Abstract
IκB kinase α (IKKα), one of the two catalytic subunits of the IKK complex involved in nuclear factor κB (NF-κB) activation, also functions as a molecular switch that controls epidermal differentiation. This unexpected function requires IKKα nuclear translocation but does not depend on its kinase activity, and is independent of NF-κB signalling. Ikkα–/– mice present with a hyperproliferative and undifferentiated epidermis characterized by complete absence of a granular layer and stratum corneum. Ikkα-deficient keratinocytes do not express terminal differentiation markers and continue to proliferate even when subjected to differentiation-inducing stimuli. This antiproliferative function of IKKα is also important for the suppression of squamous cell carcinogenesis. The exact mechanisms by which nuclear IKKα controls keratinocyte proliferation and differentiation remained mysterious for some time. Recent studies, however, have revealed that IKKα is a major cofactor in a TGFβ–Smad2/3 signalling pathway that is Smad4 independent. This pathway controls cell cycle withdrawal during keratinocyte terminal differentiation. Although these are not the only functions of nuclear IKKα, this multifunctional protein is a key regulator of keratinocyte and epidermal differentiation and a critical suppressor of skin cancer.
Keywords: epidermal differentiation, Ikkα, skin cancer, Smads, TGFβ
Introduction
The epidermis, the outermost part of the skin, is a stratified and keratinized squamous epithelium mainly composed of keratinocytes, which forms a protective barrier. Epidermal differentiation, which starts in the mouse at embryonic day (E) 12, leads to formation of several distinct cell layers characterized by their ultrastructure, mitotic state and expression of specific molecular markers (Fuchs and Byrne, 1994; Koster and Roop, 2007). The basal layer develops from the surface ectoderm at approximately E9.5 in the mouse. The p63 gene, which specifies different isoforms of a transcription factor related to tumour suppressor p53, controls basal layer formation and maintenance as well as IκB kinase α (IKKα) expression (Candi et al, 2007; Koster et al, 2007). Basal keratinocytes, including epidermal stem cells and transit-amplifying cells, are cuboidal, express cytokeratins (CKs) 5 and 14 and have a high proliferative potential (Koster and Roop, 2007). These cells form the embryonic periderm (M'Boneko and Merker, 1988), which is lost on establishment of the epidermal barrier. At E12 in the mouse, the basal cells give rise to the intermediate cell layer located between the embryonic basal layer and the periderm (Smart, 1970; Weiss and Zelickson, 1975). The intermediate cells divide several times before they withdraw from cell cycle and mature into postmitotic spinous cells (Smart, 1970; Koster and Roop, 2007). By contrast, adult basal keratinocytes directly become spinous cells when terminal differentiation is initiated, without involvement of an intermediate cell type (Koster and Roop, 2007). The spinous layer is characterized by a switch in keratin expression, from CK5 and CK14 to CK1 and CK10 (Fuchs and Green, 1980). Involucrin, a marker of early terminal differentiation is also synthesized in the upper part of this layer. The spinous cells continue their differentiation and maturation to form the granular layer, which is characterized by keratohyalin granules and expression of the late differentiation markers loricrin and filaggrin (Candi et al, 2005). Terminal differentiation gives rise to the cornified layer (stratum corneum), which consists of extremely flat, keratin-filled and anucleated keratinocytes, called corneocytes, which are mummified within a lipid matrix (Candi et al, 2005; Segre, 2006). The stratum corneum is primarily responsible for the barrier function of the skin (Elias, 2004; Segre, 2006), which is established around E17.5 in mouse (Hardman et al, 1998). Although major progress has been made in understanding the molecular changes that characterize epidermal differentiation, less is known about the signalling pathways that control these events. Here, we provide an overview of the current understanding of the central function of IKKα in the control of epidermal differentiation, homoeostasis and tumorigenesis.
Defective epidermal morphogenesis in Ikkα-deficient mice
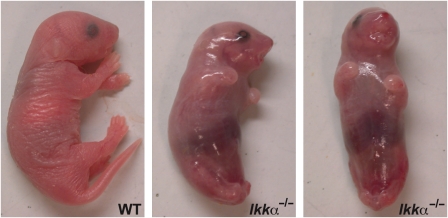
The oligomeric IKK complex is composed of two catalytic subunits, IKKα and IKKβ (DiDonato et al, 1997; Mercurio et al, 1997; Woronicz et al, 1997; Zandi et al, 1997), and a regulatory subunit named IKKγ or NEMO (nuclear factor κB (NF-κB) essential modulator) (Rothwarf et al, 1998; Yamaoka et al, 1998). This complex is the key mediator of NF-κB activation in response to proinflammatory and innate immune challenges (Rothwarf et al, 1998). Although IKKα and IKKβ share considerable sequence identity, it is IKKβ that usually serves the more critical function in the activation of classical NF-κB signalling (Tanaka et al, 1999; Li et al, 1999b, 1999c). To determine the unique functions of IKKα, several groups have disrupted the Ikkα locus in mice and were surprised to find that it has an essential function in epidermal differentiation and morphogenesis (Hu et al, 1999; Li et al, 1999a; Takeda et al, 1999). Newborn Ikkα–/– mice present with multiple morphological defects, including shiny and translucent skin, absence of erupted whiskers, shortened limbs and truncated snout and tail (Figure 1) (Hu et al, 1999; Takeda et al, 1999; Yoshida et al, 2000). These mice develop to term but die shortly after birth, probably as a consequence of a major skin barrier defect that results in severe dehydration.
Figure 1.
Macroscopical presentation of WT and Ikkα–/– mice.
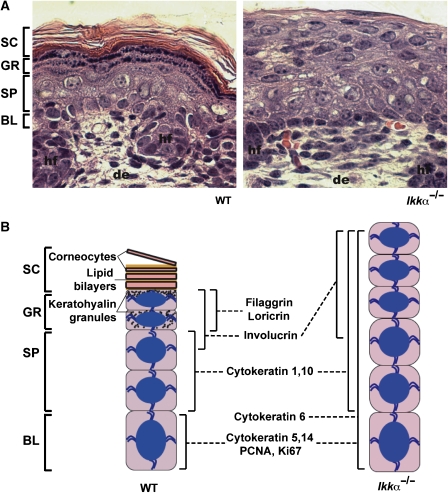
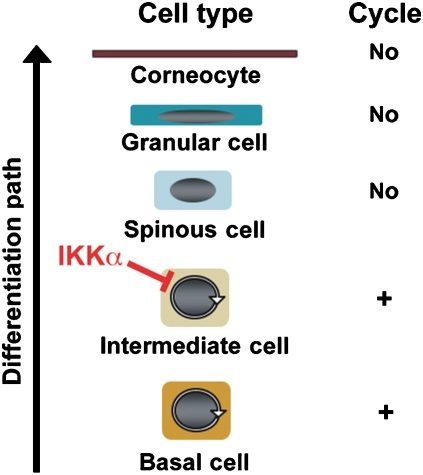
The Ikkα–/– epidermis is characterized by the presence of basal and suprabasal layers, both in a highly proliferative state, and complete absence of the granular and the cornified layers (Figure 2A) (Hu et al, 1999; Takeda et al, 1999). At the molecular level, these anomalies are characterized by the expression of CK5 and CK14 as well as proliferating cell markers, such as CK6, PCNA (proliferating cell nuclear antigen) and Ki67 in basal and suprabasal layers (Hu et al, 1999; Takeda et al, 1999). In contrast, CK1 and CK10 are appropriately expressed in the first suprabasal layers of the Ikkα–/– epidermis (Hu et al, 1999; Takeda et al, 1999). Involucrin is also synthesized in the upper part of the suprabasal layer of these mice (Takeda et al, 1999), indicating that an early step in the differentiation process still takes place, although late terminal differentiation markers, including loricrin and filaggrin, are not expressed (Hu et al, 1999; Takeda et al, 1999) (Figure 2B). It was suggested that the highly proliferative suprabasal cells of the Ikkα–/– epidermis, which express CK1 and CK10, are reminiscent of intermediate cells rather than spinous cells (Koster and Roop, 2007). Hence, the most critical function of IKKα may be induction of cell cycle exit needed for converting basal and intermediate keratinocytes to spinous cells. When this step fails, all subsequent differentiation states are aborted (Figure 3).
Figure 2.
(A) Haematoxylin and eosin staining of skin sections from WT and Ikkα–/– mice. (B) Schematic representation of normal and Ikkα-deficient epidermis with expression profile of molecular markers of proliferation and differentiation (BL: basal layer; SP: spinous layer; GR: granular layer; SC: stratum corneum; hf: hair follicle; de: dermis; magnification × 100 in (A)).
Figure 3.
Schematic representation of epidermal development stages. We propose that the main function of IKKα is to induce cell cycle exit of intermediate keratinocytes when they mature into spinous cells.
Nuclear IKKα controls terminal differentiation of keratinocytes
Control of epidermal proliferation and differentiation by IKKα does not involve its protein kinase function and is completely independent from NF-κB activation (Hu et al, 2001). Ikkα–/– keratinocytes do not exhibit a primary defect in NF-κB activation (Hu et al, 1999; Takeda et al, 1999). In contrast, Ikkα–/– keratinocytes display higher IKK and NF-κB activities than wild-type (WT) cells after incubation with either tumour necrosis factor-α or interleukin-1 (Hu et al, 2001). The observations that transgenic mice overexpressing a dominant inhibitor of NF-κB function (IκBαM) in the epidermis or mice lacking both RelA and c-Rel display epidermal hyperplasia that does not disrupt terminal differentiation and stratum corneum formation (Seitz et al, 1998; Gugasyan et al, 2004; Zhang et al, 2004), are consistent with the NF-κB-independent action of IKKα in the epidermis.
As observed in vivo, isolated Ikkα–/– keratinocytes are hyperproliferative and do not respond to differentiation-inducing signals such as confluence or high Ca2+ (Hu et al, 1999, 2001). Ikkα–/– keratinocytes, however, do differentiate in vitro when transduced by an adenovirus expressing a ‘kinase-dead' form of IKKα, indicating that the kinase function is dispensable for keratinocyte differentiation (Hu et al, 2001). Instead, IKKα needs to enter the nucleus to induce keratinocyte cell cycle arrest and terminal differentiation (Sil et al, 2004). Nuclear entry depends on a nuclear localization sequence (NLS) within the IKKα kinase domain, the disruption of which prevents the induction of keratinocyte differentiation (Sil et al, 2004). An NLS is absent from IKKβ, which cannot substitute for IKKα in keratinocyte differentiation and growth arrest. IKKα is also nuclear in basal and suprabasal cells of the epidermis (Descargues et al, 2008) and the oral epithelium (Maeda et al, 2007), findings that are consistent with the cell culture results.
The function of IKKα in the keratinocyte nucleus is linked to the production of a yet-to-be identified soluble factor or group of factors termed keratinocyte differentiation-inducing factor (kDIF) that can induce the expression of terminal differentiation markers, even in Ikkα–/– cells (Hu et al, 2001). Consistent with the existence of kDIF, transplantation of Ikkα–/– skin onto the back of immuodeficient WT mice allows the Ikkα-deficient epidermis to undergo normal differentiation, suggesting that the requirement for IKKα can be bypassed by factors produced by normal skin (Hu et al, 2001). These experiments do not exclude the possibility that another important source of kDIF or similarly acting factors are dermal fibroblasts, which are well known to produce factors that control epidermal morphogenesis (Wessells, 1977). In support of this hypothesis, a keratinocyte-specific Ikkα disruption results in a less severe epidermal differentiation defect with altered skin barrier function than the total Ikkα knockout (Gareus et al, 2007). These results were interpreted to suggest that Ikkα functions non-autonomously in the dermis to control epidermal differentiation. However, Ikkα-deficient keratinocytes from the epidermal-specific knockout mouse still fail to differentiate in vitro (Gareus et al, 2007), similar to keratinocytes from total Ikkα knockout mice (Hu et al, 2001). Collectively, these results indicate that IKKα functions within epidermal keratinocytes and probably in dermal fibroblasts to induce keratinocyte differentiation. As keratinocyte-restricted expression of IKKα, unlike the keratinocyte-specific knockout, does not result in a differentiation defect (Sil et al, 2004), it appears that the keratinocyte is the major site of IKKα action with respect to keratinocyte differentiation and epidermal-directed morphogenetic events.
IKKα is a critical component of a Smad4-independent TGFβ–Smad2/3 signalling pathway
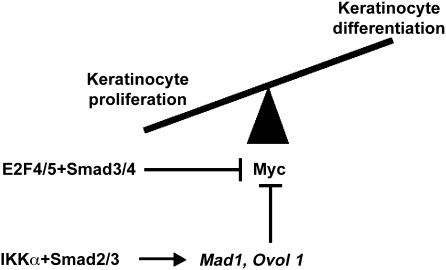
The most immediate effect of IKKα re-expression in Ikkα–/– keratinocytes is cell cycle withdrawal, which precedes the expression of differentiation markers (Hu et al, 2001). Thus, to understand the molecular function of IKKα, we searched for cell cycle-related target genes, the expression of which is IKKα dependent. This search netted several genes encoding negative cell cycle regulators, the expression of which is downregulated in Ikkα–/– keratinocytes and epidermis, including Mad1 and Ovol1 (Descargues et al, 2008), which encode negative regulators of Myc. The c-Myc oncogene is thought to influence the balance between keratinocyte proliferation and differentiation, depending on the intensity and timing of its activity (Watt et al, 2008). Mad1 is a basic region/helix-loop-helix/leucine zipper transcriptional regulator that dimerizes with Max to form Mad:Max heterodimers that antagonize the transcriptional function of Myc:Max dimers (Ayer et al, 1993; Grandori et al, 2000). Mad1–/– mice, however, are viable, phenotypically normal (Foley et al, 1998; Grandori et al, 2000) and do not show any epidermal defect. Most likely, other Mad genes, including Mad2, Mad3 and Mad4, are functionally redundant with Mad1 and compensate for its loss. Indeed Mad2 and Mad3 are also induced in keratinocytes in an IKKα-dependent manner (unpublished data). Ovol1 is a zinc-finger-containing transcription factor, which, similar to Mad1, is also expressed in differentiating suprabasal keratinocytes (Dai et al, 1998; Descargues et al, 2008). Ovol1–/– adult mice present with aberrant hair formation but normal epidermal differentiation (Dai et al, 1998). However, the suprabasal epidermis of Ovol1–/– embryos shows increased proliferation and in vitro, Ovol1–/– keratinocytes fail to exit the cell cycle in response to growth-inhibitory signals, such as high Ca2+ or TGFβ (Nair et al, 2006). This defect may be explained in part by abnormal upregulation of c-Myc, which is a direct target of Ovol1 in keratinocytes (Nair et al, 2006). Thus, IKKα controls keratinocyte proliferation and cycling through the regulation of several Myc antagonists to allow keratinocytes to embark on their differentiation pathway (Figure 4). Interestingly, another TGFβ-related mechanism involving Smad3/4 and E2F4/5 transcription factors has been shown earlier to directly inhibit c-Myc expression in keratinocytes (Chen et al, 2002) (Figure 4). Although this signalling pathway triggers antiproliferative effects of TGFβ in keratinocytes, its impact on keratinocyte differentiation is unknown.
Figure 4.
TGFβ-related signalling pathway controlling Myc activity in keratinocytes. The IKKα–Smad2/3 axis induces Mad1 and Ovol 1 expression on TGFβ stimulation. These proteins may inhibit the activity and expression of Myc, inducing in turn keratinocyte cycle exit and differentiation. Interestingly, a TGFβ–Smad3/4 signalling pathway, which is not associated with IKKα, but functions in cooperation with E2F4/5 transcription factors, has also been shown to negatively control c-Myc expression in keratinocytes (Chen et al, 2002).
Mad1 and Ovol1 are involved in the inhibition of keratinocyte proliferation induced by TGFβ family members (Vastrik et al, 1995; Gomis et al, 2006), suggesting a link between IKKα and the TGFβ signalling pathway. TGFβ family members, including TGFβs, activins and BMPs, are cytokines that control cell growth, differentiation and deposition of extracellular matrix through binding to heterodimeric cell surface receptor complexes composed of type I and II subunits, and intracellular Smad transcription factors (Shi and Massague, 2003; Feng and Derynck, 2005; Schmierer and Hill, 2007). The eight mammalian Smad proteins are divided into three distinct groups: receptor-activated Smads (R-Smads: Smad1, Smad2, Smad3, Smad5 and Smad8), a unique common Smad mediator (Co-Smads: Smad4) and inhibitory Smads (i-Smads: Smad6 and Smad7). On ligand binding, the type II receptor activates the type I receptor through its kinase domain, and the type I receptor in turn phosphorylates R-Smads. The activated R-Smads form heterodimeric complexes with Smad4, which accumulate in the nucleus and directly repress or activate specific target genes (Shi and Massague, 2003; Feng and Derynck, 2005; Schmierer and Hill, 2007).
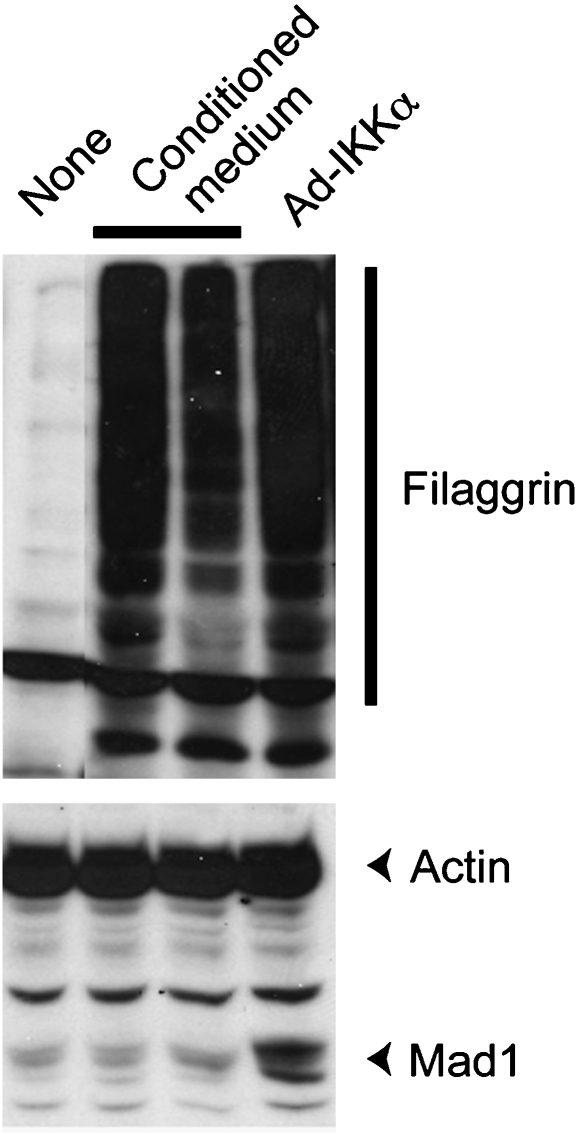
TGFβ family members, their receptors and Smad transcription factors are abundantly expressed in epidermal keratinocytes, suggesting homoeostatic and regulatory functions (He et al, 2001; Li et al, 2003). Phosphorylated Smad2 and Smad3 proteins are concentrated in the nuclei of basal and suprabasal keratinocytes, whereas nuclear Smad4 staining is more preeminent in basal cells (Descargues et al, 2008). Nonetheless, alterations of TGFβ signalling in transgenic/knockout mice have often resulted only in minor epidermal defects, thereby obscuring its exact functions in this tissue (Li et al, 2003). We recently found that IKKα interacts strongly with Smad3 and weakly with Smad2, but does not bind Smad4 (Descargues et al, 2008). IKKα associates with the C-terminal MH2 domain of Smad3 through its kinase domain (unpublished observations). The R-Smad MH2 domain is known to be essential for trans-activation, phosphorylation by type I receptors and Smad4 binding (Massague, 2000; Feng and Derynck, 2005). On stimulation by TGFβ1, IKKα and Smad3 form a transcriptional complex that accumulates in the keratinocyte nucleus to directly control the transcription of Mad1 (Descargues et al, 2008). Similar findings were made for Ovol1 (unpublished data). In Ikkα–/– keratinocytes stimulated with TGFβ1, Smad3 is no longer recruited to the Mad1 regulatory region despite its normal association with Smad4. Furthermore, nuclear staining for activated Smad2 and Smad3 is dramatically diminished in the Ikkα–/– epidermis (Descargues et al, 2008). Taken together, these results indicate that IKKα is required for nuclear accumulation and chromatin recruitment of phosphorylated Smad2 and Smad3 to IKKα-regulated genes in response to TGFβ1. Interestingly, activin A, an important regulator of epidermal differentiation (Owens et al, 2008), is involved in the induction of Mad1 expression in keratinocytes (Werner et al, 2001). As a result, one may speculate that the IKKα–Smad2/3 complex forms and activates anti-Myc genes, including Mad1, on activin A signals. As activin A also downregulates the Id1, Id2 and Id3 genes in keratinocytes (Rotzer et al, 2006), it could also be interesting to analyse whether this signalling pathway depends on IKKα. Finally, we found that kDIF functions downstream of the IKKα–Smad2/3 signalling pathway as it can induce differentiation of Ikkα–/– keratinocytes without inducing Mad1 expression (Figure 5). Exactly how kDIF functions and what it is composed of remain to be determined.
Figure 5.
Mad1 expression is not induced by kDIF-mediated keratinocyte differentiation. Conditioned medium from WT keratinocytes, which contains kDIF as shown earlier (Hu et al, 2001), failed to induce Mad1 expression in Ikkα–/– keratinocytes while leading to keratinocyte differentiation as indicated by filaggrin expression. Only the re-expression of IKKα in Ikkα–/– keratinocytes infected with adenovirus encoding this protein (Ad-IKKα) induces Mad1 expression.
The results mentioned above provided the first clear evidence for a critical function for Smad transcription factors in epidermal differentiation. Although Smad3 deficiency alone does not result in any cutaneous defect (Zhu et al, 1998; Datto et al, 1999; Yang et al, 1999) and the loss of Smad2 leads to early embryonic lethality (Nomura and Li, 1998; Waldrip et al, 1998; Weinstein et al, 1998; Heyer et al, 1999), loricrin and filaggrin expression is barely detectable in E15.5 embryos lacking both Smad3 alleles and one Smad2 allele in their epidermis (Descargues et al, 2008). Similarly, siRNA-mediated Smad2 knockdown in Smad3-deficient keratinocytes inhibited the expression of loricrin and filaggrin, as well as Mad1, in response to high Ca2+ (Descargues et al, 2008). These results suggest that Smad2 and Smad3 are functionally redundant. The importance of the TGFβ–Smad2/3–IKKα axis for proper epidermal differentiation is also underscored by the analysis of transgenic mice in which the i-Smad Smad7 is inducibly expressed in the epidermis, resulting in defective expression of loricrin and filaggrin and failed stratum corneum formation (Descargues et al, 2008). Consequently, these mutant mice display epidermal hyperplasia due to abnormal proliferation of suprabasal keratinocytes and loss of nuclear IKKα and Mad1 (Descargues et al, 2008).
Surprisingly, this new TGFβ response pathway centred around Smad2/3–IKKα complex formation is independent of Smad4. Smad4-deficient keratinocytes display normal Mad1 expression (Descargues et al, 2008) and undergo terminal differentiation in response to high Ca2+ (unpublished data). These results are consistent with the phenotype of mice with epidermal-specific deletion of Smad4 (Smad4Δ/Δ mice). These mice present with degeneration of hair follicles and dermal cysts that progress to skin tumours in old animals, but do not show any perturbated epidermal differentiation and stratum corneum formation (Yang et al, 2005; Qiao et al, 2006). Furthermore, activated Smad2 and Smad3, as well as IKKα, are normally localized in the nuclei of Smad4Δ/Δ keratinocytes, which display normal Mad1 expression (Descargues et al, 2008). Taken together, these results strongly indicate that Smad4 is not required for epidermal differentiation. The Smad4 independence of the TGFβ–Smad2/3–IKKα signalling pathway is reminiscent of another TGFβ signalling operative during erythroid development in which TIF1γ (also called TRIM33 or ectodermin) replaces Smad4 (He et al, 2006). However, TIF1γ is a RING-type ubiquitin ligase that can target Smad4 to degradation and can therefore function as a negative regulator of Smad4-dependent TGFβ signalling (Dupont et al, 2005). Hence the exact function of TIF1γ in TGFβ signalling is not fully understood, and it is not known whether it affects the IKKα-dependent pathway.
IKKα and squamous cell carcinoma
IKKα was recently identified as a tumour suppressor in squamous cell carcinoma (SCC) (Liu et al, 2006; Maeda et al, 2007). SCC is a cancer derived from squamous epithelia of the skin (epidermis), head and neck tissues (mouth, throat, oral and nasal cavities, esophagus) as well as other sites. SCC is the second most common skin cancer in Caucasians with an estimated incidence of 100 000–150 000 new cases per year in the United States (Johnson et al, 1992). Sun exposure and immune suppression increase the risk of SCC development and so does tobacco use (Rudolph and Zelac, 2004; Hampton, 2005). SCC of the oral cavity is one of the most prevalent cancers of the head and neck region with a worldwide incidence of 300 000 new cases per year, the occurrence of which is linked to tobacco use and betel nut chewing (Silverman, 2001). SCCs of the oral cavity are more aggressive than those developing from the skin, and are associated with a 5-year survival rate of about 50–55% (Silverman, 2001).
Molecular changes in SCCs are characterized by a marked heterogeneity and include activation of oncogenes, such as RAS, MYC, EGFR and Cyclin D1, as well inactivation of tumour suppressors, including p53 and p16 (Hardisson, 2003). These genetic alterations are thought to influence malignant keratinocyte behaviour and tumour progression, but the precise molecular pathogenesis of SCC is poorly understood. Interestingly, mutations in exon 15 of the IKKα locus were described in a few high-grade and poorly differentiated human SCCs of the skin and were shown to be associated with reduced IKKα expression (Liu et al, 2006). However, it seems that downregulation of IKKα due to epigenetic silencing of the IKKα locus is a more common occurrence seen in close to 30% of invasive oral SCCs (Maeda et al, 2007). It was also reported that overexpression of IKKα in the suprabasal compartment, which results in increased epidermal differentiation and reduced keratinocyte proliferation, inhibits chemically induced SCC formation and progression in mice (Liu et al, 2006). These results, together with enhanced SCC incidence in Ikkα+/– mice subjected to two-stage skin carcinogenesis and loss of Ikkα heterozygosity in the tumours, provide evidence that IKKα is a tumour suppressor in the epidermis (Liu et al, 2006; Park et al, 2007). This function of IKKα is probably mediated through the control of keratinocyte proliferation. The loss of nuclear IKKα contributes to malignant conversion of keratinocytes into less differentiated and proliferative carcinoma cells. However, a recent study has suggested that increased IKKα may be found in acantholytic SCC (ASCC) (Moreno-Maldonado et al, 2008), an histologic variant of SCC showing positive staining for CKs (Rinker et al, 2001). Unfortunately, the authors of that study have not carefully analysed whether ASCCs present with loss of nuclear IKKα, which would make their results more consistent with other studies mentioned above.
SCC and other carcinoma cells are known to overproduce TGFβ1 to modify their microenvironment through local immunosupression, extracellular matrix remodelling and neoangiogenesis, and these changes are required for tumour progression and invasiveness (Oft et al, 1996, 1998; Siegel and Massague, 2003; Li et al, 2005a, 2006). At the same time, carcinoma cells become resistant to TGFβ-induced growth arrest (Siegel and Massague, 2003; Li et al, 2005a, 2006). Altered IKKα function may contribute, at least in part, to acquired resistance to TGFβ1-induced growth arrest.
Conclusions and future directions
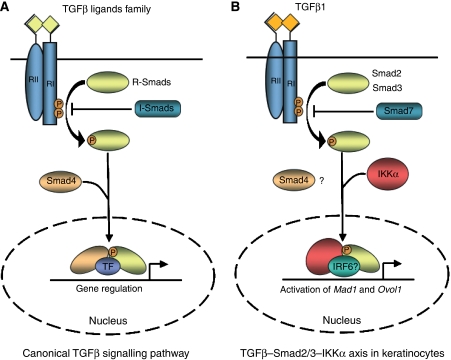
Although IKKα was first identified as a catalytic subunit of the IKK complex, which mediates NF-κB activation (DiDonato et al, 1997; Mercurio et al, 1997; Regnier et al, 1997; Zandi et al, 1997), it has quickly emerged as a multifunctional protein with several unexpected and surprising activities. In the epidermis, IKKα has turned out to be a critical regulator of keratinocyte proliferation, differentiation and oncogenic transformation, and this function is completely unrelated to its protein kinase activity or NF-κB signalling. Instead, IKKα functions as a cofactor for Smad2/3 in a Smad4-independent pathway that inhibits keratinocytes proliferation (Descargues et al, 2008) (Figure 6). In this capacity, IKKα is required for the induction of a specific subset of TGFβ-responsive genes that include the Myc antagonists Mad1 and Ovol1, but is not needed for other well-known TGFβ target genes, such as p21, p15 and p27, encoding inhibitors of cyclin-dependent kinases (CDKs) (Descargues et al, 2008). This is reminiscent of the two different classes of antiproliferative gene responses: Myc repression and inhibition of CDKs, respectively, that are induced during TGFβ-mediated cell cycle arrest (Massague et al, 2000). It is attractive to speculate that the tumour suppressive function of IKKα is exerted through this pathway as well.
Figure 6.
(A) In the canonical TGFβ signalling pathway, ligands signal through type I and II transmembrane protein kinase receptors. After TGFβ binding, type II receptor recruits and phosphorylates type I receptor, which in turn activates R-Smads. Phosphorylated R-Smads oligomerize with the co-Smad Smad4 and accumulate in the nucleus where they interact with DNA and transcription factors to regulate the expression of target genes. (B) During epidermal terminal differentiation, TGFβ1 stimulation of keratinocytes induces the formation of a complex between activated Smad2/3 and IKKα. This complex accumulates in keratinocyte nuclei independently of the presence of Smad4 and controls the transcriptional activation of Mad1 and Ovol1 probably with the cooperation of other transcription factors such as IRF6. The Smad4-independent TGFβ–Smad2/3–IKKα axis is required for cell cycle exit and induction of terminal differentiation of keratinocytes.
Other proteins may be part of the TGFβ–Smad2/3–IKKα signalling pathway, as revealed by two mouse models with functional alterations of 14-3-3σ (repeated epilation mutant mice) and IRF6, the disruption of which faithfully mimics the phenotype of Ikkα–/– mice (Herron et al, 2005; Li et al, 2005b; Ingraham et al, 2006; Richardson et al, 2006). 14-3-3σ belongs to a family of adaptors that can interact with target proteins in a sequence-specific manner, although its exact function is poorly understood (Mhawech, 2005). It was reported that IKKα may protect the 14-3-3σ locus from hypermethylation in keratinocytes by interacting with histone H3 (Zhu et al, 2007). In that study, the authors showed that 14-3-3σ is downregulated in Ikkα–/– keratinocytes, suggesting that this gene is a downstream target of IKKα (Zhu et al, 2007). The human IRF6 locus is defective in Van der Woude (VWS, OMIM: 119300) and popliteal pterygium (PPS, OMIM: 11500) syndromes, which are characterized by orofacial defects such as cleft lip and palate (Kondo et al, 2002). IRF6 belongs to a family of transcription factors that share a highly conserved helix-turn-helix DNA-binding domain and a less conserved protein-binding domain. Interestingly, this protein-binding domain is related to the C-terminal MH2 domain of Smad proteins and has been referred to SMIR (Smad and IRF) domain (Eroshkin and Mushegian, 1999). As DNA binding by Smad transcription factors depends on their association with other DNA-bound transcription factors (Derynck and Zhang, 2003; ten Dijke and Hill, 2004), one can speculate that IRF6 may be a component of the Smad2/3–IKKα transcriptional complex that accumulates in the keratinocyte nucleus to induce the obligatory cell cycle exit that precedes terminal differentiation (Figure 6). In addition, IKKα may also interact with other transcription factors, such as RARs to control epidermal barrier formation (Gareus et al, 2007). The identification of other IKKα-interacting proteins and additional IKKα target genes will provide an ever better understanding of how this critical regulator of epidermal proliferation and differentiation carries out its daily work.
Acknowledgments
This study was supported by The International Human Frontier Science Program Organization (to PD), National Institutes of Health grants (to MK) and an American Cancer Society Research Professorship (to MK).
References
- Ayer DE, Kretzner L, Eisenman RN (1993) Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72: 211–222 [DOI] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, Krammer PH, Melino G (2007) TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle 6: 274–285 [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G (2005) The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6: 328–340 [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J (2002) E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110: 19–32 [DOI] [PubMed] [Google Scholar]
- Dai X, Schonbaum C, Degenstein L, Bai W, Mahowald A, Fuchs E (1998) The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev 12: 3452–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF (1999) Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol 19: 2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE (2003) Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425: 577–584 [DOI] [PubMed] [Google Scholar]
- Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang XJ, Karin M (2008) IKKalpha is a critical coregulator of a Smad4-independent TGFbeta–Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA 105: 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M (1997) A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 388: 548–554 [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S (2005) Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121: 87–99 [DOI] [PubMed] [Google Scholar]
- Elias PM (2004) The epidermal permeability barrier: from the early days at Harvard to emerging concepts. J Invest Dermatol 122: xxxvi–xxxix [DOI] [PubMed] [Google Scholar]
- Eroshkin A, Mushegian A (1999) Conserved transactivation domain shared by interferon regulatory factors and Smad morphogens. J Mol Med 77: 403–405 [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R (2005) Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Foley KP, McArthur GA, Queva C, Hurlin PJ, Soriano P, Eisenman RN (1998) Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J 17: 774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Byrne C (1994) The epidermis: rising to the surface. Curr Opin Genet Dev 4: 725–736 [DOI] [PubMed] [Google Scholar]
- Fuchs E, Green H (1980) Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell 19: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Gareus R, Huth M, Breiden B, Nenci A, Rosch N, Haase I, Bloch W, Sandhoff K, Pasparakis M (2007) Normal epidermal differentiation but impaired skin-barrier formation upon keratinocyte-restricted IKK1 ablation. Nat Cell Biol 9: 461–469 [DOI] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, He W, Wang Q, Seoane J, Lash A, Massague J (2006) A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci USA 103: 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Gugasyan R, Voss A, Varigos G, Thomas T, Grumont RJ, Kaur P, Grigoriadis G, Gerondakis S (2004) The transcription factors c-rel and RelA control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms. Mol Cell Biol 24: 5733–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton T (2005) Skin cancer's ranks rise: immunosuppression to blame. JAMA 294: 1476–1480 [DOI] [PubMed] [Google Scholar]
- Hardisson D (2003) Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol 260: 502–508 [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C (1998) Patterned acquisition of skin barrier function during development. Development 125: 1541–1552 [DOI] [PubMed] [Google Scholar]
- He W, Cao T, Smith DA, Myers TE, Wang XJ (2001) Smads mediate signaling of the TGFbeta superfamily in normal keratinocytes but are lost during skin chemical carcinogenesis. Oncogene 20: 471–483 [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J (2006) Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 125: 929–941 [DOI] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, Grant S, Kinne J, Fisher JK, Siracusa LD (2005) A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat Genet 37: 1210–1212 [DOI] [PubMed] [Google Scholar]
- Heyer J, Escalante-Alcalde D, Lia M, Boettinger E, Edelmann W, Stewart CL, Kucherlapati R (1999) Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc Natl Acad Sci USA 96: 12595–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M (1999) Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 284: 316–320 [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M (2001) IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature 410: 710–714 [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC (2006) Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet 38: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TM, Rowe DE, Nelson BR, Swanson NA (1992) Squamous cell carcinoma of the skin (excluding lip and oral mucosa). J Am Acad Dermatol 26 (3 Part 2): 467–484 [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C et al. (2002) Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 32: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR (2007) p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA 104: 3255–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR (2007) Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol 23: 93–113 [DOI] [PubMed] [Google Scholar]
- Li AG, Koster MI, Wang XJ (2003) Roles of TGFbeta signaling in epidermal/appendage development. Cytokine Growth Factor Rev 14: 99–111 [DOI] [PubMed] [Google Scholar]
- Li AG, Lu SL, Han G, Hoot KE, Wang XJ (2006) Role of TGFbeta in skin inflammation and carcinogenesis. Mol Carcinog 45: 389–396 [DOI] [PubMed] [Google Scholar]
- Li AG, Lu SL, Han G, Kulesz-Martin M, Wang XJ (2005a) Current view of the role of transforming growth factor beta 1 in skin carcinogenesis. J Investig Dermatol Symp Proc 10: 110–117 [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, Verma IM (2005b) Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc Natl Acad Sci USA 102: 15977–15982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM (1999a) IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev 13: 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM (1999b) Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science 284: 321–325 [DOI] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M (1999c) The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med 189: 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Park E, Zhu F, Bustos T, Liu J, Shen J, Fischer SM, Hu Y (2006) A critical role for I kappaB kinase alpha in the development of human and mouse squamous cell carcinomas. Proc Natl Acad Sci USA 103: 17202–17207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'Boneko V, Merker HJ (1988) Development and morphology of the periderm of mouse embryos (days 9–12 of gestation). Acta Anat (Basel) 133: 325–336 [DOI] [PubMed] [Google Scholar]
- Maeda G, Chiba T, Kawashiri S, Satoh T, Imai K (2007) Epigenetic inactivation of IkappaB Kinase-alpha in oral carcinomas and tumor progression. Clin Cancer Res 13: 5041–5047 [DOI] [PubMed] [Google Scholar]
- Massague J (2000) How cells read TGF-beta signals. Nat Rev Mol Cell Biol 1: 169–178 [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103: 295–309 [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A (1997) IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278: 860–866 [DOI] [PubMed] [Google Scholar]
- Mhawech P (2005) 14-3-3 proteins—an update. Cell Res 15: 228–236 [DOI] [PubMed] [Google Scholar]
- Moreno-Maldonado R, Ramirez A, Navarro M, Fernandez-Acenero MJ, Villanueva C, Page A, Jorcano JL, Bravo A, Llanos Casanova M (2008) IKKalpha enhances human keratinocyte differentiation and determines the histological variant of epidermal squamous cell carcinomas. Cell Cycle 7: 2021–2029 [DOI] [PubMed] [Google Scholar]
- Nair M, Teng A, Bilanchone V, Agrawal A, Li B, Dai X (2006) Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol 173: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Li E (1998) Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393: 786–790 [DOI] [PubMed] [Google Scholar]
- Oft M, Heider KH, Beug H (1998) TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol 8: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E (1996) TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10: 2462–2477 [DOI] [PubMed] [Google Scholar]
- Owens P, Han G, Li AG, Wang XJ (2008) The role of Smads in skin development. J Invest Dermatol 128: 783–790 [DOI] [PubMed] [Google Scholar]
- Park E, Zhu F, Liu B, Xia X, Shen J, Bustos T, Fischer SM, Hu Y (2007) Reduction in IkappaB kinase alpha expression promotes the development of skin papillomas and carcinomas. Cancer Res 67: 9158–9168 [DOI] [PubMed] [Google Scholar]
- Qiao W, Li AG, Owens P, Xu X, Wang XJ, Deng CX (2006) Hair follicle defects and squamous cell carcinoma formation in Smad4 conditional knockout mouse skin. Oncogene 25: 207–217 [DOI] [PubMed] [Google Scholar]
- Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M (1997) Identification and characterization of an IkappaB kinase. Cell 90: 373–383 [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ (2006) Irf6 is a key determinant of the keratinocyte proliferation–differentiation switch. Nat Genet 38: 1329–1334 [DOI] [PubMed] [Google Scholar]
- Rinker MH, Fenske NA, Scalf LA, Glass LF (2001) Histologic variants of squamous cell carcinoma of the skin. Cancer Control 8: 354–363 [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M (1998) IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature 395: 297–300 [DOI] [PubMed] [Google Scholar]
- Rotzer D, Krampert M, Sulyok S, Braun S, Stark HJ, Boukamp P, Werner S (2006) Id proteins: novel targets of activin action, which regulate epidermal homeostasis. Oncogene 25: 2070–2081 [DOI] [PubMed] [Google Scholar]
- Rudolph R, Zelac DE (2004) Squamous cell carcinoma of the skin. Plast Reconstr Surg 114: 82e–94e [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS (2007) TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 8: 970–982 [DOI] [PubMed] [Google Scholar]
- Segre JA (2006) Epidermal barrier formation and recovery in skin disorders. J Clin Invest 116: 1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz CS, Lin Q, Deng H, Khavari PA (1998) Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc Natl Acad Sci USA 95: 2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massague J (2003) Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer 3: 807–821 [DOI] [PubMed] [Google Scholar]
- Sil AK, Maeda S, Sano Y, Roop DR, Karin M (2004) IkappaB kinase-alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature 428: 660–664 [DOI] [PubMed] [Google Scholar]
- Silverman S. Jr (2001) Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc 132 (Suppl.): 7S–11S [DOI] [PubMed] [Google Scholar]
- Smart IH (1970) Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br J Dermatol 82: 276–282 [DOI] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S (1999) Limb and skin abnormalities in mice lacking IKKalpha. Science 284: 313–316 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV (1999) Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity 10: 421–429 [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS (2004) New insights into TGF-beta–Smad signalling. Trends Biochem Sci 29: 265–273 [DOI] [PubMed] [Google Scholar]
- Vastrik I, Kaipainen A, Penttila TL, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K (1995) Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol 128: 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ (1998) Smad2 signaling in extraembryonic tissues determines anterior–posterior polarity of the early mouse embryo. Cell 92: 797–808 [DOI] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah SA (2008) MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev Cancer 8: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX (1998) Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc Natl Acad Sci USA 95: 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LW, Zelickson AS (1975) Embryology of the epidermis: ultrastructural aspects. III. Maturation and primary appearance of dendritic cells in the mouse with mammalian comparisons. Acta Derm Venereol 55: 431–442 [PubMed] [Google Scholar]
- Werner S, Beer HD, Mauch C, Luscher B, Werner S (2001) The Mad1 transcription factor is a novel target of activin and TGF-beta action in keratinocytes: possible role of Mad1 in wound repair and psoriasis. Oncogene 20: 7494–7504 [DOI] [PubMed] [Google Scholar]
- Wessells NK (1977) Tissue Interaction and Development. Menlo Park, CA: Benjamin/Cummings [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV (1997) IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278: 866–869 [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israel A (1998) Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell 93: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Yang L, Mao C, Teng Y, Li W, Zhang J, Cheng X, Li X, Han X, Xia Z, Deng H, Yang X (2005) Targeted disruption of Smad4 in mouse epidermis results in failure of hair follicle cycling and formation of skin tumors. Cancer Res 65: 8671–8678 [DOI] [PubMed] [Google Scholar]
- Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C (1999) Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J 18: 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hu Y, Karin M (2000) IkappaB kinase alpha is essential for development of the mammalian cornea and conjunctiva. Invest Ophthalmol Vis Sci 41: 3665–3669 [PubMed] [Google Scholar]
- Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M (1997) The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 91: 243–252 [DOI] [PubMed] [Google Scholar]
- Zhang JY, Green CL, Tao S, Khavari PA (2004) NF-kappaB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev 18: 17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y (2007) IKKalpha shields 14-3-3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol Cell 27: 214–227 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM (1998) Smad3 mutant mice develop metastatic colorectal cancer. Cell 94: 703–714 [DOI] [PubMed] [Google Scholar]