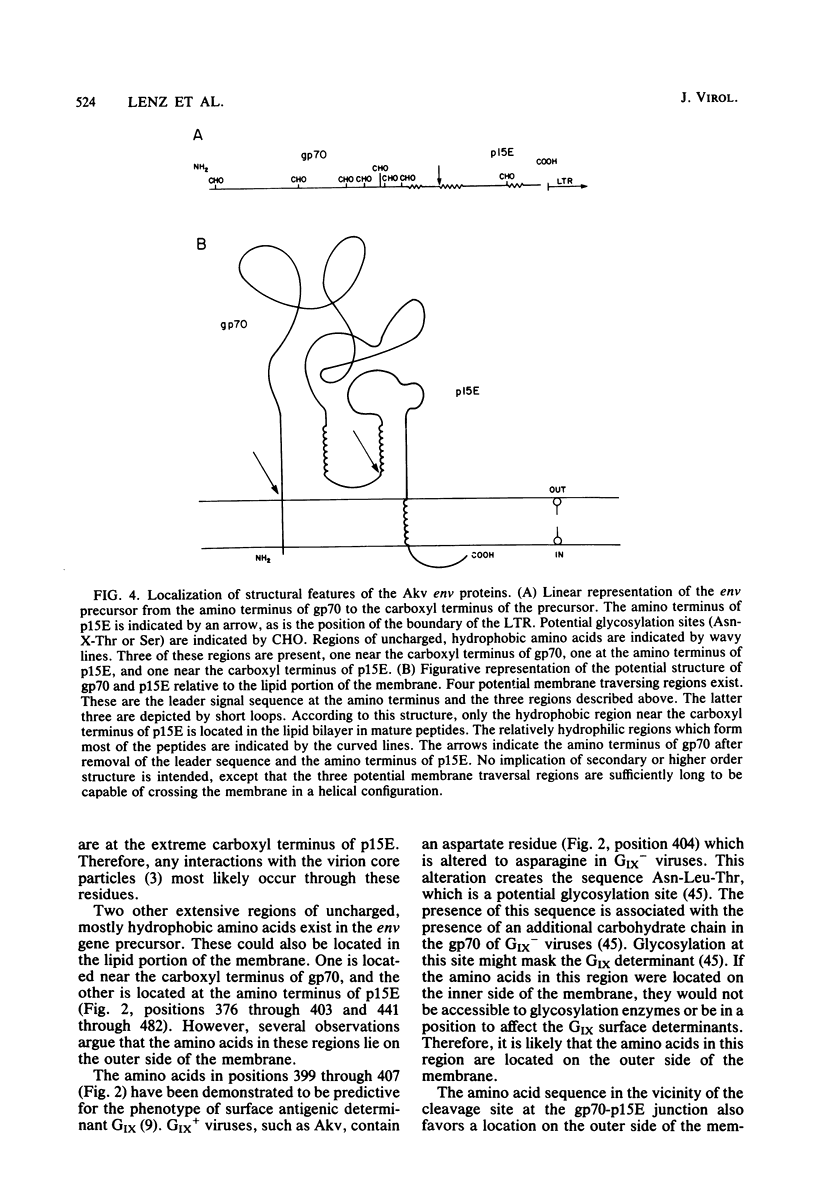
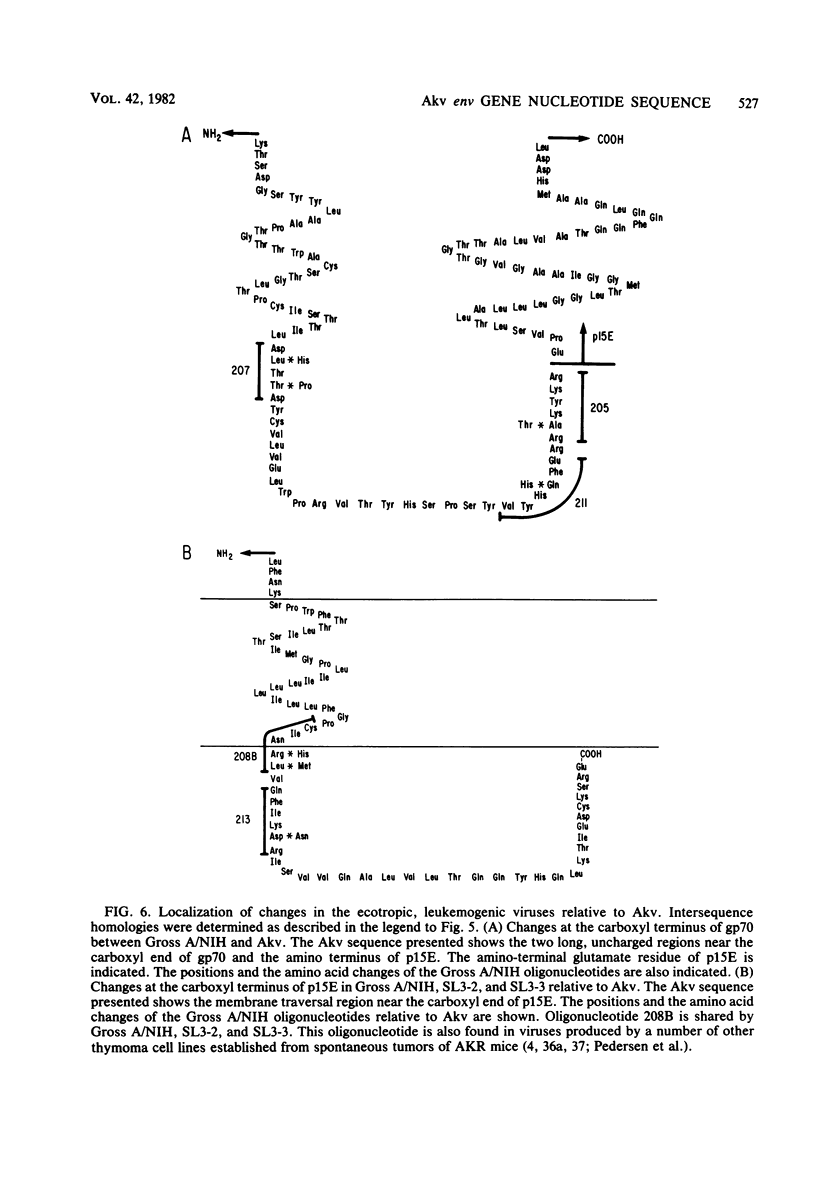
Abstract
The sequence of 2,191 nucleotides encoding the env gene of murine retrovirus Akv was determined by using a molecular clone of the Akv provirus. Deduction of the encoded amino acid sequence showed that a single open reading frame encodes a 638-amino acid precursor to gp70 and p15E. In addition, there is a typical leader sequence preceding the amino terminus of gp70. The locations of potential glycosylation sites and other structural features indicate that the entire gp70 molecule and most of p15E are located on the outer side of the membrane. Internal cleavage of the env precursor to generate gp70 and p15E occurs immediately adjacent to several basic amino acids at the carboxyl terminus of gp70. This cleavage generates a region of 42 uncharged, relatively hydrophobic amino acids at the amino terminus of p15E, which is located in a position analogous to the hydrophobic membrane fusion sequence of influenza virus hemagglutinin. The mature polypeptides are predicted to associate with the membrane via a region of 30 uncharged, mostly hydrophobic amino acids located near the carboxyl terminus of p15E. Distal to this membrane association region is a sequence of 35 amino acids at the carboxyl terminus of the env precursor, which is predicted to be located on the inner side of the membrane. By analogy to Moloney murine leukemia virus, a proteolytic cleavage in this region removes the terminal 19 amino acids, thus generating the carboxyl terminus of p15E. This leaves 15 amino acids at the carboxyl terminus of p15E on the inner side of the membrane in a position to interact with virion cores during budding. The precise location and order of the large RNase T1-resistant oligonucleotides in the env region were determined and compared with those from several leukemogenic viruses of AKR origin. This permitted a determination of how the differences in the leukemogenic viruses affect the primary structure of the env gene products.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Besmer P., Baltimore D. Mechanism of restriction of ecotropic and xenotropic murine leukemia viruses and formation of pseudotypes between the two viruses. J Virol. 1977 Mar;21(3):965–973. doi: 10.1128/jvi.21.3.965-973.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi D. P., Montelaro R. C., Frank H., Schäfer W. Assembly of type C oncornaviruses: a model. Science. 1978 Jan 13;199(4325):183–186. doi: 10.1126/science.202022. [DOI] [PubMed] [Google Scholar]
- Buchhagen D. L., Pedersen F. S., Crowther R. L., Haseltine W. A. Most sequence differences between the genomes of the Akv virus and a leukemogenic Gross A virus passaged in vitro are located near the 3' terminus. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4359–4363. doi: 10.1073/pnas.77.7.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchhagen D. L., Pincus T., Stutman O., Fleissner E. Leukemogenic activity of murine type C viruses after long-term passage in vitro. Int J Cancer. 1976 Dec 15;18(6):835–842. doi: 10.1002/ijc.2910180616. [DOI] [PubMed] [Google Scholar]
- Cloyd M. W., Hartley J. W., Rowe W. P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980 Mar 1;151(3):542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan J. E., Kindt T. J., Uehara H., Martinko J., Nathenson S. G. Primary structure of a murine transplantation antigen. Nature. 1981 May 7;291(5810):35–39. doi: 10.1038/291035a0. [DOI] [PubMed] [Google Scholar]
- Copeland N. G., Cooper G. M. Transfection by exogenous and endogenous murine retrovirus DNAs. Cell. 1979 Feb;16(2):347–356. doi: 10.1016/0092-8674(79)90011-4. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Rommelaere J., Ellis R. W., Hopkins N. Nucleotide sequences associated with differences in electrophoretic mobility of envelope glycoprotein gp70 and with GIX antigen phenotype of certain murine leukemia viruses. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1642–1645. doi: 10.1073/pnas.77.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. H., Gautsch J. W., Jensen F. C., Lerner R. A., Hartley J. W., Rowe W. P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Famulari N. G., Buchhagen D. L., Klenk H. D., Fleissner E. Presence of murine leukemia virus envelope proteins gp70 and p15(E) in a common polyprotein of infected cells. J Virol. 1976 Nov;20(2):501–508. doi: 10.1128/jvi.20.2.501-508.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Verma I. M. Size analysis and relationship of murine leukemia virus-specific mRNA's: evidence for transposition of sequences during synthesis and processing of subgenomic mRNA. J Virol. 1978 May;26(2):468–478. doi: 10.1128/jvi.26.2.468-478.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T., LaPorte P., Esty A. Nucleotide sequence studies of polyoma DNA. The Hpa II 3/5 junction to the Hpa II 4/Hae III 18 junction, encoding the origin of DNA replication and the 5' end of the early region. J Biol Chem. 1978 Sep 25;253(18):6561–6567. [PubMed] [Google Scholar]
- Garoff H., Frischauf A. M., Simons K., Lehrach H., Delius H. Nucleotide sequence of cdna coding for Semliki Forest virus membrane glycoproteins. Nature. 1980 Nov 20;288(5788):236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- Gething M. J., White J. M., Waterfield M. D. Purification of the fusion protein of Sendai virus: analysis of the NH2-terminal sequence generated during precursor activation. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2737–2740. doi: 10.1073/pnas.75.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habener J. F., Chang H. T., Potts J. T., Jr Enzymic processing of proparathyroid hormone by cell-free extracts of parathyroid glands. Biochemistry. 1977 Aug 23;16(17):3910–3917. doi: 10.1021/bi00636a029. [DOI] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Hsu M., Scheid A., Choppin P. W. Activation of the Sendai virus fusion protein (f) involves a conformational change with exposure of a new hydrophobic region. J Biol Chem. 1981 Apr 10;256(7):3557–3563. [PubMed] [Google Scholar]
- Jou W. M., Verhoeyen M., Devos R., Saman E., Fang R., Huylebroeck D., Fiers W., Threlfall G., Barber C., Carey N. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980 Mar;19(3):683–696. doi: 10.1016/s0092-8674(80)80045-6. [DOI] [PubMed] [Google Scholar]
- Karshin W. L., Arcement L. J., Naso R. B., Arlinghaus R. B. Common precursor for Rauscher leukemia virus gp69/71, p15(E), and p12(E). J Virol. 1977 Sep;23(3):787–798. doi: 10.1128/jvi.23.3.787-798.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler W., Steiner D. F., Borg J. Studies on the conversion of proinsulin to insulin. 3. Studies in vitro with a crude secretion granule fraction isolated from rat islets of Langerhans. J Biol Chem. 1973 Jul 10;248(13):4544–4551. [PubMed] [Google Scholar]
- Levy J. A. Xenotropic viruses: murine leukemia viruses associated with NIH Swiss, NZB, and other mouse strains. Science. 1973 Dec 14;182(4117):1151–1153. doi: 10.1126/science.182.4117.1151. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Rands E., Chattopadhyay S. K., Garon C. F., Hager G. L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc Natl Acad Sci U S A. 1980 Jan;77(1):614–618. doi: 10.1073/pnas.77.1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung M. L., Hering C., Hartley J. W., Rowe W. P., Hopkins N. Analysis of the genomes of mink cell focus-inducing murine type-C viruses: a progress report. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1269–1274. doi: 10.1101/sqb.1980.044.01.138. [DOI] [PubMed] [Google Scholar]
- Marshall R. D. The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem Soc Symp. 1974;(40):17–26. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGrath M. S., Weissman I. L. AKR leukemogenesis: identification and biological significance of thymic lymphoma receptors for AKR retroviruses. Cell. 1979 May;17(1):65–75. doi: 10.1016/0092-8674(79)90295-2. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Inoue A., Kita T., Nakamura M., Chang A. C., Cohen S. N., Numa S. Nucleotide sequence of cloned cDNA for bovine corticotropin-beta-lipotropin precursor. Nature. 1979 Mar 29;278(5703):423–427. doi: 10.1038/278423a0. [DOI] [PubMed] [Google Scholar]
- Naso R. B., Arcement L. J., Karshin W. L., Jamjoom G. A., Arlinghaus R. B. A fucose-deficient glycoprotein precursor to Rauscher leukemia virus gp69/71. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2326–2330. doi: 10.1073/pnas.73.7.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B. G., Hayward W. S., Robinson H. L., Fang J., Astrin S. M. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981 Feb;23(2):323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- Nowinski R. C., Hays E. F. Oncogenicity of AKR endogenous leukemia viruses. J Virol. 1978 Jul;27(1):13–18. doi: 10.1128/jvi.27.1.13-18.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. V., Stockert E., Obata Y., DeLeo A. B., Old L. J. Murine-leukemia-virus-related cell-surface antigens as serological markers of AKR ecotropic, xenotropic, and dualtropic viruses. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1255–1264. doi: 10.1101/sqb.1980.044.01.136. [DOI] [PubMed] [Google Scholar]
- Pedersen F. S., Crowther R. L., Hays E. F., Nowinski R. C., Haseltine W. A. Structure of retroviral RNAs produced by cell lines derived from spontaneous lymphomas of AKR mice. J Virol. 1982 Jan;41(1):18–29. doi: 10.1128/jvi.41.1.18-29.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen F. S., Crowther R. L., Tenney D. Y., Reimold A. M., Haseltine W. A. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumour. Nature. 1981 Jul 9;292(5819):167–170. doi: 10.1038/292167a0. [DOI] [PubMed] [Google Scholar]
- Pedersen F. S., Haseltine W. A. Analysis of the genome of an endogenous, ecotropic retrovirus of the AKR strain of mice: micromethod for detailed characterization of high-molecular-weight RNA. J Virol. 1980 Jan;33(1):349–365. doi: 10.1128/jvi.33.1.349-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A., Fleissner E. The presence of disulfide-linked gp70-p15(E) complexes in AKR murine leukemia virus. Virology. 1977 Dec;83(2):417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Porter A. G., Barber C., Carey N. H., Hallewell R. A., Threlfall G., Emtage J. S. Complete nucleotide sequence of an influenza virus haemagglutinin gene from cloned DNA. Nature. 1979 Nov 29;282(5738):471–477. doi: 10.1038/282471a0. [DOI] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Robinson H. L., Pearson M. N., DeSimone D. W., Tsichlis P. N., Coffin J. M. Subgroup-E avian-leukosis-virus-associated disease in chickens. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1133–1141. doi: 10.1101/sqb.1980.044.01.122. [DOI] [PubMed] [Google Scholar]
- Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980 Jun;20(2):303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Rommelaere J., Faller D. V., Hopkins N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc Natl Acad Sci U S A. 1978 Jan;75(1):495–499. doi: 10.1073/pnas.75.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M. R., Tung J. S., Hopkins N., Robbins P. W. Relationship of GIX antigen expression to the glycosylation of murine leukemia virus glycoprotein. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6420–6424. doi: 10.1073/pnas.77.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E., Donoghue D. J., Baltimore D. Analysis of a 5' leader sequence on murine leukemia virus 21S RNA: heteroduplex mapping with long reverse transcriptase products. Cell. 1978 Mar;13(3):435–451. doi: 10.1016/0092-8674(78)90318-5. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Cloyd M. W., Hartley J. W. Status of the association of mink cell focus-forming viruses with leukemogenesis. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1265–1268. doi: 10.1101/sqb.1980.044.01.137. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Pincus T. Quantitative studies of naturally occurring murine leukemia virus infection of AKR mice. J Exp Med. 1972 Feb 1;135(2):429–436. doi: 10.1084/jem.135.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest J. P., Feldmann R. J. Membrane proteins: amino acid sequence and membrane penetration. J Mol Biol. 1974 Aug 25;87(4):853–858. doi: 10.1016/0022-2836(74)90090-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M., Marchesi V. T. Amino-acid sequence and oligosaccharide attachment sites of human erythrocyte glycophorin. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2964–2968. doi: 10.1073/pnas.72.8.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. The assembly of proteins into biological membranes: The membrane trigger hypothesis. Annu Rev Biochem. 1979;48:23–45. doi: 10.1146/annurev.bi.48.070179.000323. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]


