triton X-100, 0.2 mm [32P]PA, 10,000 cpm/nmol) for 20 min at 30 °C, and PAP1 activity was measured by following the release of water-soluble (32)Pi from chloroform-soluble [32P]PA. The assays were carried out in triplicate. The PAP1 activity was normalized to reflect the relative nominal protein levels and measured by scanning five different low exposures of Western blot films and averaging the band intensities using ImageJ software.
RESULTS
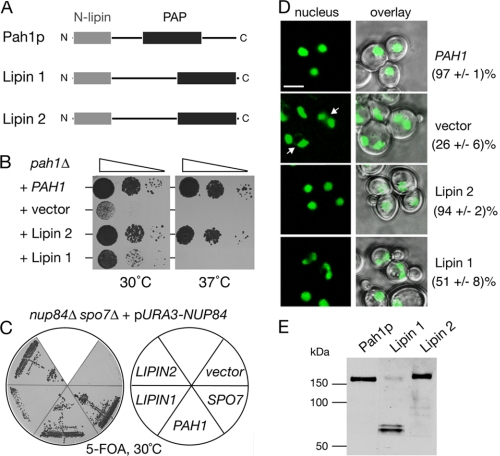
Mammalian Lipins Are Orthologues of the Yeast Pah1p—To address whether the function of lipins is conserved throughout evolution, we asked whether the mammalian lipin 1 and 2 could function in the distantly related unicellular eukaryote Saccharomyces cerevisiae. The human lipin 1 (corresponding to the short murine α form) and mouse lipin 2 open reading frames were placed under the control of the PAH1 promoter and were expressed in a yeast pah1Δ deletion mutant that lacks the endogenous yeast lipin PAH1 gene. Lipin 2 fully rescued the temperature-sensitive growth defect of the pah1Δ strain (Fig. 1B). The pah1Δ mutant cells contain enlarged and irregularly shaped nuclei often consisting of two interconnected lobes within a single cell and also accumulate nuclear/ER membranes into the cytoplasm. These structural defects are efficiently rescued by the expression of lipin 2 in pah1Δ cells, as judged by the reappearance of round nuclei to the same extent as in pah1Δ cells expressing PAH1 (Fig. 1D). Lipin 1 could rescue the growth defect of pah1Δ at 30 °C but not at 37 °C (Fig. 1B). Consistent with this, expression of lipin 1 in pah1Δ cells resulted in a partial restoration of nuclear morphology (Fig. 1D). To compare the expression levels of the two lipins and Pah1p in the pah1Δ cells, we expressed them as C-terminal GFP fusions. The fusion proteins maintain their function because they are still able to rescue the pah1Δ growth defect as efficiently as the untagged proteins.4 The lipin 2-GFP is expressed in yeast at similar levels as the Pah1p-GFP fusion (Fig. 1E). Although an intact lipin1-GFP can be also detected, most of this fusion protein is broken down to a ∼70 kDa C-terminal product, predicted to contain the haloacid dehalogenase-like domain. No other major degradation products were detected in the lipin 1-GFP sample.4 The breakdown of the lipin 1 fusion could explain why it partially rescues the pah1Δ defects.
FIGURE 1.
The function of lipin 1 and 2 is evolutionarily conserved from yeast to mammals. A, primary structure of Pah1p, lipin 1, and lipin 2. The conserved N-terminal domain and the C-terminal PAP domain are indicated. B, lipin 2 rescues the temperature-sensitive growth defect of the pah1Δ mutant. pah1Δ cells were transformed with a 2 μ (high copy) vector containing the indicated genes, spotted on YEPD plates, and grown at 30 or 37 °C for 36 h. C, lipins can rescue the nup84Δ spo7Δ synthetic lethality. The nup84Δ spo7Δ double deletion strain carrying a URA3-containing vector expressing NUP84 was transformed with the indicated plasmids. Transformants were grown on plates containing 5-fluoroorotic acid for 3 days. No growth indicates synthetic lethality. D, lipin 2 rescues the nuclear structure defects of the pah1Δ cells. The pah1Δ mutant expressing an intranuclear reporter (PUS1-GFP) was transformed with the same plasmids as in B and visualized by confocal microscopy. Arrows point to cells containing irregularly shaped/multilobed nuclei. The percentage of cells containing a single round nucleus is given. Two different transformants per strain were analyzed and for each one the number of cells counted was 200. Bars, 5 μm. E, expression of lipin 1 and lipin 2 in yeast. Protein extracts from pah1Δ cells expressing PAH1-GFP, lipin 1-GFP or lipin 2-GFP fusions and grown at 30 °C to early logarithmic phase were analyzed by Western blot using anti-GFP antibodies.
Next, we used a second assay to investigate whether lipins can function in yeast. The combined knock-out of both the nucleoporin NUP84 and the phosphatase subunit SPO7, which dephosphorylates Pah1p, causes lethality (19). The synthetic lethality of the nup84Δ spo7Δ strain can be rescued by the overexpression of PAH1 (10). If lipins can provide the function of Pah1p in yeast, they should also be able to rescue the synthetic lethality of the nup84Δ spo7Δ mutant. To test this hypothesis, we used a nup84Δ spo7Δ double mutant complemented with a centromeric plasmid with the URA3 marker, containing the NUP84 gene (Fig. 1C). Because this strain cannot survive the loss of the URA3-NUP84 plasmid, it will die when grown on medium containing the drug 5-fluoroorotic acid that is toxic to cells expressing the URA3 gene. If a functional or partially functional PAH1 allele is introduced, then cells grown under non-selective conditions can afford the loss of the URA3-NUP84 plasmid and can grow in the presence of 5-fluoroorotic acid. Consistent with the complementation data described above, lipin 2 expression can efficiently rescue the synthetic lethality of the nup84Δ spo7Δ mutant, whereas lipin 1 displays a weaker rescuing activity (Fig. 1C). Taken together, these data show that the function of lipins is evolutionarily conserved from budding yeast to mammals.
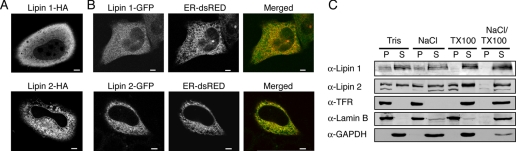
Distinct Intracellular Localizations for Lipin 1 and 2—In contrast to the PA phosphatases of the lipid phosphate phosphatase (LPP) family and other enzymes involved in phospholipid biosynthesis, lipins do not contain predicted transmembrane domains, suggesting that their interaction with membrane surfaces is more transient. Antibodies against lipin 1 and 2 failed to visualize the endogenous proteins in HeLa M cells using a variety of different fixation procedures.4 Therefore, to address their localization, we transfected HeLa M cells with lipin 1 and 2 as either HA or GFP-tagged fusions. In fixed cells, lipin 1-HA shows a predominantly cytosolic staining, whereas lipin 2-HA shows a mixed soluble and reticular staining and a nuclear envelope-associated pool that is reminiscent of ER proteins (Fig. 2A). Lipin 2-HA did not show any significant co-localization with a mitochondrial or a Golgi marker (supplemental Fig. S1). Consistent with its reticular distribution, in live cells a proportion of lipin 2-GFP fusion shows co-localization with an endoplasmic reticulum marker, whereas lipin 1-GFP shows again a diffuse cytosolic distribution (Fig. 2B).
FIGURE 2.
Subcellular localization of lipin 1 and 2. A, localization of lipin 1-HA and lipin 2-HA fusions in formaldehyde fixed HeLa M cells. Expression of lipin-HA fusions was driven by the cytomegalovirus promoter. Bars, 5 μm. B, localization of lipin 1-GFP and lipin 2-GFP fusions in live HeLa M cells. Live HeLa M cells transiently expressing a dsRed-ER reporter and lipin 1-GFP (upper panel) or lipin 2-GFP (lower panel) were visualized by confocal microscopy. Expression of the tagged lipins was driven by the cytomegalovirus promoter. Bars, 5 μm. C, fractionation of endogenous lipin 1 and 2 in HeLa M cells. The cells were lysed in either buffer alone or buffer containing 1 m NaCl, 1% Triton X-100, or both. The extracts were incubated for 20 min at 4 °C and then centrifuged at 100,000 × g for 1 h. Equal volumes from the pellet (P) and supernatant (S) were loaded on an 8% SDS-PAGE and immunoblotted using the indicated antibodies.
To investigate the subcellular distribution of the endogenous lipins, we lysed HeLa M cells in the presence or absence of high salt, nonionic detergent or both and followed their distribution in soluble and insoluble fractions using antibodies against lipin 1 and 2. The majority of endogenous lipin 1 was recovered in the soluble fraction, even when cells were lysed in a buffer lacking salt (Fig. 2C). Lipin 1 displays multiple electrophoretic forms (Fig. 2C, see also later) with the faster migrating species partitioning mostly in the pellet and the slower migrating bands found in the soluble fraction. Interestingly, and unlike lipin 1, which is solubilized in the presence of detergent, the insoluble pool of lipin 2 is resistant to extraction even in the presence of 1 m NaCl or 1% Triton X-100. The insoluble pool of lipin 2 can be solubilized only with a combination of both high salt and nonionic detergent (Fig. 2C). Thus, although both lipins dephosphorylate PA in phospholipid bilayers, they exhibit distinct affinities for membranes in HeLa M cells.
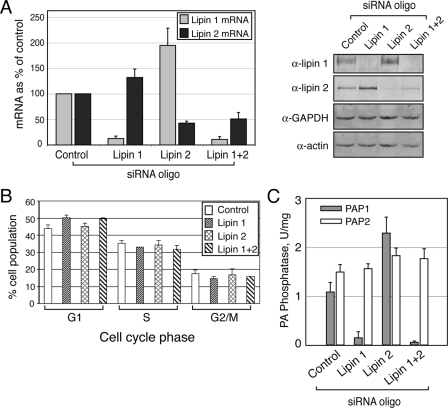
Reciprocal Regulation of Lipin 1 and 2 Controls Cellular PAP1 Activity in HeLa M Cells—To investigate their cellular functions, we depleted lipin 1 and lipin 2 in HeLa M cells using siRNA. To this end we used two different siRNA oligonucleotides for each lipin that could, independently, deplete efficiently the respective proteins 72 h after their transfection (Fig. 3A and data not shown). Notably, depletion of either lipin resulted in an increase of both the mRNA and protein levels of the other lipin (Fig. 3A), suggesting that the knock-down cells activate a compensatory mechanism. Cell cycle analysis shows that depletion of lipin 1 for 72 h causes a 6.3% increase in the relative number of cells in G1 phase, whereas depletion of lipin 2 has no effect (Fig. 3B).
FIGURE 3.
Depletion of lipin 1 and 2 in HeLa M cells. A, siRNA down-regulates lipin 1 and 2 mRNA and protein expression. The cells were transfected with either a nontargeting (Control), or lipin 1, lipin 2, or lipin 1 and 2 small interfering RNA duplexes. 72 h after transfection, mRNA (left panel) or protein (right panel) levels were determined by real time PCR and immunoblotting, respectively, using the indicated antibodies. B, cell cycle profiles of cells from A were determined by flow cytometry as described under “Experimental Procedures.” The significance of the difference between control and lipin 1 siRNA-treated cells found in the G1 phase is indicated by p < 0.00005. The data are averages from eight independent experiments. C, lipin 1 is the major PAP1 enzyme in HeLa M cells. Control and lipin siRNA-treated cells were lysed 72 h after the knock-down, and PAP assays were performed as described under “Experimental Procedures.” The results shown were determined from triplicate enzyme assays ± S.D. and were reproduced in two independent experiments.
Because both lipin 1 and 2 are expressed in HeLa M cells, we set out to compare their relative contribution in the regulation of the cellular Mg2+-dependent PA phosphatase activity. To do this, we silenced lipin 1, lipin 2, or both in HeLa M cells, prepared extracts 72 h after transfection of the respective siRNAs and measured the PAP1 and PAP2 activities. Importantly, lipin 1-depleted extracts show an 86% reduction in the total cellular PAP1 activity, indicating that lipin 1 is the major PAP1 enzyme in these cells (Fig. 3C). Surprisingly, knock-down of lipin 2 has the opposite effect and results in a dramatic increase in the cellular PAP1 levels (Fig. 3C). This could be explained by the fact that lipin 1 levels increase significantly in the lipin 2-depleted cells, leading to an increase in the total PAP1 activity. The combined lipin 1 and 2 knock-down led to a further small but significant decrease of the PAP1 activity (Fig. 3C). On the other hand, the Mg2+-independent PA phosphatase (PAP2) activity did not change in any of the lipin-depleted cells (Fig. 3C). Taken together, the complementary up-regulation of each lipin in the respective knock-down and its effect on cellular PAP1 activity strongly suggest that lipin 1 is the more active enzyme, whereas the activity of lipin 2 is more tightly regulated in HeLa M cells.
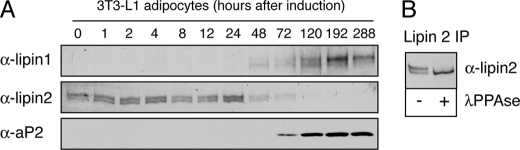
Reciprocal Regulation of Lipin 1 and 2 in Differentiating Adipocytes—The reciprocal regulation of the levels of the two lipins in HeLa M cells prompted us to test how their expression is coordinated during adipogenesis where PAP1 activity is required for the biosynthesis of the storage lipid triacylglycerol. To address this question, 3T3-L1 preadipocytes were induced to differentiate and protein extracts were prepared at various time points afterward. As seen in Fig. 4A, lipin 2 is expressed in preadipocytes and also during the early stages of adipogenesis before its levels decrease almost to undetectable levels in mature adipocytes. Lipin 1 shows a reciprocal pattern with no detectable expression in preadipocytes and a gradual increase after 48 h of differentiation, which coincides with a decrease in lipin 2 levels (Fig. 4A). Lipin 1 is expressed first as a low and subsequently as a higher molecular weight protein band on SDS-PAGE that persists in mature adipocytes, probably corresponding to the two isoforms of lipin 1 generated by alternative splicing (12). Interestingly, lipin 2 is also expressed as a high and low molecular weight protein band on SDS-PAGE, but in this case the higher band is due to phosphorylation as it resolves to the lower form following phosphatase treatment of the immunoprecipitated protein (Fig. 4B).
FIGURE 4.
Reciprocal pattern of lipin 1 and 2 protein expression during adipogenesis. A, 3T3-L1 preadipocytes were induced to differentiate for 12 days. At the indicated time points, the cell lysates were prepared, protein concentrations were measured by Bradford assay, and equal protein amounts/time point were analyzed by SDS-PAGE followed by immunoblotting using lipin 1 and 2 antibodies. Adipocyte differentiation was monitored by following αP2 expression. B, lipin 2 is phosphorylated in 3T3-L1 cells. Endogenous lipin 2 was immunoprecipitated (IP) from extracts of 3T3-L1 preadipocytes. Immune pellets were incubated with or without λ-phosphatase (λPPase) and analyzed by SDS-PAGE followed by immunoblotting using lipin 2 antibodies.
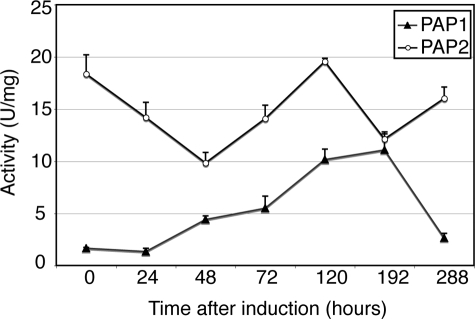
The fact that lipin 1 is the major PAP1 enzyme in HeLa M cells suggests that the change in the ratio between the protein levels of lipin 1 and 2 during adipogenesis could contribute to the maintenance of high biosynthetic PAP1 activity required for bulk triglyceride production. To test this hypothesis, we quantified the PAP1 activity during the differentiation of 3T3 preadipocytes. Indeed, cellular PAP1 levels increase steadily 24 h after induction of differentiation and peak at the 192-h time point, overlapping significantly with the induction pattern of lipin 1 (Fig. 5).
FIGURE 5.
PAP1 activity increases during adipogenesis. 3T3-L1 preadipocytes were induced to differentiate for 12 days. At the indicated time points, the cell lysates were prepared, and the PAP1 and PAP2 activities were measured as described under “Experimental Procedures.” The values for each time point were determined from triplicate enzyme assays ± S.D. A unit of PA phosphatase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min. Specific activity was defined as units/mg protein.
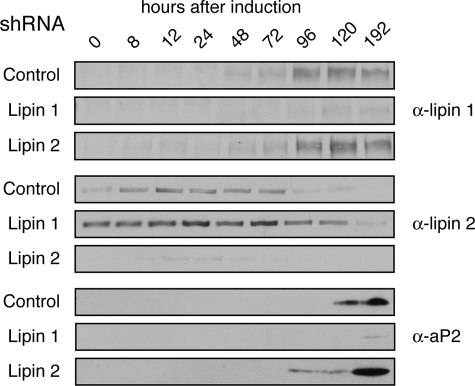
To examine whether expression of the different lipins is co-regulated during adipocyte differentiation, we silenced lipin 1 or lipin 2 by infecting 3T3-L1 preadipocytes with retroviruses carrying shRNAs specifically targeting their mRNA sequences. For each lipin, two cell lines were constructed, each expressing a different shRNA (see “Experimental Procedures”), and the results described below were reproduced with both. Infection of the cells with shRNA targeting either lipin 1 or lipin 2 resulted in an efficient decrease in the levels of the respective protein (Fig. 6). Notably, in lipin 1 knock-down cells, lipin 2 protein levels significantly increased at all time points examined and persisted for longer during differentiation (Fig. 6). Despite the increased lipin 2 levels, depletion of lipin 1 inhibited the differentiation of preadipocytes to mature adipocytes as judged by the failure to induce aP2 expression (Fig. 6) or to accumulate intracellular lipid (triacylglycerol/steryl esters), followed by Nile Red staining.4 In contrast, depletion of lipin 2 did not inhibit adipogenesis. Indeed, knock-down of lipin 2 led to a more rapid and robust induction of aP2, suggesting that adipogenesis may be accelerated in these cells (Fig. 6). Together, these data are consistent with the reciprocal regulation of the two lipins observed in HeLa M cells and also indicate that they perform distinct functions in differentiating adipocytes.
FIGURE 6.
shRNA-mediated silencing of lipin 1 and 2 in differentiating adipocytes. 3T3-L1 preadipocytes stably transfected with retroviral vectors expressing shRNA targeting lipin 1 (Lipin 1), lipin 2 (Lipin 2) or a control sequence (Control), were induced to differentiate for 8 days. At the indicated time points, the cell lysates were prepared, the protein concentrations were measured by Bradford assay, and equal protein amounts/time point were analyzed by SDS-PAGE followed by immunoblotting using the indicated antibodies.
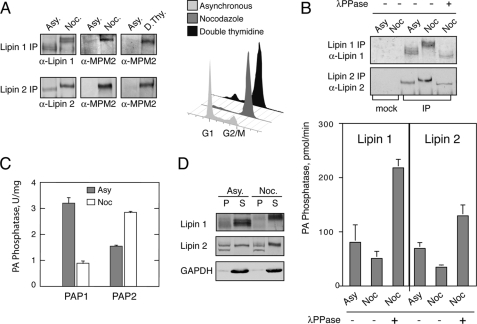
Mitotic Phosphorylation of Lipin 1 and 2 Inhibits Their PA Phosphatase Activity—We next asked whether the function of the two lipins could also be regulated by post-translational modifications. The fact that the yeast lipin Pah1p is phosphorylated during mitosis (10) and that the mammalian lipins can functionally replace the yeast enzyme (Fig. 1) prompted us to test whether lipin 1 and 2 are also mitotically phosphorylated. To address this possibility, endogenous lipin 1 and 2 were immunoprecipitated from asynchronous or mitotic HeLa M cells, generated by nocodazole arrest, and analyzed by Western blotting. As seen in Fig. 7A, both lipins undergo mobility shifts that reduce their electrophoretic mobility in the nocodazole-arrested samples. This mobility shift is due to phosphorylation because it can be reversed in a concentration-dependent manner after incubation of the immunoprecipitated proteins with λ-phosphatase (Fig. 7B and data not shown). Importantly, the hyperphosphorylated lipin 1 and 2 forms are recognized by MPM2, a monoclonal antibody that is specific for phospho-Ser/Thr-Pro sites, the minimal Cdk1 phosphorylation motif (20) (Fig. 7A).
FIGURE 7.
Mitotic phosphorylation of lipin 1 and 2 regulates their PAP1 activity. A, lipins are phosphorylated on Cdk1 motifs during mitosis. Left panel, Endogenous lipin 1 and lipin 2 were immunoprecipitated (IP) from extracts of asynchronous (Asy) or mitotic HeLa M cells synchronized either by nocodazole arrest (Noc) or double thymidine arrest at the G1/S boundary followed by release and collection during mitosis (8–10 h post-release) (D. Thy). Immune pellets were analyzed by Western blotting using anti-lipin 1, anti-lipin 2, and anti-phospho-Ser/Thr-Pro (MPM2) antibodies. Right panel, flow cytometry of the cells used for the lipin immunoprecipitations. B, mitotic phosphorylation of lipins inhibits their PAP1 activity. Upper panel, immunoprecipitated lipin 1 and 2 from asynchronous or nocodazole-treated HeLa M cells with or without incubation with λ-phosphatase (λPPase) were analyzed by Western blotting as indicated. Mock immunoprecipitations were performed using the preimmune lipin 1 and 2 sera. Lower panel, immune pellets from the above immunoprecipitations were assayed for PAP1 activity as described under “Experimental Procedures.” The results shown were determined from triplicate enzyme assays ± S.D. The data for the asynchronous and nocodazole samples are the averages from two independent experiments. C, total PAP1 activity decreases during mitosis. Lysates from asynchronous or mitotic cells were assayed for PAP1 and PAP2 activity as described under “Experimental Procedures.” The results shown were determined from triplicate enzyme assays ± S.D. D, mitotic phosphorylation of lipins regulates their membrane recruitment. Lysates from asynchronous or mitotic cells were centrifuged at 100,000 × g. Equal volumes from the pellet (P) and supernatant (S) were immunoblotted using the indicated antibodies.
To independently confirm that lipins are mitotically phosphorylated, we immunoprecipitated them from mitotic cells synchronized with a different procedure. HeLa M cells were synchronized at the G1/S boundary by double thymidine block and released to allow progression into M phase, and then lipins were immunoprecipitated. Consistent with the results obtained from the nocodazole treated cells, mitotic HeLa M cells synchronized by the double thymidine block also result in MPM2-specific phosphorylation of lipin 1 and 2 (Fig. 7A). Taken together, these data demonstrate that lipin 1 and 2 are mitotically phosphorylated on Cdk1 consensus sites.
Next we examined the effect of the mitotic phosphorylation on the function of lipins. We first asked whether phosphorylation could regulate the PA phosphatase activity of lipin 1 and 2. To address this, endogenous lipin 1 and 2 were immunoprecipitated from asynchronous or nocodazole-arrested HeLa M cells with or without λ-phosphatase treatment, and the immune pellets were then assayed for Mg2+-dependent PA phosphatase activity. As seen in Fig. 7B, mitotic phosphorylation of both lipin 1 and 2 decreases their PA phosphatase activity, whereas dephosphorylation stimulates it. Consistent with a role of phosphorylation in inhibiting the activities of the lipins, the total Mg2+-dependent PAP activity (PAP1) in the extracts of nocodazole-arrested HeLa M cells decreased when compared with the activity of asynchronous cells (Fig. 7C). On the other hand, the total Mg2+-independent activity (PAP2), which is mediated by the LPP family of phosphatases, responded in the opposite way and showed increased levels in the nocodazolederived extracts (Fig. 7C).
Phosphorylation also regulates membrane recruitment of lipin 1 and 2 in HeLa M cells. As seen in Figs. 2C and 7D, the slow migrating forms of lipin 1 are significantly enriched in the soluble fractions of both the asynchronous and mitotic cell lysates. Similarly, in the nocodazole-treated cells, only the phosphorylated lipin 2 becomes soluble. Taken together, these data show that mitotic phosphorylation of lipin 1 and 2 is a major determinant of their function in HeLa M cells, by controlling both their PAP1 activity and membrane recruitment.
DISCUSSION
In contrast to yeasts, nematodes, and flies that express only one lipin, mammals express three lipins that display overlapping expression in many different tissues. This observation raises two interesting questions: first, whether there are functional differences between the lipins, and second, how cells coordinate the activities of different lipins during growth and development.
Our results show that there are two major differences between lipin 1 and 2 in HeLa M cells. First, the two lipins show distinct intracellular localization, with lipin 1 exhibiting a more soluble distribution and lipin 2 exhibiting a detergent- and salt-insoluble pool. In this context it is interesting to notice that although the murine lipin 1α is primarily intranuclear in HEK293 and 3T3-L1-derived adipocytes (11, 12), we found that its apparent human homologue displays a cytosolic localization in HeLa M cells. The basis for this difference is not clear. Lipin 2 displays also a distinct ER-associated pool. Because the various lipin paralogues exhibit overlapping expression in many different tissues (14), their distinct localization raises the interesting possibility of the compartmentalization of the PAP1 reaction and thus control of PA, DAG, and phospholipid homeostasis, on different organelles. Because lipins have very similar domain organization, their targeting to different intracellular compartments might be mediated by interaction with different binding partners. Second, the levels of lipin 1, but not lipin 2, correlate with the cellular PAP1 activity in HeLa M cells. Thus, siRNA-mediated depletion of lipin 1 leads to a dramatic decrease of PAP1 activity. This is consistent with the absence of PAP1 activity in tissues from the fld mouse that expresses an inactive mutant lipin 1 allele (13, 14). On the other hand, depletion of lipin 2 has the opposite effect and leads to an increase of both lipin 1 expression and PAP1 activity levels. Thus, lipin 1 is the major PAP1 enzyme in HeLa M cells. Higher lipin 2 protein levels in lipin 1 siRNA and shRNA experiments in HeLa M and 3T3-L1 cells, respectively, do not compensate the relevant knock-down phenotypes, indicating that the two lipins perform non-redundant functions in lipid biosynthesis.
The different functions of the two lipins could explain the temporal regulation of their expression during adipogenesis. Cellular models of adipogenesis suggest that preadipocytes first become committed to differentiation, followed by a limited proliferation and a final maturation process when they acquire their specialized adipogenic properties, including lipid accumulation (21). Lipin 1 is not expressed in the preadipocytes, which suggests that lipin 2 and possibly lipin 3 are able to provide the necessary PAP1 activity prior to differentiation. On the other hand, expression of a highly active PAP1 enzyme is needed to support bulk triglyceride production at the final stages of differentiation. Interestingly, depletion of lipin 2 correlates with an earlier appearance of both the adipogenic marker aP2 (Fig. 6) and lipid accumulation.4 Thus, the levels of lipin 2 are able to indirectly influence adipogenesis. This mirrors the situation in HeLa M cells, where lipin 2 knock-down increases cellular PAP1 activity and underscores the significance of the temporal regulation of lipin 1 expression.
Another important finding of this work is that phosphorylation of lipins inhibits their PAP1 activity during mitosis. What could be the function of this inhibition? Phospholipids accumulate in the S phase, whereas their synthesis is inhibited during the G2/M phase, when cellular metabolism reaches the lowest rate in the cell cycle (22). The decrease of the PAP1 activity caused by the mitotic phosphorylation of lipin 1 and 2 could contribute to the inhibition of phospholipid accumulation prior to cell division. Interestingly, CCT1α, the enzyme catalyzing the rate-limiting step in the biosynthesis of phosphatidylcholine, is also inactivated by hyperphosphorylation late in the cell cycle (22). These data are consistent with a model where biosynthetic production of phospholipids is inhibited at multiple steps during cell division. Phospholipid biosynthesis is also developmentally regulated, and recent studies found that transcription of lipin 1 is induced in cells that undergo ER proliferation (23, 24). It will be interesting to test whether dephosphorylation-dependent activation of lipins accompanies some of these transcriptional changes.
Contrary to the results presented here, a recent study found that dephosphorylation of adipocyte lipin 1 does not have any effect on its PAP1 activity (13). It is possible that the kinases responsible for the phosphorylation of lipin 1 in adipocytes and in mitotic HeLa M cells are not the same. Accordingly, phosphorylation of lipin 1 at different sites could have different functional consequences in either cell line. Alternatively, these differences could be due to the different responses of the two lipin 1 isoforms to phosphorylation. Adipocytes, which were used by Harris et al. (13) as a source of lipin 1, express mostly the longer β form (12), whereas HeLa M cells used to immunoprecipitate lipin 1 in our assays express two forms that differ slightly in their electrophoretic mobilities (Fig. 7B), presumably representing the α and β forms. It is possible that dephosphorylation of only one isoform (α) is responsible for the increase in PAP1 activity.
Proteins that regulate the phosphorylation state of lipins are expected to influence PAP1 activity and consequently lipid and membrane biogenesis. A recent study reported that Dullard, the mammalian orthologue of the protein phosphatase Nem1p that regulates Pah1p activity and nuclear membrane growth in yeast (10), localizes to the inner side of the nuclear envelope and dephosphorylates lipin 1 in BHK cells (25). Because Pah1p regulates nuclear membrane biogenesis in yeast, this finding raises the intriguing possibility that lipins could perform a similar function in mammalian cells. Future experiments will address these possibilities.
In summary, we provide the first evidence that the two related PA phosphatases lipin 1 and 2 display differences in their localization, expression pattern, and activity. Such a differential regulation of the PAP1 activity could explain why higher eukaryotes evolved multiple lipin paralogues. This also suggests that although lipin 1 is the major PAP1 enzyme in HeLa M cells and murine adipocytes, lipin 2 and 3 have as of yet undetermined important roles. The future determination of the specific functions of each lipin is important to understand how these enzymes control the biosynthetic production of lipids, lipid signaling, cell differentiation, and membrane growth.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140 (to G. M. C.). This work was also supported by a Wellcome Trust career development fellowship (to S. S.), by the United States Public Health Service, and by a British Heart Foundation intermediate fellowship (to J. J. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: PA, phosphatidate; DAG, diacylglycerol; ER, endoplasmic reticulum; siRNA, small interfering RNA; shRNA, small hairpin RNA; PAP1, Mg2+-dependent phosphatidate phosphatase; PAP2, Mg2+-independent PA phosphatase; HA, hemagglutinin; GFP, green fluorescent protein; PBS, phosphate-buffered saline; BSA, bovine serum albumin; HRP, horseradish peroxidase.
N. Grimsey and S. Siniossoglou, unpublished data.
References
- 1.Lykidis, A., and Jackowski, S. (2001) Prog. Nucleic Acids Res. Mol. Biol. 65 361-393 [DOI] [PubMed] [Google Scholar]
- 2.van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008) Nat. Rev. Mol. Cell Biol. 9 112-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wymann, M. P., and Schneitter, R. (2008) Nat. Rev. Mol. Cell Biol. 9 162-176 [DOI] [PubMed] [Google Scholar]
- 4.Walton, P. A., and Possmayer, F. (1984) Biochim. Biophys. Acta 796 364-372 [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Munoz, A., Hatch, G. M., Martin, A., Jamal, Z., Vance, D. E., and Brindley, D. N. (1992) FEBS Lett. 301 103-106 [DOI] [PubMed] [Google Scholar]
- 6.Brindley, D. N. (2004) J. Cell Biochem. 92 900-912 [DOI] [PubMed] [Google Scholar]
- 7.Athenstaedt, K., and Daum, G. (2006) Cell Mol. Life Sci. 63 1355-1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, R. A., and Lee, D. P. (2004) Prog. Lipid Res. 43 134-176 [DOI] [PubMed] [Google Scholar]
- 9.Han, G.-S., Wu, W. I., and Carman, G. M. (2006) J. Biol. Chem. 281 9210-9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 24 1931-1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterfy, M., Phan, J., Xu, P., and Reue, K. (2001) Nat. Genet. 27 121-124 [DOI] [PubMed] [Google Scholar]
- 12.Peterfy, M., Phan, J., and Reue, K. (2005) J. Biol. Chem. 280 32883-32889 [DOI] [PubMed] [Google Scholar]
- 13.Harris, T. E, Huffman, T. A., Chi, A., Shabanowitz, J., Hunt, D. F., Kumar, A., and Lawrence, J. C., Jr. (2007) J. Biol. Chem. 282 277-286 [DOI] [PubMed] [Google Scholar]
- 14.Donkor, J., Sariahmetoglu, M., Dewald, J., Brindley, D. N., and Reue, K. (2007) J. Biol. Chem. 282 3450-3457 [DOI] [PubMed] [Google Scholar]
- 15.Phan, J., and Reue, K. (2005) Cell Metab. 1 73-83 [DOI] [PubMed] [Google Scholar]
- 16.Finck, B. N., Gropler, M. C., Chen, Z., Leone, T. C., Croce, M. A., Harris, T. E., Lawrence, J. C., Jr., and Kelly, D. P. (2006) Cell Metab. 4 199-210 [DOI] [PubMed] [Google Scholar]
- 17.Rochford, J. J., Semple, R. K., Laudes, M., Boyle, K. B., Christodoulides, C., Mulligan, C., Lelliott, C. J., Schinner, S., Hadaschik, D., Mahadevan, M., Sethi, J. K., Vidal-Puig, A., and O'Rahilly, S. (2004) Mol. Cell Biol. 24 9863-9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne, V. A., Grimsey, N., Tuthill, A., Virtue, S., Gray, S. L., Nora, E. D., Semple, R. K., O'Rahilly, S., and Rochford, J. J. (2008) Diabetes, 57 2055-2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siniossoglou, S., Santos-Rosa, H., Rappsilber, J., Mann, M., and Hurt, E. (1998) EMBO J., 17 6449-6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, F. M., Tsao, T. Y., Fowler, S. K., and Rao, P. N. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 2926-2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen, E. D., and MacDougald, O. A. (2006) Nat. Rev. Mol. Cell Biol. 7 885-896 [DOI] [PubMed] [Google Scholar]
- 22.Jackowski, S. (1994) J. Biol. Chem. 269 3858-3867 [PubMed] [Google Scholar]
- 23.Fagone, P., Sriburi, R., Ward-Chapman, C., Frank, M., Wang, J., Gunter, C., Brewer, J. W., and Jackowski, S. (2007) J. Biol. Chem. 282 7591-75605 [DOI] [PubMed] [Google Scholar]
- 24.Sriburi, R., Bommiasamy, H., Buldak, G. L., Robbins, G. R., Frank, M., Jackowski, S., and Brewer, J. W. (2007) J. Biol. Chem. 282 7024-7034 [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y., Gentry, M. S., Harris, T. E., Wiley, S. E., Lawrence, J. C., Jr., and Dixon, J. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6596-6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.