Abstract
Francisella novicida U112 phospholipids, extracted without hydrolysis, consist mainly of phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, and two lipid A species, designated A1 and A2. These lipid A species, present in a ratio of 7:1, comprise 15 % of the total phospholipids, as judged by 32Pi labeling. Although lipopolysaccharide is detectable in F. novicida U112, less than 5 % of the total lipid A is covalently linked to it. A1 and A2 were analyzed by electrospray ionization and matrix-assisted laser desorption ionization mass spectrometry, gas chromatography/mass spectrometry and NMR spectroscopy. Both compounds are disaccharides of glucosamine, acylated with primary 3-hydroxystearoyl chains at positions 2, 3, and 2′, and a secondary palmitoyl residue at position 2′. Minor isobaric species and some lipid A molecules containing a 3-hydroxypalmitoyl chain in place of 3-hydroxystearate are also present. The 4′- and 3′-positions of A1 and A2 are not derivatized, and Kdo is not detectable. The 1-phosphate groups of both A1 and A2 are modified with an α-linked galactosamine residue, as shown by NMR spectroscopy and gas chromatography/mass spectrometry. An α-linked glucose moiety is attached to the 6′-position of A2. The lipid A released by mild acid hydrolysis of F. novicida lipopolysaccharide consists solely of component A1. F. novicida mutants lacking the arnT gene do not contain a galactosamine residue on their lipid A. Formation of free lipid A in F. novicida might be initiated by an unusual Kdo hydrolase present in the membranes of this organism.
Lipopolysaccharide (LPS) makes up the outer leaflet of the outer membranes of most Gram-negative bacteria (1-3). It consists of a hydrophobic moiety known as lipid A, a non-repeating core oligosaccharide, and a distal repeating oligosaccharide, termed the O-antigen (1-3). Lipid A of wild-type E. coli is a hexa-acylated disaccharide of glucosamine that is phosphorylated at the 1- and 4′-positions (1-3) (Fig. 1A). It is recognized by the TLR4/MD2 receptor of the innate immune system (4-8), which triggers an inflammatory response and helps to clear localized infections. However, a more generalized response to lipid A in the context of a systemic infection, accompanied by massive over-production of cytokines, can lead to Gram-negative septic shock and death (9, 10).
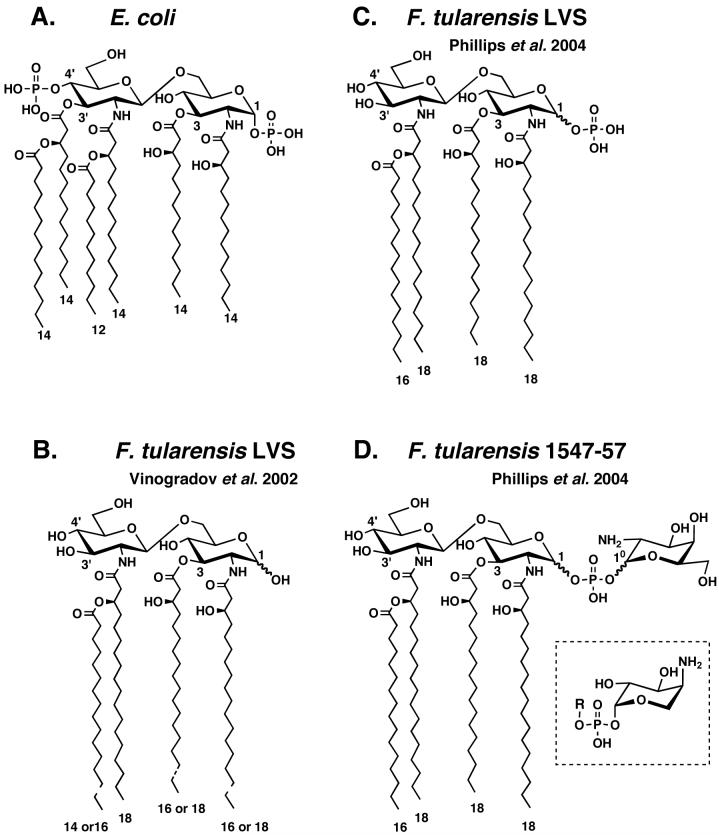
Figure 1. E. coli lipid A compared to structures proposed for F. tularensis lipid A.
Panel A. The major lipid A species of wild-type E. coli K-12 is a hexa-acylated disaccharide of glucosamine, substituted with phosphate groups at the 1- and 4′-positions (3, 38). Key positions are numbered. Panel B. F. tularensis LVS lipid A was first reported as a tetra-acylated disaccharide of glucosamine lacking both phosphate residues and the 3′ acyl chain (18). Dotted lines indicate possible acyl chain length heterogeneity. Panel C. A more recent study of LVS lipid A demonstrated the presence of a phosphate group at the 1 position but not the anomeric configuration (22). Panel D. In the same study (22), the lipid A of F. tularensis 1547-57 was shown to be a tetra-acylated disaccharide of glucosamine substituted with a novel galactosamine-phosphate group at position 1. The anomeric configurations of the proximal glucosamine and galactosamine-1-phosphate units were not determined (22). The inset shows the structure of the 4-amino-4-deoxy-l-arabinose-1-phosphate substituent found on lipid A in polymyxin-resistant mutants of E. coli and Salmonella (36, 40, 50).
Wild-type E. coli cells contain ∼ 0.15 lipid A residues per glycerophospholipid molecule (11). All of the lipid A residues are covalently attached to LPS, whereas free lipid A and lipid A precursors are not usually detectable by 32Pi labeling (12) or mass spectrometry (MS) (13). Heptose-deficient mutants of E. coli are viable but synthesize a minimal LPS substructure consisting of Kdo2-lipid A, which is fully active as a TLR4/MD2 agonist (14). However, free lipid A and lipid A precursors do not accumulate in these mutants (15).
Francisella tularensis, a highly infectious Category A human pathogen, causes tularemia and is of concern because of its potential for bio-terrorism (16). The LPS of F. tularensis has been isolated (17), and the structures of its O-antigen, core and lipid A domains have been partially characterized (18-22). Some differences and ambiguities in the structures reported for F. tularensis lipid A are noteworthy (Fig. 1). For instance, Vinogradov et al. claimed that lipid A from the live vaccine strain (LVS) of F. tularensis is not phosphorylated at either the 1 or the 4′ positions (Fig. 1B) (18). In contrast, Phillips et al. reported the presence of a phosphate group at the 1 position of F. tularensis LVS lipid A (Fig. 1C) and, in addition, described an unprecedented galactosamine phosphate substituent at the 1 position of lipid A isolated from F. tulalrensis 1547-57 (Fig. 1D) (22). Because NMR spectroscopy was not utilized by Phillips et al. (22), the anomeric configurations of the galactosamine and the proximal glucosamine residues could not be determined (Figs. 1C and 1D).
Strains of Francisella appear to make less LPS than E. coli, as judged by phenol-water extraction (18, 23), and the bioactivity of Francisella LPS is very low (24, 25). We now report that F. tularensis subsp. novicida U112, an attenuated mouse pathogen, does in fact synthesize lipid A in amounts comparable to E. coli (11) but that most of this material is not glycosylated with core or O-antigen sugars. Consequently, it is not recovered by the traditional phenol-water method for purifying LPS (18). Instead, the F. novicida glycerophospholipid fraction, extracted with chloroform/methanol according to the method of Bligh and Dyer (13, 26), contains two prominent lipid A species, designated A1 and A2. These substances were purified and analyzed by MS and NMR spectroscopy. The ratio of A1 to A2 is about 7:1. Both A1 and A2 are disaccharides of glucosamine, acylated predominantly with 3-hydroxystearoyl chains at the 2, 3, and 2′-positions, and a secondary palmitoyl chain at the 2′-position (Fig. 1). Both A1 and A2 contain a phosphate moiety at the 1-position, which is further derivatized with a galactosamine residue, as confirmed by gas chromatography/mass spectrometry (GC/MS). NMR studies demonstrate unequivocally that the anomeric carbons of the galactosamine and proximal glucosamine units of A1 and A2 have the alpha configurations. We further show that the incorporation of the galactosamine residue into F. novicida lipid A is dependent upon its arnT gene orthologue (27, 28). Neither A1 nor A2 contains a phosphate group at the 4′-position, presumably because of the periplasmic 4′-phosphatase encoded by lpxF in Francisella (29). The difference between A1 and A2 is the presence of an unusual α-linked glucose moiety at the 6′-position in A2. While compound A1 is probably identical to the lipid A species released by mild acid hydrolysis from F. tularensis LPS (Fig. 1D), a glucose-modified derivative of F. tularensis lipid A has not been described.
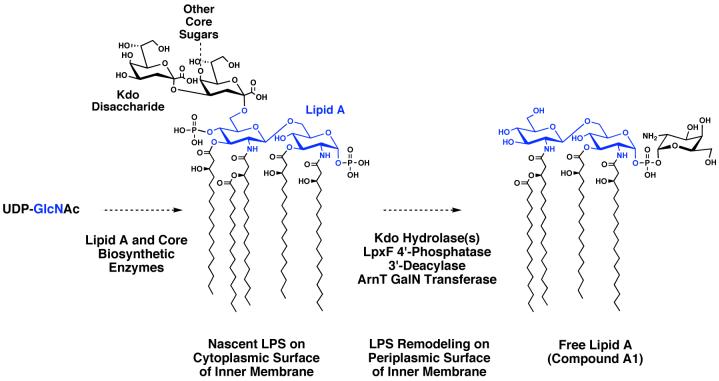
The discovery of large quantities of free lipid A associated with low levels of LPS in wild-type F. novicida is without precedent in LPS biochemistry and might play a role in the pathogenesis of Francisella infections. We show that lipid A from the LVS strain of F. tularensis also is present mostly in the free form. We propose that free lipid A may arise from nascent LPS by a novel system of extra-cellular remodeling enzymes, which include a Kdo hydrolase (15), the 4′-phosphatase LpxF (29), a galactosamine transferase encoded by F. novicida arnT, and a 3′-O-deacylase (Scheme 1).
Scheme 1. A proposed enzymatic pathway for the processing of nascent LPS to generate free lipid A molecules in Francisella novicida.
The glucosamine residues are shown in blue. The galactosamine unit incorporated by ArnT is shown in black. The genes encoding the Kdo hydrolase and 3′-deacylase, as well as their sub-cellular localization, are unknown.
MATERIALS AND METHODS
Materials and reagents
Glass backed 0.25 mm Silica Gel 60 thin layer chromatography plates were from E. Merck, Darmstadt, Germany. Chloroform, ammonium acetate, and sodium acetate were obtained from EMD Science, Gibbstown, NJ, while pyridine, methanol, and formic acid were from Mallinckrodt, Phillipsburg, NJ. Trypticase soy broth, yeast extract and tryptone were purchased from Difco, Detroit, MI. DEAE cellulose was purchased from Whatman, Florham Park, NJ. 32Pi was purchased from NEN Life Science Products. Tri-Sil Reagent was from Pierce, Rockford, IL. The methanol-HCl kit was from Supelco, St. Louis, MO. D2O, CD3OD and CDCl3 were from Sigma-Aldrich, St. Louis, MO. F. novicida U112 was obtained from Dr. F. Nano, University of Victoria, British Columbia, Canada. The F. tularensis LVS strain was purchased from the American Type Culture Collection, Rockville, MD.
Construction of a F. novicida mutant with a deletion in its arnT orthologue
The single orthologue of the arnT gene (27) present in the F. novicida genome (30) was amplified by PCR with 2 kb of flanking F. novicida genomic DNA. The forward primer (5′-CGTGCGGCCGCACTTATTACGACCTTCTTCGGTATTA-3′) was designed with a clamp region, a NotI restriction site (underlined), and a match to the coding strand of chromosomal DNA about 2 kb upstream of F. novicida arnT. The reverse PCR primer (5′-ACGCTCGAGATGTTTGATATTGTTTTTTATTAGCATATT-3′) was designed with a clamp region, a XhoI restriction site (underlined), and a match to the anti-coding strand of chromosomal DNA about 2kb downstream of F. novicida arnT. The PCR was performed using PfuTurbo polymerase and F. novicida genomic DNA as the template. Amplification was carried out in a 100 μl reaction mixture containing 200 ng template, 200 ng primers and 2 units of polymerase. The reaction was started at 94 °C for 2 min, followed by 30 cycles of denaturation (30 seconds at 94 °C), annealing (30 seconds at 58 °C), and extension (5 min at 72 °C). After the 30th cycle, a 10 min extension time was used. The reaction product was analyzed on a 1% agarose gel. The desired band was excised and gel-purified. The PCR product was then digested using XhoI and NotI, and ligated into the expression vector pWSK29 (31), which had been similarly digested. The ligation mixture was eletroporated into E. coli DY330 (32), and colonies were selected for those with the appropriate inserts on LB plates containing 100 μg/ml ampicillin. Plasmid pWSK29-5000F was isolated in this manner, and the insert was confirmed by DNA sequencing.
Next, the F. novicida arnT orthologue (30) in the plasmid pWSK29-5000F was replaced with a kanamycin resistance gene. Tn903 from plasmid pWKS130 was amplified by PCR. It was flanked on its 5′ and 3′ ends by 50 bp of the chromosomal DNA that is immediately upstream or downstream of F. novicida arnT. The forward and reverse primers are 5′-TAATTCCCGCTAAATTTATTCCATACTTTTGCCAGATAAATACTACTATCATGAGCCATATTCAACGGGAAACG-3′ and 5′-TAATCTAAAGATCATAAAAAAATAGTTAG-TTCTGTTTATTTTTAAATCAGTTAGAAAAACT-CATCAAGCATCAAATG-3′, respectively. The sequences belonging to the kanamycin resistance cassette are underlined. The PCR was performed using Pfu polymerase. Amplification was carried out in a 100 μl reaction mixture containing 100 ng of template, 250 ng primers and 2 units of Pfu polymerase. The reaction was started at 94 °C for 1 min, followed by 25 cycles of denaturation (45 seconds at 94 °C), annealing (45 seconds at 55 °C), and extension (60 seconds at 72 °C). After the 25th cycle, a 10 min extension time was used. The PCR product was resolved on a 1.0% agarose gel and purified. The purified PCR product was electroporated into E. coli DY330/pWSK29-5000F, which had been grown at 30 °C until A600 reached 0.5 and then induced at 42 °C for 20 min. After electroporation, recombination occurred between the homologous sequences on the linear PCR product and the arnT gene on the plasmid, resulting in replacement of the arnT orthologue with the kanamycin resistance gene. The desired plasmid, designated pWSK29-5000K, was then isolated from kanamycin-resistant transformants, selected on LB agar containing 20 μg/ml kanamycin and 100 μg/ml ampicillin, and digested with ApaI and SacII. About 100 ng of the linear insert DNA was gel-purified, and the replacement of F. novicida arnT with the kanamycin resistance gene was verified by DNA sequencing.
The final step was to replace the arnT orthologue present in the genome of F. novicida with the kanamycin resistance gene. F. novicida U112 cells were grown in 100 ml of Chamberlain’s medium (33) to A600 of 0.5. Cells were harvested by centrifugation and resuspended in 5 ml of transformation buffer (34). Next, 1 ml of the cell suspension was mixed with 100 ng of the linear insert DNA, prepared as described above, and shaken at 100 rpm at 37 °C for 30 min. Then 5 ml of Chamberlain’s medium was added, and the mixture was shaken at 37 °C for 2 hrs at 250 rpm. The cells were harvested and resuspended in 1 ml of TSB-C medium (3% trypticase soy broth and 0.1% cysteine). Approximately 100 μl of the cell suspension was spread onto a TSB-C plate containing 10 μg/ml kanamycin. Genomic DNA was isolated from a kanamycin-resistant transformant, and the replacement of the F. novicida arnT gene with the kanamycin resistance gene was confirmed by DNA sequencing.
Isolation of Phospholipids and Lipid A from 32P Labeled Cells
F. novicida was grown in TSB-C medium, and E. coli was grown in LB broth (1% tryptone, 0.5% yeast extract and 1% NaCl). Typically, 20 ml cells, inoculated from an overnight culture to A600 = 0.02, were grown in the presence of 5 μCi/ml 32Pi to A600 of 1.2. The cells were collected by centrifugation and washed with phosphate-buffered saline (35). The cell pellets were resuspended in 3 ml of a single-phase Bligh-Dyer mixture (26) consisting of chloroform, methanol and water (1:2:0.8, v/v/v), incubated at room temperature for 60 min, and centrifuged to remove insoluble debris. The supernatant (containing mostly glycerophospholipids and the free lipid A) was removed and converted to a two-phase Bligh-Dyer system (26) by adding chloroform and water to generate a mixture consisting of chloroform, methanol and water (2:2:1.8, v/v/v). The insoluble debris pellets from the initial extractions (containing intact LPS, proteins and nucleic acids) were resuspended in 3 ml of 12.5 mM sodium acetate, pH 4.5, with 1% SDS, heated at 100 °C for 30 min (36), cooled to room temperature and then converted to a two-phase Bligh-Dyer system by the addition of chloroform and methanol.
The solvent phases from both sets of extractions were individually mixed and separated by low-speed centrifugation. The upper phases were washed once with a fresh pre-equilibrated Bligh-Dyer lower phases. The appropriate lower phases were pooled and dried under a stream of nitrogen. The 32P-labeled lipids were re-dissolved in chloroform and methanol (4:1, v/v) and spotted onto a Silica Gel 60 TLC plate (10, 000 cpm per lane), which was developed with chloroform, pyridine, 88% formic acid and water (50:50:16:5, v/v/v/v). After drying, the plates were exposed to a PhosphorImager Screen overnight, and the 32P-labeled lipids were detected with a Molecular Dynamics Storm PhosphorImager.
Large Scale Purification of Free Lipid A from F. novicida U112
Two liters of F. novicida U112 cells were grown at 37 °C in TSB-C, harvested by centrifugation, and washed with phosphate-buffered saline. About 10 g wet cells were obtained. The cells were extracted for 1 hr at room temperature with 1.8 l of a single-phase Bligh-Dyer mixture and centrifuged to remove insoluble debris. The initial supernatant (containing most of the glycerophospholipids and free lipid A) was converted to a two-phase Bligh-Dyer system. The debris pellet (containing LPS) was washed once with a single-phase Bligh-Dyer mixture and then resuspended in 360 ml of 12.5 mM sodium acetate, pH 4.5, containing 1% SDS (36). The suspension was heated in a boiling water bath for 30 min to release covalently linked lipid A from intact LPS (36). The cooled suspension was converted to a two-phase Bligh-Dyer mixture. The lower phases from the initial supernatant and the hydrolyzed LPS were dried separately by rotary evaporation. About 260 mg lipids were obtained from the initial supernatant, and 4 mg of lipids were obtained from the hydrolyzed debris pellets.
The dried lipids from the initial supernatant and the debris pellets were dissolved in chloroform, methanol and water (2:3:1, v/v/v). Each sample was applied to a 4 ml DEAE-cellulose column in the acetate form equilibrated with the same solvent (36, 37). The run-through was saved. Each column was washed with 10 column volumes of chloroform, methanol and water (2:3:1, v/v/v). The various lipid components were then eluted stepwise with 5 column volumes each of chloroform, methanol and ammonium acetate (2:3:1, v/v/v) with ammonium acetate concentrations of 60 mM, 120 mM, 240 mM, 360 mM and 480 mM (36). Fractions equal to one column volume were collected, and 20 μl of each fraction was spotted onto a TLC plate to monitor the lipid A and phospholipid elution profile. The plates were developed in the solvent of chloroform, methanol, acetic acid and water (25:15:4:4 v/v/v/v), and the lipids were visualized by spraying the plates with 10% sulfuric acid in ethanol, followed by charring. All F. novicida lipid A species were found to emerge in the DEAE cellulose run-through together with phosphatidylethanolamine and phosphatidylcholine, as judged by TLC and ESI/MS. The run-through fractions were converted to two-phase Bligh-Dyer mixtures. The lower phases were recovered and dried by rotary evaporation. At this stage, 100 mg lipids were recovered from the initial supernatant, and 2 mg were obtained from the hydrolyzed debris pellets.
To purify the lipid A species further, preparative thin layer chromatography was employed. Lipids obtained from the run-through fractions of the DEAE column were dissolved in chloroform, methanol (4:1,v/v) and applied to a TLC plate, which was developed with chloroform, methanol, pyridine, acetic acid, water (25:10:5:4:3, v/v/v/v/v). While the plates were drying, the lipid A bands could be seen transiently as white zones. These bands were marked with a pencil and scraped off after the plates were dry. The silica chips were extracted with a single-phase Bligh-Dyer mixture for 1 hour at room temperature. The suspension was centrifuged. The supernatant was passed through a small column fitted with a small glass wool plug and converted into a two-phase Bligh-Dyer system. The two phases were separated by centrifugation, and the lower phase was dried. About 9.8 mg of lipid A component A1 and 1.4 mg of component A2 were purified from the initial supernatant, but only ∼ 0.1 mg of A1 was recovered from the hydrolyzed debris pellets.
Lipid A species from the arnT knockout mutant were isolated by the same procedure described above, except that the cells were grown at 30 °C rather than 37 °C, and the lipid A was retained by the DEAE cellulose column, given the loss of the galactosamine substituent in the mutant.
To remove the ester-linked 3-hydroxyacyl chains of A1 and A2, the purified compounds were treated with 10% aqueous triethylamine for 4 hours at room temperature. The partially deacylated lipids, designated HA1 and HA2, were extracted from the reaction mixture by the Bligh-Dyer method and purified by preparative thin layer chromatography.
MALDI/TOF MS
Matrix-assisted laser desorption ionization time-of-flight (MALDI/TOF) mass spectra were acquired using an AXIMA-CFR from Kratos Analytical (Manchester, UK), equipped with a nitrogen laser (337 nm), 20 kV extraction voltage, and time delayed extraction. The samples were prepared for MALDI/TOF MS analysis by depositing 0.3 μl of the lipid sample dissolved in chloroform and methanol (4:1, v/v), followed by adding 0.3 μl of a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, v/v) as the matrix. The samples were left to dry at room temperature before the mass spectra were acquired in linear mode. Each spectrum was the average of 100 laser shots.
Electrospray Ionization (ESI) MS and MS/MS Analysis
All MS/MS spectra were acquired on a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (ABI/MDS-Sciex, Toronto, Canada) equipped with an ESI source. Lipid A samples were dissolved in chloroform and methanol (2:1, v/v) at about 25 μg/ml and subjected to ESI/MS in the negative ion mode (14). Nitrogen was used as collision gas for MS/MS experiments (14). Data acquisition and analysis were performed using the instrument’s Analyst QS software.
GC/MS Analysis
The purified lipid A species of F. novicida were hydrolyzed in acidic methanol, N-acetylated, and then converted to trimethylsilyl ethers. Mannose, galactose, glucosamine, galactosamine and glucose standards were processed in parallel with the lipid A samples. Typically, 0.5-1.0 mg sample was thoroughly dried in a Reacti-vial equipped with a Teflon-lined screw cap. Samples were hydrolyzed by adding 300 μl of 1 M HCl in methanol and heated at 80 °C for 15 hours. The reaction mixtures were then cooled, and the solvents were removed under a stream of nitrogen. Next, 200 μl of anhydrous methanol, 40 μl of pyridine, and 40 μl of acetic anhydride were added to the vial. The reaction mixtures were mixed and incubated overnight at room temperature. The solvents were evaporated under a stream of nitrogen. Finally, silylation of free OH groups was achieved by adding 200 μl of Tri-Sil reagent to the dried samples, mixing and incubating at room temperature for an hour. The samples were dried under a gentle stream of nitrogen, and re-dissolved in 100 μl of hexane and transferred to new vials for GC/MS analysis.
GC/MS was performed using a Finnigan Trace MS coupled with a Trace GC 2000 gas chromatography. The column is a 30 m RTX-5MS (0.25 mm internal diameter and 0.25 mm phase thickness) from Restek (Bellefonte, PA). The column oven temperature was held at 100 °C for 3 min, increased to 325 °C at a rate of 20 °C/min, and then held at 325 °C for 3 min. The injector was operated in the split mode (1:20 split), and the temperature of the injection port was kept at 200 °C. Helium was the carrier gas with a constant flow rate of 1 ml/min. The instrument was operated in the electron ionization mode with the electron energy set at 70 eV.
NMR spectroscopy
Typically, 1-2 mg samples of F. novicida lipid A were dissolved in 0.3 ml of CDCl3, CD3OD and D2O (2:3:1, v/v/v) in a 3 mm NMR tube. Proton and carbon chemical shifts are reported relative to internal tetramethylsilane (TMS) at 0.00 ppm. The 2H signal of CD3OD was used as a field frequency lock with the residual signal of CD3OD serving as the secondary reference at 49.5 ppm for carbon spectra. 1H NMR spectra were recorded at the Duke NMR Center on Varian Inova 800 or 600 NMR spectrometers, equipped with Varian cryogenic probes. 1H NMR spectra at 800 MHz were obtained with a 7.2 kHz spectral window, a 67° pulse field angle (4.5 μsec), a 4.5 sec acquisition time, and a 1 sec relaxation delay. The spectra were digitized using 64k points to obtain a digital resolution of 0.225 Hz/pt. Two-dimensional NMR experiments (COSY, NOESY, TOCSY, HMQC and HMBC) were performed at 800 MHz as previously described (38-40). Zero-quantum correlation (ZQCOSY) 2D NMR was coded into Varian NMR software based on Mueller’s sequence (41). 1H-decoupled 31P NMR spectra were recorded at 202.3 MHz on a Varian Inova 500 spectrometer with a spectral window of 12143.3 Hz digitized into 25,280 data points (digital resolution of 1Hz/point or ∼0.005 ppm/point), a 60° pulse flip angle (8 μs), and a 1.6-s repeat time. 31P chemical shifts were referenced to 85% H3PO4 at 0.00 ppm. Inverse decoupled difference spectra were recorded as 1H-detected 31P-decoupled heteronuclear NMR experiments, as previously described (38, 40).
RESULTS
Free Lipid A Among the Phospholipids of F. novicida
F. novicida U112 and E. coli W3110 were labeled uniformly with 32Pi until late log phase. Lipids were extracted directly from intact cells by the method of Bligh and Dyer, or after pH 4.5 hydrolysis at 100 °C of the intact LPS present in the Bligh-Dyer insoluble debris pellet, as previously described (36). Radioactive lipids were quantified by TLC and PhosphorImager analysis. E. coli phospholipids were recovered by the direct extraction (Fig. 2, lane 1), whereas E. coli lipid A was released from the insoluble debris pellet only after hydrolysis at pH 4.5 (36) (data not shown). Hydrolyzing the F. novicida debris pellet at pH 4.5 yielded very little lipid A (about 100 times less than from E. coli). However, several unknown radioactive substances were recovered together with the glycerophospholipids by direct Bligh-Dyer extraction of F. novicida cells (Fig. 2, lane 2). The rapidly-migrating lipids (Fig. 2, lane 2) are phosphatidylethanolamine, phosphatidylglycerol, cardiolipin and phosphatidylcholine, as judged by ESI/MS/MS (not shown). The slowly-migrating unknowns, designated compounds A1 and A2 (Fig. 2, lane 2), were purified on a larger scale and confirmed as lipid A derivatives by MS and NMR spectroscopy (see below). A1 and A2 make up 15 - 20% of the total 32P-labeled lipids recovered by direct extraction of intact F. novicida cells, as judged by PhosphorImager analysis. This is about the same ratio of lipid A to glycerophospholipids as is found in E. coli, except that E. coli lipid A must first be released from LPS by mild acid hydrolysis (11, 36). The fact that over 95 % of A1 and A2 is extracted directly from F. novicida with a single-phase Bligh-Dyer mixture demonstrates that F. novicida lipid A is not covalently linked to LPS. The amount of intact LPS present in F. novicida is low compared to E. coli, where it represents about 3.4 % of the dry weight (23). As in F. novicida, most of the lipid A in the F. tularensis LVS strain is in the free form (data not shown).
Figure 2. Phospholipids extracted from 32Pi labeled E. coli versus F. novicida U112.
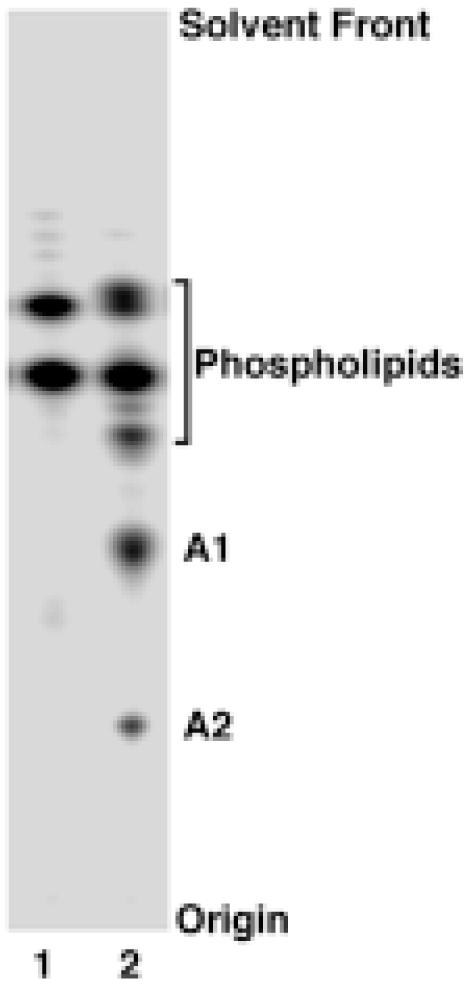
Lipids were extracted from uniformly labeled E. coli (lane 1) or F. novicida (lane 2) by the method of Bligh and Dyer (26) with the solvent chloroform, methanol and water (1:2:0.8, v/v/v). Lipids were separated by TLC using chloroform, pyridine, 88% formic acid and water (50:50:16:5, v/v/v/v) and visualized with a PhosphorImager. About 10,000 cpm were loaded. Glycerophospholipids were identified by migration with standards and by ESI/MS/MS (not shown). A1 and A2, which did not match known standards, are novel free lipid A molecules, as discussed in the text.
To purify larger amounts of A1 and A2, 2 l of F. novicida U112 were grown to late log phase. Lipids were extracted from intact cells with a single-phase Bligh-Dyer mixture or after pH 4.5 hydrolysis of the extracted, insoluble debris pellet. Portions of the crude lipids were separated on a TLC plate, which was sprayed with 10% sulfuric acid and charred on a hot plate. Consistent with the 32P-labeling, almost all the lipids were recovered by direct extraction of the intact cells, and only a small amount was recovered after hydrolysis of the debris pellet. The pattern of lipids on the charred plate (not shown) was virtually the same as that determined by 32P-labeling (Fig. 2, lane 2), showing that F. novicida does not synthesize lipid A species lacking phosphate groups, as is the case in Rhizobium etli and Rhizobium leguminosarum (39, 42, 43). Only compound A1 was observed amongst the lipids released by pH 4.5 hydrolysis of the debris pellet, indicating A1 as the sole hydrophobic anchor for the intact LPS in F. novicida cells.
The crude lipids extracted directly from the intact cells were subjected to anion exchange chromatography on a DEAE-cellulose column (36). A1 and A2 eluted together with phosphatidylethanolamine, lyso-phosphatidylethanolamine and phosphatidylcholine in the run-through, indicating that they possess no net negative charge. Compounds A1 and A2 were then further purified by preparative TLC (Fig. 3A). From 2 l of cells, about 9.8 mg of A1 and 1.4 mg of A2 were recovered by direct extraction, but only ∼ 0.1 mg of A1 was released from the Bligh-Dyer insoluble debris pellet by hydrolysis at pH 4.5.
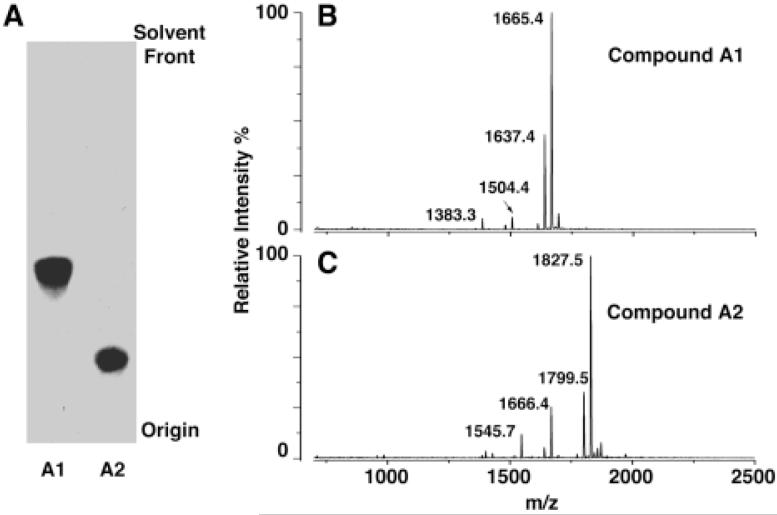
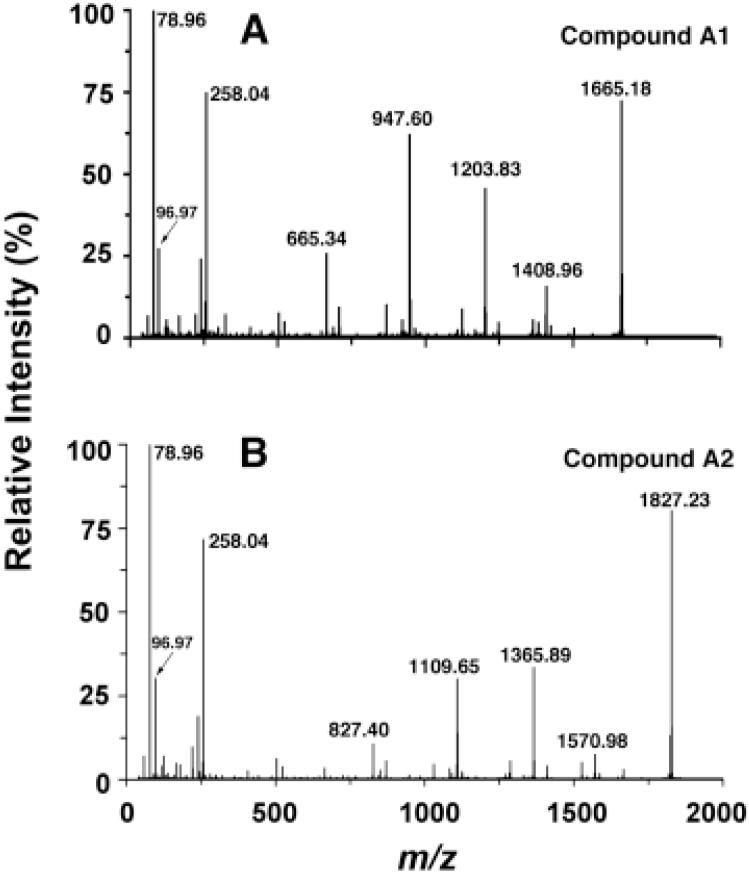
Figure 3. Purification and negative ion MALDI/TOF MS of A1 and A2.
Panel A. Compounds A1 and A2 were purified from a 2 l culture of F. novicida, as described in the Experimental Procedures. About 10 μg A1 and 5 μg A2 were loaded onto a TLC plate and separated using the solvent chloroform, methanol, acetic acid and water (25:15:4:4, v/v/v/v). Lipids were detected by spraying the plate with 10 % sulfuric acids in ethanol, followed by charring. Panel B. Negative ion MALDI/TOF mass spectrum of compound A1. Panel C. Negative ion MALDI/TOF mass spectrum of A2. Some fatty acid chain length heterogeneity and partial fragmentation is evident in both spectra.
Negative Ion MALDI/TOF MS of Compounds A1 and A2
A1 of F. novicida was analyzed by MALDI/TOF MS in the negative ion mode (Fig. 3B). The peaks at m/z 1665.4 and 1637.4 are interpreted as [M-H]- molecular ions, which differ by 28 amu, i.e. two methylene units. The ion at m/z 1504.4 may be derived from the base peak ion at m/z 1665.4 by loss of a 161 amu moiety, possibly a hexosamine unit (22). The m/z 1383.3 ion is derived from the m/z 1665.4 ion by loss of a 3-hydroxystearoyl group.
The negative ion MALDI/TOF mass spectrum of A2 (Fig. 3C) shows a similar pattern as A1. The peaks at m/z 1827.5 and 1799.5 could be interpreted as molecular ions [M-H]-, differing by 28 mass units. The peaks at m/z 1666.4 and 1545.7 might be derived from these parent ions by loss of a hexosamine or a 3-hydroxystearoyl moiety, respectively. All the peaks in the spectrum of A2 are 162 amu larger than the corresponding peaks of A1, suggesting that A2 contains an additional 162 amu substituent, possibly a hexose residue.
GC/MS Analysis of Compounds A1 and A2
The labile hexosamine unit, observed in the MALDI/TOF mass spectra of A1 and A2, could be attached to the 1-phosphate group, as proposed for lipid A of F. tularensis 1547-57 (Fig. 1D). To test this idea, A1 was hydrolyzed in 0.1 M HCl at 100 °C for 30 min. A hexosamine moiety attached to the 1-phosphate group of a lipid A molecule would be cleaved under these conditions. The hydrolysis mixture was then converted into a two-phase Bligh/Dyer system by the addition of chloroform and methanol. The phases were separated and dried. The materials in the dried upper and lower phases were hydrolyzed in acidic methanol, N-acetylated, converted to trimethylsilyl ethers and analyzed by GC/MS. Glucose, mannose, galactose, glucosamine and galactosamine standards were processed in parallel. The products recovered from the upper phase of the A1 hydrolysis mixture yielded peaks at 11.46 and 11.52 min, as did a galactosamine standard, confirming that the labile hexosamine unit of A1 is indeed galactosamine (data not shown). The products recovered from the lower phase yielded peaks at 10.59, 11.52 and 12.40 min (data not shown); their electron impact mass spectra were searched against the National Institute of Standards and Technology (NIST) library of mass spectra and identified as derivatives of palmitate, 3-hydroxypalmitate and 3-hydroxystearate, respectively. The presence of both 3-hydroxypalmitate and 3-hydroxystearate is consistent with the fatty acid chain length heterogeneity seen in the negative ion MALDI/TOF mass spectrum of A1 (Fig. 3B).
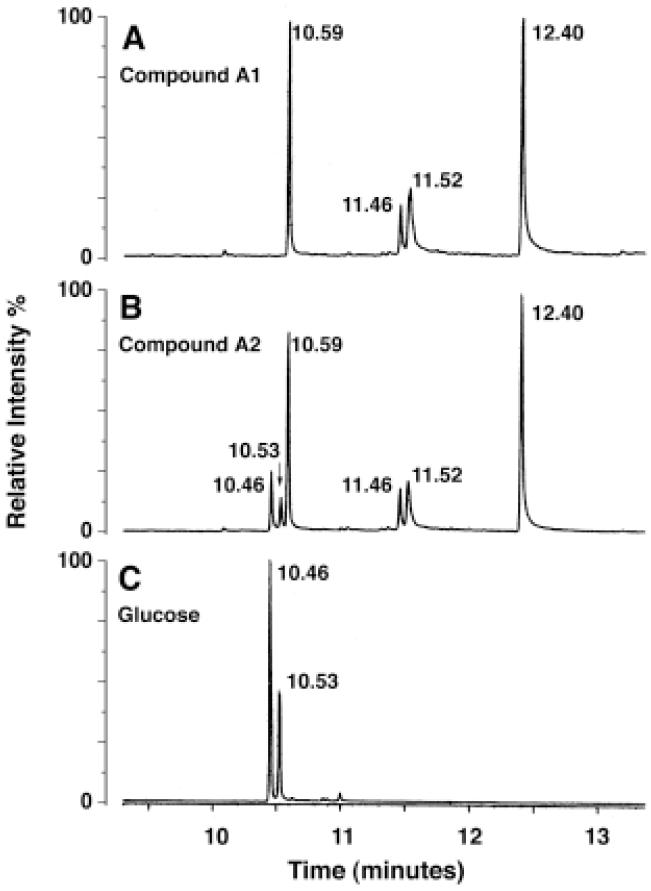
To characterize the additional hexose moiety present in A2, intact A1 and A2 were hydrolyzed in acidic methanol, N-acetylated, and converted to trimethylsilyl ethers, followed by GC/MS analysis (Fig. 4). Standards of glucose, mannose, galactose, glucosamine and galactosamine were processed in parallel. Peaks at 11.46 and 11.52 min were observed in the profiles of both A1 and A2, confirming that they both contain galactosamine. Peaks at 10.59, 11.52 and 12.40 min were observed with both A1 and A2 (Fig. 4A and B), the electron impact mass spectra of which matched with derivatives of palmitate, 3-hydroxypalmitate and 3-hydroxystearate, respectively. However, the peaks seen at 10.46 and 10.53 min in A2 (Fig. 4B) were not present in A1 (Fig. 4A), suggesting that they were derived from the additional hexose unit present in A2. Identical peaks at 10.46 and 10.53 min were seen with the glucose standard (Fig. 4C), but not with the other standards, showing that the additional hexose unit in A2 must be glucose.
Figure 4. GC/MS analysis of compounds A1 and A2.
The trimethysilyl ether derivatives of the N-acetylated methyl glycosides and the fatty acid methyl esters, obtained by methanol-HCl hydrolysis of compounds A1 (Panel A), A2 (Panel B) and a glucose standard (Panel C), were separated and detected by electron impact ionization MS.
ESI/MS/MS of Compounds A1 and A2
The molecular ions [M-H]- of A1 and A2 (at m/z 1665.18 and 1827.23, respectively) were further analyzed by high resolution ESI/MS/MS (Fig. 5). The peaks at m/z 78.96 [PO3]- and 96.97 [H2PO4]- confirm the presence of a phosphate group. The prominent peak at m/z 258.04, observed in both spectra (Fig. 5), has the ion mass expected for a phosphorylated hexosamine, suggesting the galactosamine moiety is connected to the phosphate group (Fig. 1D). Fragment ions derived from the parent ions by neutral loss of palmitic acid were observed at m/z 1408.96 for A1 and at m/z 1570.98 for A2 (Fig. 5), respectively. Fragment ions derived by neutral loss of both hydroxystearic acid and the hexosamine unit were observed at m/z 1203.83 for A1 and m/z 1365.89 for A2, and fragment ions arising from neutral loss of palmitic acid, hydroxystearic acid, and the hexosamine unit were observed at m/z 947.60 for A1 and m/z 1109.65 for A2. The further cleavage of an N-linked hydroxyacyl chain could generate the fragment ions at m/z 665.34 for A1 and m/z 827.40 for A2. Taken together with the GC/MS data (Fig. 4), the ESI/MS/MS spectra show that A1 and A2 are very similar in structure, both containing phosphate, galactosamine, palmitate and hydroxystearate residues. The phosphate and galactosamine units are connected to each other, as demonstrated by the peak at m/z 258.34.
Figure 5. Negative ion ESI/MS/MS analysis of A1 and A2.
Collision-induced dissociation mass spectra of [M-H]- at m/z 1665.18 (Panel A) and m/z 1827.33 (Panel B) were obtained using an ABI QSTAR-XL quadrupole time-of-flight tandem mass spectrometer (14).
Mild Alkaline Hydrolysis of A1 and A2
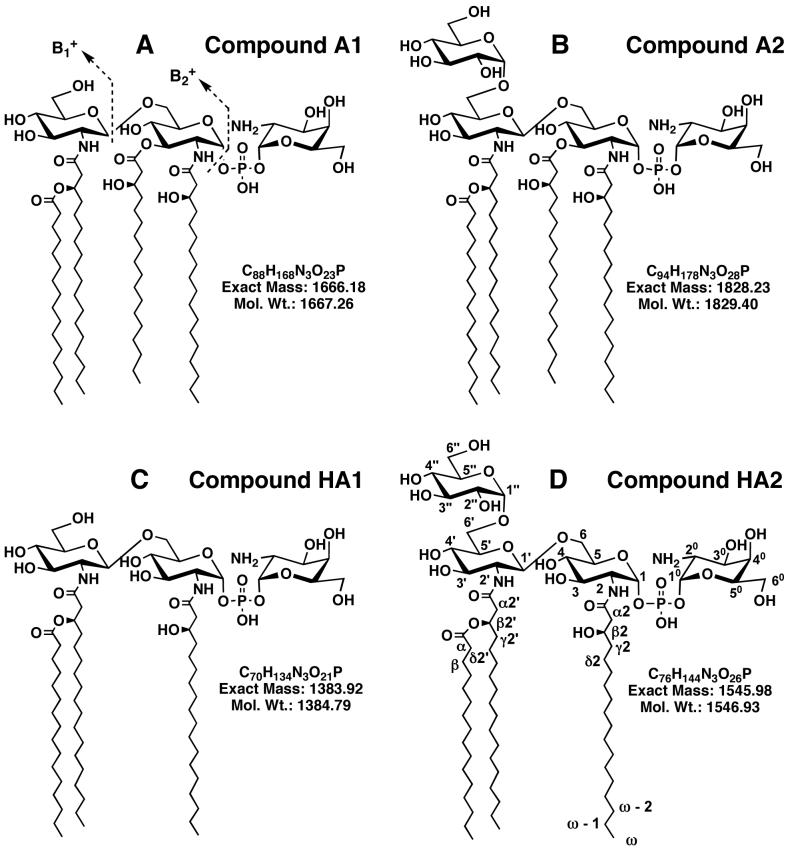
The ester-linked 3-hydroxyacyl chains of lipid A are very susceptible to mild alkaline hydrolysis (44). To determine the locations of the ester-linked fatty acids, A1 and A2 were treated with 10% aqueous triethylamine for 4 h at 25 °C. The products, HA1 and HA2, were purified and analyzed by negative ion MALDI/TOF MS. The molecular ions of HA1 and HA2 are each 282 amu smaller than those of A1 and A2, respectively (not shown), demonstrating that only one base-labile 3-hydroxystearoyl moiety is present in each compound. When analyzed by TLC in chloroform, pyridine, 88% formic acid and water (50:50:16:5, v/v/v/v), HA1 and HA2 migrate with Rfs that are similar to those of A1 and A2, respectively. These findings suggest that the base-labile 3-hydroxystearoyl moiety is attached to proximal glucosamine 3-position in both A1 and A2 (Fig. 1), as deacylation of the 3′-position usually causes a larger shift in the Rf than deacylation of the 3-position (44).
Positive Ion MALDI/TOF MS
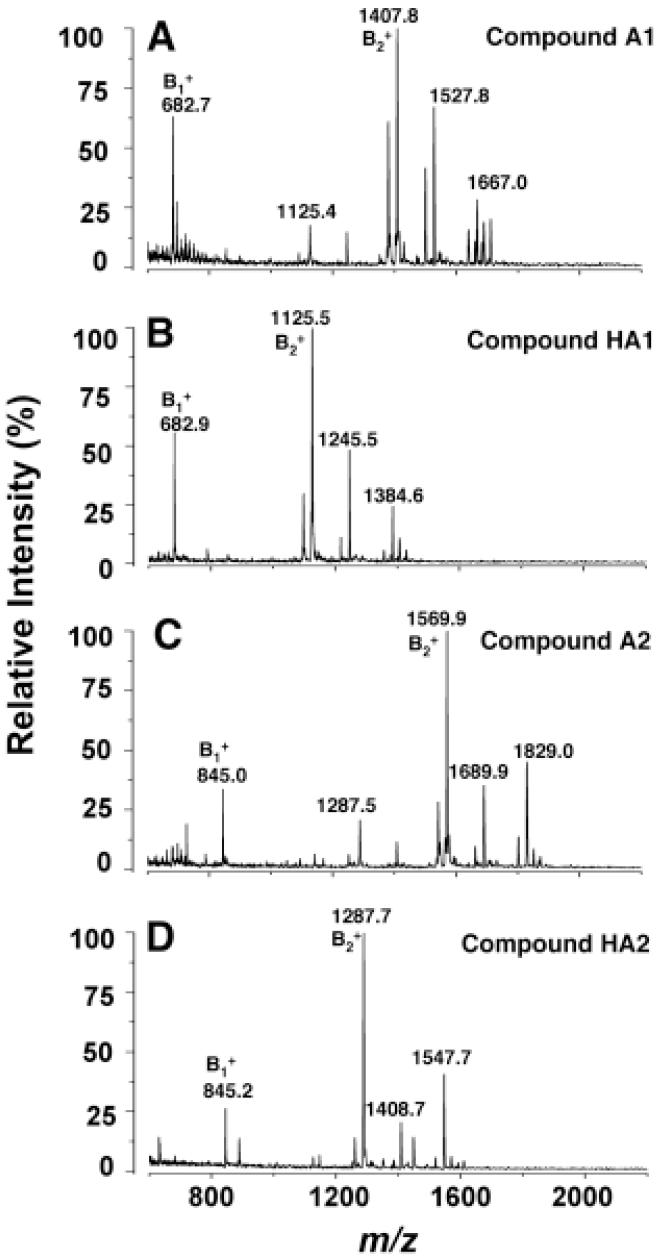
Compounds A1, HA1, A2 and HA2 were analyzed by positive ion MALDI/TOF MS (Fig. 6). Parent ions, interpreted as [M+H]+, were observed at m/z 1667.0 for A1, m/z 1384.6 for HA1, m/z 1829.0 for A2 and m/z 1547.7 for HA2 (Fig. 6). Variable amounts of sodium adducts [M+Na]+ were also detected (not labeled in Fig. 6). A prominent B1+ ion peak (45) near m/z 683, derived from the distal glucosamine unit, was present in the spectra of both A1 and HA1 (Fig. 6A and 6B), confirming that the ester-linked 3-hydroxystearoyl moiety is attached to the proximal glucosamine unit at position 3 in A1. Prominent B1+ ions near m/z 845.0 were observed for both A2 and HA2 (Fig 6C and 6D), demonstrating that the base-labile 3-hydroxystearoyl acid moiety is connected to the 3-position in A2 as in A1. However, the difference in the masses of the B1+ ions of A1 versus A2 or HA1 versus HA2 is 162 amu, showing that the additional glucose residue present in A2 is linked to the distal glucosamine unit.
Figure 6. Positive ion MALDI/TOF MS of A1, HA1, A2 and HA2.
Mild alkaline hydrolysis with triethylamine was used to generate HA1 and HA2 from A1 and A2, respectively (44). This procedure removes only the unsubstituted ester-linked 3-hydroacyl chains. The proposed structures of these compounds and the characteristic cleavages that give rise to the B1+ and B2+ ions are summarized in Fig. 7. Some fatty acid chain length heterogeneity was observed, as in the negative ion spectra (Fig. 3).
Neutral loss of the galactosamine moiety from the 1-phosphate group of each of the four compounds gave rise to product ions mainly as sodium adducts at m/z 1527.8 for A1, m/z 1245.5 for HA1, m/z 1689.9 for A2, m/z 1408.7 for HA2 (Fig. 6). The B2+ ions (45), which are derived from the parent ions by loss of the galactosamine 1-phosphate group, were observed at m/z 1407.8 for A1, m/z 1125.5 for HA1, m/z 1569.9 for A2, and m/z 1287.7 for HA2. The peaks at m/z 1125.4 in the A1 spectrum and at m/z 1287.5 in the A2 spectrum could arise by loss of the labile, ester-linked hydroxystearoyl moiety from the B2+ ion of each compound. As in the negative ion spectra (Fig. 4), some fatty acid chain length heterogeneity was observed in the positive ion spectra, though less on the distal unit, as judged by the masses of the B1+ ions (Fig. 6). The proposed structures of A1, HA1, A2 and HA2, which are based on the MS analysis together with the NMR data discussed below, are shown in Fig. 7.
Figure 7. Proposed structures of free lipid A species purified from F. novicida U112.
Compounds A1 (Panel A) and A2 (Panel B) are converted by mild alkaline hydrolysis with aqueous triethylamine to compounds HA1 (Panel C) and HA2 (Panel D) respectively. The anomeric configurations and the location of glucose residue at position 6′ in A2 and HA2 were determined by NMR spectroscopy, as discussed in the text. Positions are labeled according to the scheme shown in Panel D. The characteristic cleavages that give rise to the B1+ and B2+ ions during MS (45) are shown in Panel A.
1H and 31P NMR Analysis of Compounds A1, A2, HA1 and HA2
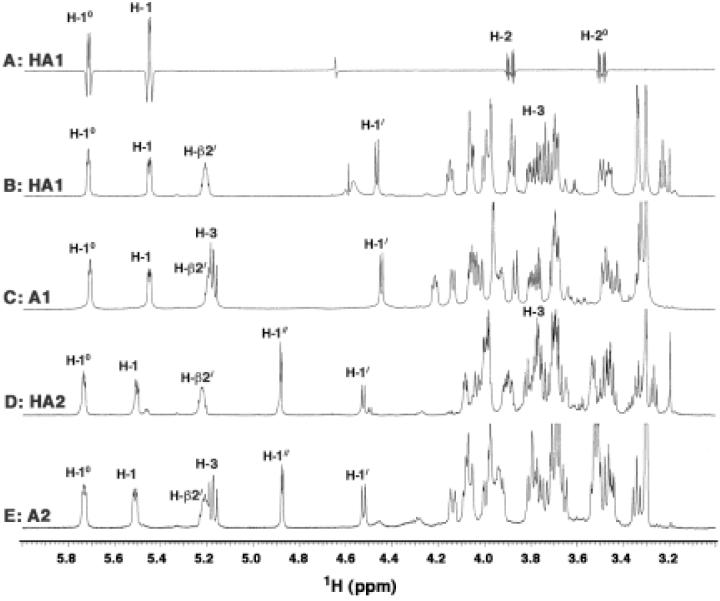
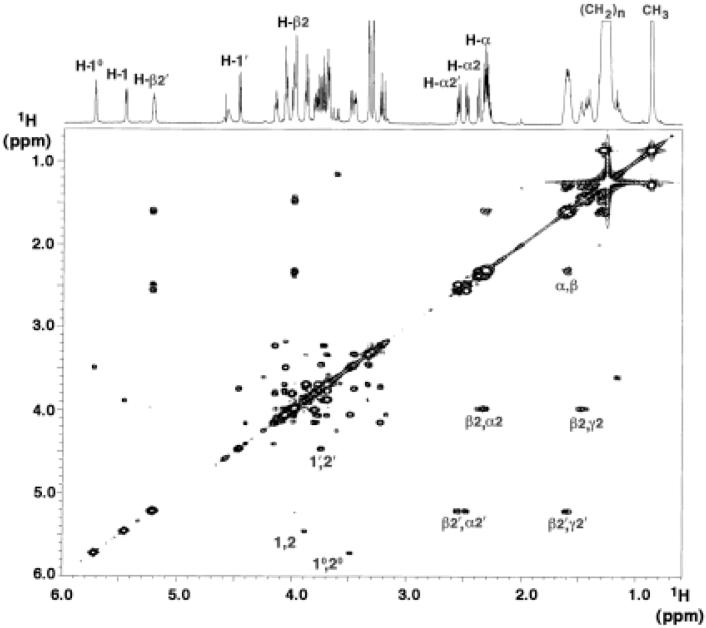
The 800 MHz 1H NMR spectra of compounds HA1, HA2, A1 and A2 dissolved in CDCl3, CD3OD and D2O (2:3:1, v/v/v) reveal relatively well-resolved resonances in the sugar (3.2-5.8 ppm) (Fig. 8) and acyl chain regions (0.8-2.7 ppm) (shown for HA1 in Fig. 9), similar to previous NMR spectra for E. coli and R. leguminosarum lipid A in the same solvent (38-40). The numbering scheme for the relevant protons is shown in Fig. 7D. The full assignments of the NMR chemical shifts and coupling constants for HA1 and HA2 (Table 1) were made from 2D COSY, ZQCOSY, TOCSY, HMQC and HMBC spectra.
Fig. 8. 1H NMR spectra of HA1, A1, HA2 and A2 and 31P-decoupled 1H NMR difference spectrum of HA1.
Panel A. Difference 500 MHz 1H NMR spectrum (38, 40) of F. novicida HA1 obtained by subtraction of 1H spectra obtained with selective on- and off-resonance decoupling of the 31P NMR signal at -2.18 ppm. The difference spectrum reveals the simultaneous appearance of doublets at 5.45 ppm (H-1) and 5.72 ppm (H-10) and double-doublets at 3.90 ppm (H-2) and 3.49 ppm (H-20). This pattern demonstrates that the single phosphorus group in compound HA1 is a monophosphodiester linkage that connects C-1 of the proximal glucosamine unit with C-10 of the capping galactosamine (Fig. 7). The observation of the H-2 and H-20 double-doublets in the difference spectrum allows estimates of the J1,2 and J2,3 coupling constants (Table 1). Panels B to E. Partial 1D 800 MHz 1H NMR spectra of compounds HA1, A1, HA2 and A2 respectively in CDCl3:CD3OD:D2O (2:3:1, v/v/v), highlighting key sugar and β-hydroxymethine proton resonances. The numbering scheme for each position is defined in Fig. 7 and Table 1.
Figure 9. An 800 MHz 1H-1H COSY analysis of compound HA1.
Key correlations within the sugar rings and acyl chains are labeled. The acyl chain cross-peaks distinguish the unsubstituted β-hydroxyacyl chain from the esterified β-hydroxyacyl (acyloxyacyl) chain. The numbering scheme for each position is defined in Fig. 7 and Table 1.
Table 1.
1H and 13C NMR Assignments of Compounds HA1 and HA2 from F. novicida
1H and 13C chemical shifts at 25 °C in CDCl3,CD3OD,D2O (2:3:1, v/v/v) are relative to an internal TMS standard and are assigned based on 2D NMR experiments as discussed in the text. The numbering scheme is shown in Fig. 7
| Position | HA1 | HA2 | HA1 | HA2 |
|---|---|---|---|---|
| δH [(mult), J(Hz)] | δH [(mult), J(Hz)] | δC [mult] | δC [mult] | |
| 10 | 5.72 [dd,3.5,5.4] | 5.74 [dd,3.6,5.8] | 93.5 [dd] | 93.3 [dd] |
| 20 | 3.49 [dt,3.3,10.7] | 3.53 [dt,3.3,10.7] | 52.4 [dd] | 52.3 [dd] |
| 30 | 4.06 [m] | 4.00 [m] | 67.0 [d] | 67.5 [d] |
| 40 | 3.98 [m] | 3.98 [m] | 69.3 [d] | 69.3 [d] |
| 50 | 4.07 [m] | 4.01 [m] | 73.0 [d] | 74.0 [d] |
| 6a0 | 3.77 [m] | 3.78 [m] | 62.6 [t] | 62.2 [t] |
| 6b0 | 3.69 [m] | 3.70 [m] | ||
| 1 | 5.46 [dd,3.3,7.4] | 5.51 [dd,3.1,7.9] | 94.0 [dd] | 94.8 [dd] |
| 2 | 3.90 [m] | 3.90 [m] | 54.5 [dd] | 54.7 [dd] |
| 3 | 3.72 [m] | 3.74 [m] | 70.5 [d] | 70.9 [d] |
| 4 | 3.23 [dd] | 3.2 [dd] | 71.5 [d] | 71.6 [d] |
| 5 | 4.15 [m] | 4.08 [m] | 73.4 [d] | 73.4 [d] |
| 6a | 4.00 [m] | 4.04 [m] | 72.0 [t] | 71.1 [t] |
| 6b | 3.80 [dd,7.3,12.0] | 3.80 [dd,7.3,12.0] | ||
| 1′ | 4.47 [d,8.6] | 4.54 [d,8.6] | 103.6 [d] | 103.1 [d] |
| 2′ | 3.75 [m] | 3.77 [m] | 56.0 [d] | 56.1 [d] |
| 3′ | 3.46 [m] | 3.49 [m] | 74.5 [d] | 75.2 [d] |
| 4′ | 3.33 [m] | 3.45 [m] | 70.7 [d] | 71.0 [d] |
| 5′ | 3.34 [m] | 3.54 [m] | 77.0 [d] | 75.6 [d] |
| 6′a | 3.88 [m] | 3.92 [m] | 61.3 [t] | 66.7 [t] |
| 6′b | 3.70 [m] | 3.77 [m] | ||
| 1” | 4.88 [d,3.7] | 98.8 [d] | ||
| 2” | 3.47 [m] | 72.9 [m] | ||
| 3” | 3.69 [m] | 74.5 [m] | ||
| 4” | 3.34 [m] | 71.4 [d] | ||
| 5” | 3.70 [m] | 72.9 [d] | ||
| 6”a | 3.82 [m] | 62.1 [t] | ||
| 6”b | 3.70 [m] | |||
| α2a | 2.39 [dd,3.1,14.2] | 2.39 [dd,3.1,14.2] | 44.2 [t] | 44.2 [t] |
| α2b | 2.35 [m] | 2.35 [m] | ||
| β2 | 3.98 [m] | 3.98 [m] | 69.0 [d] | 69.3 [d] |
| γ2 | 1.45 [m] | 1.45 [m] | 37.4 | |
| δ2 | 1.3 [m] | 1.3 [m] | ||
| CO2 | 175.1 | 174.8 | ||
| α2′a | 2.5 [dd,7.6,14.5] | 2.56 [dd,7.6,14.5] | 41.8 [t] | 41.6 [t] |
| α2′b | 2.49 [dd,5.6,14.5] | 2.49 [dd,5.6,14.5] | ||
| β2′ | 5.22 [m] | 5.23 [m] | 72.0 [d] | 71.8 [d] |
| γ2′ | 1.61 [m] | 1.61 [m] | 34.5 | |
| δ2′ | 1.3 | 1.3 | ||
| CO2′ | 173.4 | 173.2 | ||
| α CH2 | 2.31 [m] | 2.31 [m] | 35.2 [t] | 35.3 [t] |
| β CH2 | 1.61 [m] | 1.61 [m] | 25.5 | |
| γ CH2 | 1.3 | 1.3 | ||
| CO | 175.7 | 175.6 | ||
| (CH2)n | 1.24 [m] | 1.24 [m] | 30.4 [m] | 30.4 [m] |
| ω-2 CH2 | 1.26 [m] | 1.26 [m] | 32.7 [t] | 32.7 [t] |
| ω-1 CH2 | 1.26 [m] | 1.26 [m] | 23.4 [t] | 23.4 [t] |
| ω CH3 | 0.88 [t, ∼7] | 0.88 [t, ∼7] | 14.0 [q] | 14.5 [q] |
31P NMR spectroscopy at 202 MHz of HA1, HA2, A1 and A2 revealed a single 31P resonance around -2 ppm (not shown), indicating that each contains a single phosphorus atom. 1H-observed, 31P-decoupled experiments (38, 40) were implemented to identify the linkage site of the phosphate group on the glucosamine disaccharide backbone. The difference 1H NMR spectrum derived from selective on- and off-resonance decoupling of the -2.18 ppm 31P signal of HA1 revealed the simultaneous simplification of the H-1 and H-10 signals (at 5.46 and 5.72 ppm respectively) into two doublets, and the H-2 and H-20 resonances (at 3.90 and 3.49 ppm respectively) into two double-doublets (Fig. 8A). This indicates that the single phosphate group, present in HA1, bridges the proximal glucosamine and the capping galactosamine residue, consistent with the fragments ions seen by MS. Difference 1H NMR spectra confirmed that the single phosphate moiety present in HA2, A1 and A2 likewise connected the proximal glucosamine and capping galactosamine residue (not shown).
The 1D 1H NMR spectra of HA1 (Fig. 8B) and A1 (Fig. 8C) reveal three resolved anomeric resonances, consistent with the presence of three sugars. However, the NMR spectra of HA2 and A2 (Fig. 8D and E, respectively) show four resolved anomeric signals, consistent with the presence of the additional glucose moiety in A2 and HA2. The α CH2 signals of HA1 (Fig. 9) and HA2 (not shown) integrate to six protons, while the α CH2 resonances of A1 and A2 integrate to eight protons (not shown). The integration is consistent with the presence of three acyl chains in HA1 and HA2, and four acyl chains in A1 and A2, respectively (Fig. 7). All four lipids show one esterified β-hydroxymethine proton near 5.2 ppm (Fig. 8B, 8C, 8D and 8E), consistent with the presence of a single acyloxyacyl group (Fig. 7) (38, 39). The downfield shift of the H-3 signal to near 5.16 ppm in A1 and A2 (Fig. 8C and 8E) confirms the presence of an acyl chain at position 3 in these compounds (Fig. 7).
1H-1H COSY and NOESY Analysis
The anomeric 5.46 ppm double-doublet (H-1) and the 4.47 ppm doublet (H-1′) of compound HA1 are at similar shift positions to those of E. coli lipid A (38, 40). The low field position and double-doublet character of H-1 are typical of an α anomeric proton in a pyranose 1-phosphate (38-40). The anomeric double-doublet at 5.72 ppm (designated H-10) (Figs. 8 and 9) is not seen in E. coli lipid A (38). H-10 connects to six other sugar resonances (Table 1 and Fig. 9), consistent with the presence of a galactosamine residue. The low field position and double-doublet character of H-10 indicates that the 10-position is phosphorylated (Figs. 7-9). H-4′ of compound HA1 (3.33 ppm) (Table 1) resonates 0.8 ppm upfield of H-4′ of E. coli lipid A (4.17 ppm) (38), consistent with the proposal that the 4′-position of compound HA1 is not phosphorylated (Fig. 7). The presence of O-acyl chains of positions 3 and 3′ in E. coli lipid A (Fig. 1A) shifts the corresponding H-3 and H-3′ signals downfield to ∼5.2 ppm (38, 40). In contrast, the H-3 and H-3′ signals of compound HA1 appear in the sugar region at 3.72 and 3.46 ppm (Table 1), respectively, indicating that there is no O-acyl substituent at positions 3 and 3′ of HA1. This pattern is very similar in HA2 (Table 2).
The small J1,2, J10,20 and J1”,2” couplings (3.2 - 3.7 Hz) (Table 1) provide further evidence that the proximal glucosamine unit, the galactosamine residue and the glucose residue of HA2 (Fig. 7) are in the α-anomeric configuration. Likewise, the proximal glucosamine and galactosamine units of HA1 are α-anomers. The large J1′,2′ coupling (8.6 Hz) (Table 1) shows that the distal glucosamine unit of both HA1 and HA2 is in the β-configuration (Fig. 7). The NOE enhancements from H-1′ to H-6a and H-6b (and from H-1” to H6a′ and H6b′) in the NOESY spectrum of HA2 (Supplementary Fig. 1) provide independent evidence for the proposed 1′-6 and 1”-6′ glycosidic linkages in HA2 (Fig. 7D). The NOE enhancement from H-1′ to H6a and H6b is also seen with HA1 (data not shown). The NOE enhancement from H-1 to H-10 in both HA2 (Supplementary Fig. 1) and HA1 (not shown) confirms the proposed monophosphodiester linkage between the proximal glucosamine and the capping galactosamine residues (Fig. 7C and D).
Two sets of α/β cross-peak pairs are seen in the COSY (Fig. 9) and TOCSY (not shown) spectra of F. novicida compound HA1, indicating the presence of only two β-hydroxyacyl chains (Fig. 7C). The cross-peak pairs near 2.3 ppm and 4.0 ppm (designated α2/β2) arise from the α-protons adjacent to the β-oxymethine of the unsubstituted β-hydroxyacyl chain. The other α/β cross-peak pairs near 2.5 ppm and 5.2 ppm (designated α2′/β2′) are considerably more downfield, consistent with a 2′ acyloxyacyl moiety in HA1 (Fig. 7C). Our results are consistent with the acylation patterns deduced from previous NMR studies of the lipid A obtained from LPS of the LVS strain of F. tularensis (18).
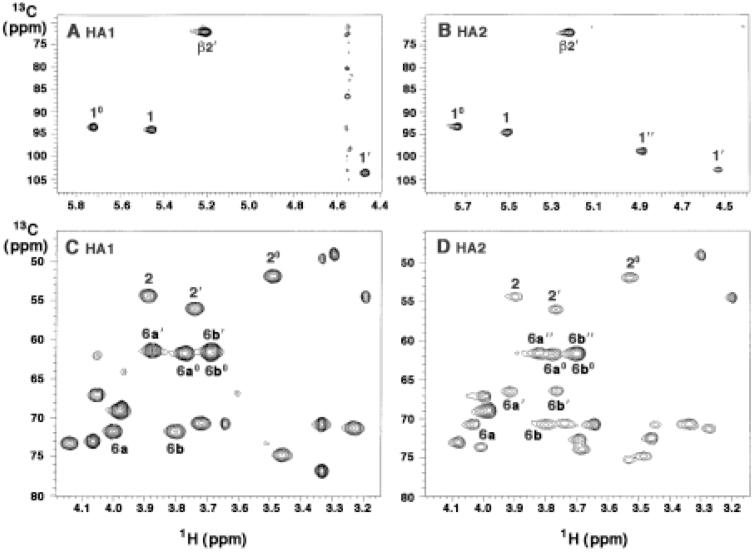
13C-NMR Analysis of HA1 and HA2
The HMQC spectra of HA1 and HA2 (Fig. 10) reveal the following key features: 1) The HMQC analysis confirms the presence of three anomeric protons in HA1 and four anomeric protons in HA2 (Fig. 10A and 10B). The C-10, C-1 and C-1′ chemical shift positions support the proposed α-, α- and β-configurations respectively for the capping galactosamine, proximal glucosamine and distal glucosamine units in both HA1 and HA2 (46). 2) Nitrogen-substituted carbons of amino sugars usually resonate around 52 to 54 ppm (46). The HMQC spectra of compounds HA1 and HA2 (Fig. 10C and 10D) reveal three methine resonances at 54.5 ppm (C-2), 56.0 ppm (C-2′) and 52.4 ppm (C-20), indicating that HA1 and HA2 contain three amino sugars (Fig. 7C and D). 3) The HMQC of HA1 (Fig. 10C) confirms the presence of a glycosylated C-6 (at 72 ppm) and a nonglycosylated C-6′ and C-60 (signals at 61.3 and 62 ppm, respectively) (46-48). However, in the HMQC of HA2 (Fig. 10D), C6′ shifts from a nonglycosylated position to a glycosylated position at 66.7 ppm, in agreement with the NOESY data (Supplementary Fig. 1), which likewise indicates that the glucose moiety present in HA2 is linked to the 6′-position.
Figure 10. Partial 800 MHz HMQC spectra of HA1 and HA2.
Panels A and B show the 1H/13C cross peaks in the anomeric region for HA1 and HA2 respectively. The numbering for each position is defined in Fig. 7 and Table 1. This analysis confirms the presence and anomeric linkage assignments of the three sugars in HA1 and four sugars in HA2. Panels C and D show the remaining sugar region of HA1 and HA2 respectively. The numbering scheme for the C-20, C-2 and C-2′ aminomethines (demonstrating the presence of three amino sugars in each compound) and for the C6 methylene cross peaks is shown in Fig. 7.
HA1 and HA2 gave excellent HMBC responses in both the sugar and acyl chain regions (Supplementary Fig. 2). Strong cross peaks from H-1” to C-6′ and from H-6a′ to C-1” were observed in the HMBC of compound HA2 (Supplementary Fig. 2B), thus confirming the attachment of the glucose unit to the 6′ position of HA2 (Fig. 7D). Linkage of a glucose residue to position 6′ is without precedent in lipid A biochemistry, as Kdo is normally attached at this important site.
Origin of the Galactosamine Group in Francisella Lipid A
d-galactosamine is similar in structure and charge to 4-amino-4-deoxy-l-arabinose (Fig. 1D). The latter is synthesized and attached to lipid A in polymyxin-resistant mutants of E. coli and Salmonella (49, 50). The inner membrane enzyme ArnT transfers the 4-amino-4-deoxy-l-arabinose residue from the donor substrate undecaprenyl phosphate-4-amino-4-deoxy-l-arabinose to the 1 and/or 4′ phosphate groups of lipid A (27, 28). We reasoned that a membrane protein with sequence similarity to ArnT might be responsible for the attachment of the galactosamine residue to lipid A in F. novicida. A deletion mutant of the single arnT orthologue present in F. novicida (30) synthesizes lipid A molecules lacking the galactosamine moiety found in the wild-type, as judged by the loss of the 161 amu substituent (Fig. 11A versus 11B). Loss of the galactosamine modification causes the cells to grow more slowly above 30 °C and is associated with a shift to shorter fatty acyl chains on lipid A (more of the molecular species at m/z 1476.014 versus m/z 1504.045) (Fig. 11B). Interestingly, the structure of lipid A present in this arnT deletion mutant of F. novicida is nearly identical to that reported by Phillips et al. for the LVS strain of F. tularensis (Fig. 1C) (22).
Figure 11. Absence of the galactosamine substituent on lipid A in the arnT mutant of F. novicida.
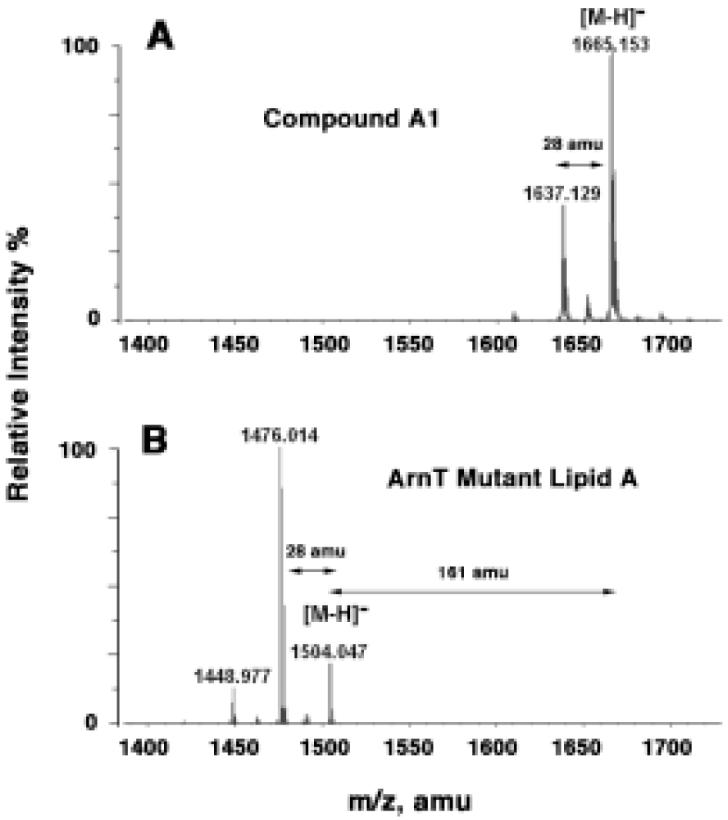
Panel A. Negative ion ESI mass spectrum of compound A1 from wild-type F. novicida U112. Panel B. Negative ion ESI mass spectrum of the major free lipid A species isolated from the arnT knockout of F. novicida. For ESI/MS, lipids were dissolved in chloroform and methanol (2:1, v/v) at about 25 μg/ml.
DISCUSSION
Virtually all Gram-negative bacteria, including sequenced strains of Francisella, encode orthologues of the enzymes that assemble Kdo2-lipid A (3, 30), the outer membrane lipid to which the other core and O-antigen sugars of LPS are attached. It is therefore remarkable that over 95 % of the lipid A present in F. novicida is recovered in the “free” form, i.e. lacking the usual Kdo, core and O-antigen sugars. Because it is not extensively glycosylated, F. novicida lipid A is extracted together with the glycerophospholipids by the method of Bligh and Dyer. Small amounts of LPS are indeed detectable in F. novicida, as well as other strains of Francisella (18, 22), and their lipid A has been partially characterized by other investigators (Fig. 1). However, the possibility that Francisella lipid A was mostly present in the “free” form was not considered (17, 18, 22), and the glycerophospholipid fraction was not examined.1 As shown in Fig. 2, compounds A1 and A2 make up about 15 % of the total phospholipid, the same ratio of lipid A to glycerophospholipids found in E. coli (11). The key distinction is that all the lipid A of E. coli is covalently linked to LPS and can only be released by mild acid hydrolysis at 100 °C (11, 36). In wild-type E. coli, none of the lipid A is present in the “free” form (36), as is also the case in all other, well-characterized Gram-negative bacteria.
Component A1 (Fig. 7) of the free lipid A in F. novicida has the same structure as proposed by Phillips et al. for the lipid A moiety covalently linked to LPS in F. tularensis 1547-57 (Fig. 1D) (22). Compound A2 (Fig. 7) was not reported by Phillips et al., consistent with our observation that it is not present in the lipid A released from F. novicida LPS by mild acid hydrolysis (data not shown). Given the unprecedented glucose substituent linked to the 6′ position of A2, attachment of the Kdo and core sugars would be precluded (3). The GC/MS analysis (Fig. 6) validates the presence of galactosamine in both A1 and A2, and an additional glucose substituent in the latter. Our NMR data (Figs. 8-10 and Supplementary Figs.) unequivocally demonstrate the alpha anomeric configurations for the galactosamine, the glucose and the proximal glucosamine units (Fig. 7). The axial disposition of the phosphate group attached to the galactosamine moiety of A1 and A2 is the same as that seen with the 4-amino-4-deoxy-l-arabinose substituent (Fig. 1D, inset), which is attached to lipid A in polymyxin-resistant mutants of E. coli and Salmonella (40, 50). Both galactosamine and 4-amino-4-deoxy-l-arabinose may neutralize the negative charge of the proximal phosphate group, thereby conferring resistance to cationic anti-microbial peptides.
The presence of all the genes encoding the enzymes that synthesize Kdo2-lipid A (30), in conjunction with the proposed structures of A1 and A2, raises the possibility that Francisella cells first assemble conventional core-lipid A molecules on the inner surface of their inner membrane (Scheme 1), and then dismantle and modify them, perhaps after they are flipped to the periplasmic surface of the inner membrane by the ABC transporter MsbA (51, 52). A Kdo hydrolase (Scheme 1) that might initiate this process is present in F. novicida (15). The active site of the 4′-phosphatase (Scheme 1), encoded by lpxF, is located on the outer surface of the inner membrane (29). Mutants of F. novicida lacking the 4′-phosphatase2 synthesize free lipid A molecules containing the 4′-phosphate residue, demonstrating that LpxF alone accounts for its removal in living cells. How the 3′ acyl chain is removed (Scheme 1) to generate A1 and A2 remains unclear. In Salmonella, the 3′ acyl chain can be cleaved by the outer membrane enzyme LpxR (53), but an LpxR orthologue sequence is not present in the Francisella genome. The absence of the 4′-phosphate and 3′-hydroxyacyl group in Francisella lipid A (Figs. 1 and 7), in conjunction with the relatively long acyl chains, may account for the low bioactivity of Francisella LPS (24, 25).
The Francisella genome (30) encodes one orthologue of ArnT, the enzyme that uses an undecaprenyl phosphate linked donor substrate to transfer 4-amino-4-deoxy-l-arabinose to lipid A on the outer surface of the inner membrane in E. coli and Salmonella (27, 28). Given the structural similarity of galactosamine and 4-amino-4-deoxy-l-arabinose (Fig. 1D), we inactivated the F. novicida arnT gene and examined the structure of its lipid A. As shown in Fig. 11, the galactosamine residue is absent in the lipid A of the F. novicida arnT mutant. In fact, this construct synthesizes free lipid A molecules with the structure proposed for LVS lipid A by Phillips et al. (Fig. 1C) (22), suggesting that the LVS strain may also harbor a mutation in its arnT gene. As in F. novicida, most of the lipid A in the LVS strain is recovered in the free form (data not shown). The absence of the galactosamine moiety might sensitize the LVS strain to some cationinc anti-microbial peptides. The identification of the endogenous galactosamine donor, possibly a polyisoprene phosphate derivative by analogy to the 4-amino-4-deoxy-l-arabinose pathway (28), and the development of an in vitro assay for F. novicida ArnT (Scheme 1) should shed light on the mechanism of galactosamine transfer to lipid A. An in vitro assay for glucose addition to the distal residue to make A2 (Fig. 7) also remains to be developed.
Given that F. novicida contains a distinct lipid A 1-phosphatase, encoded by LpxE (15), it is surprising that all the lipid A molecules present in our arnT deletion mutant retained their 1-phosphate group (Fig. 11). Heterologous expression of lpxE in E. coli removes over 90 % of the lipid A 1-phosphate moiety (15). However, LpxE, unlike LpxF, requires the presence of the Kdo disaccharide for catalytic activity (15, 29). If the rate of Kdo removal by the Francisella Kdo hydrolase (15) is much faster than the rate of cleavage of the 1-phosphate group by LpxE, one might not expect to see much lipid A 1-dephosphorylation. Although easily assayed in vitro (15), the gene encoding the F. novicida Kdo hydrolase and its sub-cellular localization are unknown. A Kdo hydrolase has also recently been reported to be present in strains of Helicobacter, but the gene encoding this enzyme is unknown (54).
An alternative explanation for the failure of LpxE to cleave the 1-phosphate group is the possibility that MsbA exports nascent lipid A more rapidly than it can be glycosylated by KdtA (3) in F. novicida. Accordingly, the active site of LpxE, which faces the outer surface of the inner membrane (15), would not be presented with its requisite substrate. If this scenario is correct, it should be possible to delete kdtA in F. novicida (55). This idea is consistent with the recent discovery that over-expression of MsbA in E. coli bypasses the normal requirement for Kdo biosynthesis (56). In such constructs, only the tetra-acylated lipid A precursor, lipid IVA, is synthesized (56) and then flipped to the outer surface of the inner membrane by the excess MsbA (56). Consequently, no core domain is incorporated, as is the case with compounds A1 and A2 (Fig. 7) of F. novicida. Whatever the explanation, some kind of lipid A molecule is nevertheless required for the growth of F. novicida, because it is sensitive to the LpxC inhibitor CHIR-090, which blocks the second step of lipid A biosynthesis (57).
Further elucidation of the enzymology and genetics of lipid A modification in Francisella (Scheme 1) may prove useful for the development of effective vaccines, which are not currently available for this important pathogen (16). As noted above, F. novicida 4′ phosphatase mutants synthesize free lipid A molecules containing the 4′-phosphate moiety.2 Such mutants are hypersensitive to polymyxin, are greatly attenuated in virulence in a mouse infection model2, and might provide protection against tularemia.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Francis Nano (University of Victoria, British Columbia, Canada) for providing us with F. novicida U112.
Funding: This research was supported by NIH grants R37-GM-51796 to C. R. H. Raetz, GM-54882 to R. J. Cotter, and NCI P30-CA-14236 to A. A. Ribeiro at the Duke NMR Spectroscopy Center. The mass spectrometry facility in the Department of Biochemistry of the Duke University Medical Center and Dr. Z. Guan were supported by the LIPID MAPS Large Scale Collaborative Grant number GM-069338 from NIH.
ABBREVIATIONS
- COSY
correlation spectroscopy
- ESI/MS
electrospray ionization/mass spectrometry
- GC/MS
gas chromatography/mass spectrometry
- HMQC
heteronuclear multiple quantum coherence spectroscopy
- HMBC
heteronuclear multiple bond coherence spectroscopy
- Kdo
3-deoxy-D-manno-octulosonic acid
- LPS
lipopolysaccharide
- MALDI/TOF
matrix-assisted laser desorption ionization/time-of-flight
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- NOESY
nuclear Overhauser effect spectroscopy
- TLC
thin layer chromatography
- TOCSY
total correlation spectroscopy
Footnotes
Vinogradov et al. (18) noted the presence of a “minor amount”... of “free lipid A” in their smooth LPS preparation but did not examine the phospholipid fraction.
X. Wang, A. A. Ribeiro, Z. Guan, S. N. Abraham and C. R. H. Raetz, in preparation.
SUPPORTING INFORMATION AVAILABLE
The NOESY analysis of HA2 and the HMBC spectra of HA1 and HA2 are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Raetz CRH. Biochemistry of endotoxins. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- (2).Brade H, Opal SM, Vogel SN, Morrison DC. Endotoxin in Health and Disease. Marcel Dekker, Inc.; New York: 1999. p. 950. [Google Scholar]
- (3).Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- (5).Gangloff M, Gay NJ. MD-2: the Toll ‘gatekeeper’ in endotoxin signalling. Trends Biochem. Sci. 2004;29:294–300. doi: 10.1016/j.tibs.2004.04.008. [DOI] [PubMed] [Google Scholar]
- (6).Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- (7).Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- (8).Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- (9).Lien E, Ingalls RR. Toll-like receptors. Crit. Care Med. 2002;30:S1–11. [PubMed] [Google Scholar]
- (10).Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- (11).Galloway SM, Raetz CRH. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- (12).Raetz CRH. Molecular genetics of membrane phospholipid synthesis. Annu. Rev. Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- (13).Guan Z, Breazeale SD, Raetz CRH. Extraction and identification by mass spectrometry of undecaprenyl diphosphate-MurNAc-pentapeptide-GlcNAc from Escherichia coli. Anal. Biochem. 2005;345:336–339. doi: 10.1016/j.ab.2005.07.002. [DOI] [PubMed] [Google Scholar]
- (14).Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr., Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- (15).Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin. Microbiol. Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further application of the procedure. Methods Carbohydr. Chem. 1965;5:83–91. [Google Scholar]
- (18).Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- (19).Prior JL, Prior RG, Hitchen PG, Diaper H, Griffin KF, Morris HR, Dell A, Titball RW. Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J. Med. Microbiol. 2003;52:845–851. doi: 10.1099/jmm.0.05184-0. [DOI] [PubMed] [Google Scholar]
- (20).Vinogradov E, Perry MB. Characterisation of the core part of the lipopolysaccharide O-antigen of Francisella novicida (U112) Carbohydr. Res. 2004;339:1643–1648. doi: 10.1016/j.carres.2004.04.013. [DOI] [PubMed] [Google Scholar]
- (21).Vinogradov E, Conlan WJ, Gunn JS, Perry MB. Characterization of the lipopolysaccharide O-antigen of Francisella novicida (U112) Carbohydr. Res. 2004;339:649–654. doi: 10.1016/j.carres.2003.12.013. [DOI] [PubMed] [Google Scholar]
- (22).Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Novel modification of lipid A of Francisella tularensis. Infect. Immun. 2004;72:5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Neidhardt FC. Chemical composition of Escherichia coli. In: Neidhardt F, editor. Escherichia coli and Salmonella typhimurium. I. ASM Publications; Washington, D. C: 1987. pp. 3–6. [Google Scholar]
- (24).Sandstrom G, Sjostedt A, Johansson T, Kuoppa K, Williams JC. Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol. Immunol. 1992;5:201–210. doi: 10.1111/j.1574-6968.1992.tb05902.x. [DOI] [PubMed] [Google Scholar]
- (25).Ancuta P, Pedron T, Girard R, Sandstrom G, Chaby R. Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect. Immun. 1996;64:2041–2046. doi: 10.1128/iai.64.6.2041-2046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bligh EG, Dyer JJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- (27).Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. An inner membrane enzyme in Salmonella typhimurium and Escherichia coli that transfers 4-amino-4deoxy-l-arabinose to lipid A. Induction in polymyxin resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- (28).Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CRH. Accumulation of a polyisoprene-linked amino sugar in polymyxin resistant mutants in Salmonella typhimurium and Escherichia coli. Structural characterization and possible transfer to lipid A in the periplasm. J. Biol. Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- (29).Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4’-phosphatase LpxF. J. Biol. Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, Karp PD, Larsson E, Liu Y, Michell S, Prior J, Prior R, Malfatti S, Sjostedt A, Svensson K, Thompson N, Vergez L, Wagg JK, Wren BW, Lindler LE, Andersson SG, Forsman M, Titball RW. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- (31).Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- (32).Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chamberlain RE. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tyeryar FJ, Lawton WD. Factors affecting transformation of Pasteurella novicida. J. Bacteriol. 1970;104:1312–1317. doi: 10.1128/jb.104.3.1312-1317.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Dulbecco R, Vogt M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhou Z, Lin S, Cotter RJ, Raetz CRH. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- (37).Raetz CRH, Kennedy EP. Function of cytidine diphosphate-diglyceride and deoxycytidine diphosphate-diglyceride in the biogenesis of membrane lipids in Escherichia coli. J. Biol. Chem. 1973;248:1098–1105. [PubMed] [Google Scholar]
- (38).Ribeiro AA, Zhou Z, Raetz CRH. Multi-dimensional NMR structural analyses of purified lipid X and lipid A (endotoxin) Magn. Res. Chem. 1999;37:620–630. [Google Scholar]
- (39).Que NLS, Ribeiro AA, Raetz CRH. Two-dimensional NMR spectroscopy and structures of six lipid A species from Rhizobium etli CE3; detection of an acyloxyacyl residue in each component and origin of the aminogluconate moiety. J. Biol. Chem. 2000;275:28017–28027. doi: 10.1074/jbc.M004009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhou Z, Ribeiro AA, Raetz CRH. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-l-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 2000;275:13542–13551. doi: 10.1074/jbc.275.18.13542. [DOI] [PubMed] [Google Scholar]
- (41).Mueller L. Mapping of spin-spin coupling via zero-quantum coherence. J. Mag. Res. 1984;59:326–331. [Google Scholar]
- (42).Bhat UR, Forsberg LS, Carlson RW. The structure of the lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. A unique, non-phosphorylated lipid A containing 2-amino-2-deoxy-gluconate, galacturonate, and glucosamine. J. Biol. Chem. 1994;269:14402–14410. [PubMed] [Google Scholar]
- (43).Que NLS, Lin S, Cotter RJ, Raetz CRH. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli; demonstration of a conserved distal and a variable proximal portion. J. Biol. Chem. 2000;275:28006–28016. doi: 10.1074/jbc.M004008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Basu SS, White KA, Que NL, Raetz CRH. A deacylase in Rhizobium leguminosarum membranes that cleaves the 3-O-linked beta-hydroxymyristoyl moiety of lipid A precursors. J. Biol. Chem. 1999;274:11150–11158. doi: 10.1074/jbc.274.16.11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Costello CE, Vath JE. Tandem mass spectrometry of glycolipids. Methods in Enzymology. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- (46).Agrawal PK. NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry. 1992;31:3307–3330. doi: 10.1016/0031-9422(92)83678-r. [DOI] [PubMed] [Google Scholar]
- (47).Agrawal PK, Bush CA, Qureshi N, Takayama K. Structural analysis of lipid A and Re-lipopolysaccharides by NMR spectroscopic methods. Adv. Biophys. Chem. 1994;4:179–236. [Google Scholar]
- (48).van Halbeek H. In: Encyclopedia of NMR. Grant DM, Harris RK, editors. Wiley; Chichester: 1996. pp. 1107–1137. [Google Scholar]
- (49).Nummila K, Kilpeläinen I, Zähringer U, Vaara M, Helander IM. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 1995;16:271–278. doi: 10.1111/j.1365-2958.1995.tb02299.x. [DOI] [PubMed] [Google Scholar]
- (50).Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CRH. Lipid A modifications in polymyxin resistant Salmonella typhimurium. PmrA dependent 4-amino-4-deoxy-l-arabinose and phosphoethanolamine addition. J. Biol. Chem. 2001;276:43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- (51).Doerrler WT, Reedy MC, Raetz CRH. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- (52).Doerrler WT, Gibbons HS, Raetz CRH. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- (53).Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Stead C, Tran A, Ferguson D, Jr., Cotter R, McGrath S, Trent S. A novel 3-deoxy-D-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J. Bacteriol. 2005;187:3374–3383. doi: 10.1128/JB.187.10.3374-3383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Belunis CJ, Clementz T, Carty SM, Raetz CRH. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 1995;270:27646–27652. doi: 10.1074/jbc.270.46.27646. [DOI] [PubMed] [Google Scholar]
- (56).Meredith TC, Aggarwal P, Mamat U, Lindner B, Woodard RW. Redefining the Requisite Lipopolysaccharide Structure in Escherichia coli. ACS Chem. Biol. 2006;1:33–42. doi: 10.1021/cb0500015. [DOI] [PubMed] [Google Scholar]
- (57).McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CRH. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry. 2005;44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.