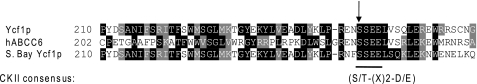Abstract
The yeast vacuolar membrane protein Ycf1p and its mammalian counterpart, MRP1, belong to the ABCC subfamily of ATP-binding cassette (ABC) transporters that rid cells of toxic endogenous and xenobiotic compounds. Like most members of the ABCC subfamily, Ycf1p contains an N-terminal extension in addition to its ABC “core” domain and transports substrates in the form of glutathione conjugates. Ycf1p is subject to complex regulation to ensure its optimal function. Previous studies showed that Ycf1p activity is stimulated by a guanine nucleotide exchange factor, Tus1p, and is positively regulated by phosphorylation in its ABC core domain at residues Ser-908 and Thr-911. Here we provide evidence that phosphorylation of Ser-251 in the Ycf1p N-terminal extension negatively regulates activity. Mutant Ycf1p-S251A exhibits increased resistance to cadmium in vivo and increased Ycf1p-dependent transport of [3H]estradiol-β-17-glucuronide in vitro as compared with wild-type Ycf1p. Activity is restored to the wild-type level for Ycf1-S251E. To identify kinase(s) that negatively regulate Ycf1p function, we conducted an integrated membrane yeast two-hybrid (iMYTH) screen and identified two kinase genes, CKA1 and HAL5, deletion of which increases Ycf1p function. Genetic evidence suggests that Cka1p may regulate Ycf1p function through phosphorylation of Ser-251 either directly or indirectly. Overall, this study provides compelling evidence that negative, as well as positive, regulation of Ycf1p is mediated by phosphorylation.
Transporters of the ATP-binding cassette (ABC)3 superfamily are expressed in all organisms, from microbes to mammals, and transport chemically diverse compounds across cellular membranes (1, 2). ABC transporters are divided into seven subfamilies (A–G) based on conserved sequences within their nucleotide-binding domains (NBDs). Recent interest has focused on the ABCC subfamily of ABC transporters, of which the prototype member is the mammalian multidrug resistance-associated protein 1 (MRP1), also called ABCC1. Mutations in several members of the ABCC subfamily cause human diseases, including cystic fibrosis, pseudoxanthoma elasticum, and Dubin-Johnson syndrome resulting from mutations in ABCC7 (CFTR), ABCC6 (MRP6), and ABCC2 (MRP2), respectively (1–3), whereas overexpression of MRP1 and other ABCC proteins is associated with multidrug resistance in a variety of tumor cell lines (1, 2, 4, 5).
Members of the ABCC subfamily have two distinguishing features in addition to their sequence homology. First, many of them have the unique ability to transport substrates in the form of glutathione conjugates or complexes (4, 6). Second, whereas all ABC transporters contain an ABC “core” domain (two membrane-spanning domains (MSDs) and two NBDs connected by a cytosolic domain; see Fig. 1A), a hallmark of the ABCC subfamily is that most of its members contain a significant N-terminal extension (NTE) (4, 6). The NTE comprises five membrane spans (MSD0) and a cytosolic loop (L0) (Fig. 1A). We and others have shown that MSD0 plays a role in localization for yeast and human MRPs and that the L0 is required for transport function (7–12). Some evidence indicates the L0 may directly interact with transporter substrates such as GSH and leukotriene C4 (7, 11, 13, 14). It has also been proposed that L0 plays a key role in dimerization of MRP1, which in turn is important for protein function (15).
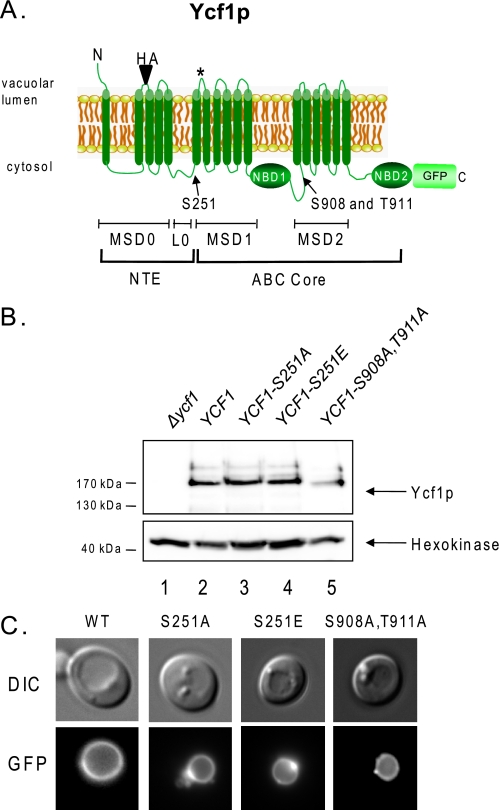
FIGURE 1.
Ycf1p is phosphorylated at Ser-251. A, the structure of Ycf1p consists of an NTE containing membrane-spanning domain MSD0 and a cytosolic linker (L0) and an ABC core region containing two membrane-spanning domains (MSD1 and MSD2) and two nucleotide-binding domains (NBD1 and NBD2). Ycf1p undergoes processing in vivo at the site indicated by an asterisk in MSD1, giving rise to N- and C-terminal fragments that together are required for Ycf1p activity (9). Ycf1p phosphorylation at Ser-908 and Thr-911 is required for transport activity (35), and phosphorylation within the NTE at Ser-251 negatively regulates transport activity, as shown in this study. B, WT and mutant forms of GFP-tagged Ycf1p were examined by Western blot analysis using anti-GFP antibody, and hexokinase was probed with an anti-hexokinase antibody as the loading control. The C-terminal portion of Ycf1p (labeled Ycf1p) contains the GFP tag, as shown in A, and is detected in this analysis. Ycf1p expression was quantitated and normalized to hexokinase expression using ImageQuant software and the Bio-Rad Versidoc system. The amount of Ycf1p expressed relative to WT for S251A, S251E, and S908A,T911A is 1, 1.1, and 0.47, respectively C, vacuole localization of GFP-tagged versions of WT and mutant Ycf1p was examined by fluorescence microscopy. Yeast strains used in B and C are SM5270, SM5280, SM5506, SM5507, and SM5508.
The substrate specificity, biochemical requirements for transporter function, and factors involved in transcriptional regulation of ABCC transporters have been relatively well studied (4, 6, 16, 17). In contrast, the post-translational regulation of the ABCCs has in general not been well characterized. As the importance of the ABCC transporters in detoxification and drug resistance becomes evident, a better understanding of how the function of ABCC proteins is regulated in vivo will be important. For ABCC7 (CFTR), phosphorylation of sites within the “R” domain of the ABC core has been suggested to positively and negatively regulate chloride channel function (18–21). CFTR is an atypical ABCC transporter, as it does not contain an N-terminal extension. To date, phosphorylation for other members of the ABCC subfamily of mammalian transporters has not been reported.
To examine whether phosphorylation can regulate the activity of a prototypical ABCC transporter, we used Saccharomyces cerevisiae. Yeast represent a highly tractable system for genetic and biochemical analysis of transporters. The best studied ABCC subfamily member in yeast is Ycf1p (Fig. 1A) (9, 22–26). Ycf1p is homologous to human MRP1, and in a ycf1Δ yeast mutant, human MRP confers functional complementation, providing compelling evidence for their mechanistic conservation (27). Ycf1 resides on the vacuolar membrane and detoxifies yeast of xenobiotics, including heavy metals (cadmium, lead, mercury, arsenite), oxidants (diamide), and endogenous toxins (the intermediate in the adenine biosynthesis pathway that is glutathione conjugated and accumulates in an ade2 mutant) (24–26, 28–33). Detoxification involves the Ycf1p-dependent transport of these compounds into the vacuole where they are sequestered from their targets in the cytosol or nucleus (25, 26). Recent evidence from our laboratory and others suggests that there are multiple layers of transcriptional as well as post-transcriptional regulation of Ycf1p, presumably reflecting regulated control mechanisms (16, 17, 34, 35).
To identify regulatory interactors for a protein of interest, protein-protein interaction studies, such as yeast two-hybrid analysis, GST pulldowns, and co-immunoprecipitation, are typically carried out. However, the highly hydrophobic nature of transmembrane proteins such as Ycf1p complicates the use of these standard methods (36, 37). We recently reported the development of iMYTH, a modified version of the split ubiquitin membrane yeast two-hybrid system, showing that it is a powerful method for identifying physiological regulators of membrane proteins such as the ABC transporters (34, 37). Using iMYTH in a library screening format, we identified potential Ycf1p interaction partners. One of these, Tus1p, a guanine nucleotide exchange factor for the small GTPase Rho1p, is a Rho1p-dependent positive regulator of Ycf1p transport activity both in vivo and in an in vitro transport assay (34). In the present study, we have extended our iMYTH analysis of Ycf1p and identified two kinases that appear to function as negative regulators of Ycf1p, discussed in detail below.
An important resource for yeast researchers is a growing number of large scale proteomic studies aimed at identifying the yeast “phospho-proteome.” We found that Ycf1p is among the vast repertoire of yeast phosphoproteins reported in three separate high throughput studies (38–40). These studies have variously reported that two, three, or four residues within the ABC core domain of Ycf1p are phosphorylated, including Ser-903, Ser-908, Thr-911, and Ser-914. Strong experimental evidence indicates that phosphorylation of Ser-908 and Thr-911 plays a positive role in regulating Ycf1p function, because mutation of either residue to alanine results in almost complete loss of transporter activity, whereas mutation to glutamate to mimic phosphorylation restores activity (26, 35). One additional phosphorylated residue in Ycf1p, Ser-251, was reported in two of the high throughput studies (39, 40). Notably, Ser-251 lies outside of the ABC core and is within the L0 domain of the NTE of Ycf1p. Because L0 is required for Ycf1p transport activity, and the analogous region in mammalian MRP1 has been implicated in the binding of reduced glutathione (GSH) and an MRP1 substrate (leukotriene C4) as well as in influencing dimerization, an interesting possibility is that phosphorylation of Ser-251 may directly regulate Ycf1p function.
Here we have examined the issue of whether phosphorylation within the NTE regulates Ycf1p function. Surprisingly, our results suggest that phosphorylation of the NTE at Ser-251 negatively regulates Ycf1p function, in contrast to the positive regulation observed for phosphorylation of the ABC core region at Ser-908 and Thr-911. We also identified two kinase genes (CKA1 and HAL5) among the Ycf1p iMYTH interactors. Ycf1p transport activity is increased in cka1Δ and hal5Δ mutants, as compared with a WT strain, in a Ycf1p-dependent manner. Further genetic evidence suggests that Cka1p might negatively regulate Ycf1p function through phosphorylation of Ser-251. These experiments, together with previous studies, suggest that the transport function of Ycf1p is intricately controlled, as it is subject to both positive and negative regulation by phosphorylation, and involves distinct regions of the protein, the ABC core, and NTE, respectively.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Growth Conditions—Yeast strains used in this study are listed in Table 1. Standard dropout media (SC) were prepared as described previously (41). The plates used for cadmium spot tests were prepared by adding cadmium (CdCl2) to the indicated concentrations in SC medium immediately before pouring plates as described (9, 23). Cultures were grown at 30 °C. The yeast medium used in iMYTH screening is described in Iyer et al. (36).
TABLE 1.
Yeast strains used in this study
| Straina | Relevant genotype | Reference |
|---|---|---|
| SM4460b | MATa Δmet 15 Δleu2 Δura3 Δhis3 | Mason et al. (23) |
| SM5270 | Δycf1::KanMX | Paumi et al. (34) |
| SM5280 | SM5270 with pSM1753 [2μ YCF1-GFP URA3] | Paumi et al. (34) |
| SM5506 | SM5270 with pSM2243 [2μ YCF1-S251A-GFP URA3] | This study |
| SM5507 | SM5270 with pSM2244 [2μ YCF1-S251E-GFP URA3] | This study |
| SM5508 | SM5270 with pSM2245 [2μ YCF1-S908A, T911A-GFP URA3] | This study |
| SM5509 | Δhal5::KanMX | Open Biosystems |
| SM5510 | Δcka1::KanMX | Open Biosystems |
| SM5511 | SM4460 with pSM1774 [2μ YCF1-HAloop-GFP URA3]c | This study |
| SM5512 | SM4460 with pSM2246 [2μ YCF1-S251A-HAloop-GFP URA3]c | This study |
| SM5513 | SM5509 with pSM1774 [2μ YCF1-HAloop-GFP URA3]c | This study |
| SM5514 | SM5510 with pSM1774 [2μ YCF1-HAloop-GFP URA3]c | This study |
| SM5515 | Δhal5::KanMX Δycf1::URA3 | This study |
| SM5516 | Δcka1::KanMX Δycf1::URA3 | This study |
| SM5519 | SM5510 with pSM1753 [2μ YCF1-GFP URA3] | This study |
| SM5520 | SM5510 with pSM2243 [2μ YCF1-S251A-GFP URA3] | This study |
| SM5521 | SM5510 with pSM2244 [2μ YCF1-S251E-GFP URA3] | This study |
| SM5522 | SM5509 with pSM1753 [2μ YCF1-GFP URA3] | This study |
| SM5523 | SM5509 with pSM2243 [2μ YCF1-S251A-GFP URA3] | This study |
| SM5524 | SM5510 with pSM2244 [2μ YCF1-S251E-GFP URA3] | This study |
| SM5546 | SM5270 with pSM2247 [2μ YCF1-S251A,S908A,T911A-GFP URA3] | This study |
| L40 | MATa, trp1, leu2, his3, LYS2::lexA-HIS3, URA3::lexA-lacZ | Paumi et al. (34) |
| THY AP4 | MATa, ura3, leu2,lexA::lacZ::trp1, lexA::HIS3, lexA::ADE2 | Paumi et al. (34) |
| AP4-YCF1-CT | THY.AP4 with YCF1::CT-KanMX | Paumi et al. (34) |
| AP4-SHO1-CT | THY-AP4 with SHO1::CT-KanMX | Paumi et al. (34) |
The double knock-out strains SM5515 (Δycf1::URA3 Δhal5::KanMX) and SM5516 (Δycf1::URA3 Δcka1::KanMX) were created by standard homologous recombination as follows. The single deletions Δhal5::KanMX and Δcka1::KanMX in the BY4741 background were obtained from the MATa yeast genome deletion collection (Open Biosystems). We confirmed the replacement of each gene with the KanMX cassette by standard PCR using a forward primer within the KanMX cassette and a reverse primer within the 3′ region of each gene. Each confirmed deletion was given an SM strain designation (SM5509 and SM5510) to signify that the appropriate genotype was confirmed. A PCR product containing the URA3 coding sequence flanked by 60 bp of homology to the 5′ upstream and 3′ downstream coding sequence of YCF1 was gel-purified according to the manufacturer's directions (Qiagen), transformed into SM5509 (Δhal5::KanMX) and SM5510 (Δcka1::KanMX), and double disruptants were selected on SC-URA G418-containing plates, yielding strains SM5515 and SM5516, respectively.
Plasmids—Plasmids used in this study are listed in Table 2. Phosphorylation site mutants of Ycf1p were created in the starting plasmid pSM1753 (2μ YCF1-GFP URA3) using standard PCR-mediated mutagenesis (Stratagene QuikChange II-XL). In brief, primers harboring the mutation of interest were designed using the QuikChange Web site primer design program (Stratagene), and PCR-mediated mutagenesis was carried out. The product of the PCR reaction was digested for 1–3 h with DpnI. A 10-μl aliquot of the DpnI digest was ligated (Takara Co.) and transformed into the DH5α bacterial cloning strain. DH5α clones containing mutant plasmid constructs were selected on LB agar plates containing carbenicillin, purified, and sequenced for verification, which resulted in the creation of pSM2243, pSM2244, and pSM2245, containing S251A, S251E, and S908A,T911A, respectively. To construct the YCF1 triple mutant, S251A,S908A,T911A, a restriction fragment containing the S251A mutation from pSM2243 was co-transformed with linearized pSM2245 into yeast, and homologous recombination generated the recombinant plasmid pSM2246.
TABLE 2.
Plasmid used in this study
| Plasmid | Relevant genotype | Reference |
|---|---|---|
| pSM1753 | [2μ YCF1-GFP URA3] | Mason et al. (23) |
| pSM1774a | [2μ YCF1-HALoop-GFP URA3] | Mason et al. (23) |
| pSM2243 | [2μ YCF1-S251A-GFP URA3] | This study |
| pSM2244 | [2μ YCF1-S251E-GFP URA3] | This study |
| pSM2245 | [2μ YCF1-S908A, T911A-GFP URA3] | This study |
| pSM2246 | [2μ YCF1-S251A-HAloop-GFP URA3]a | This study |
| pSM2306 | [2μ YCF1-S251A, S908A, T911A-GFP URA3] | This study |
| pPR3-N | Contains the CYC1 promoter followed by the multiple cloning site and a hemagglutinin epitope. This is the “prey” vector that was used to clone the NubG-X cDNA library. Contains a TRP1 marker. | Dualsystems |
| pPR3-N-HAL5 | Contains the prey HAL5 found from a cDNA NubG-X screen. Contains a TRP1 marker. | This study |
| pPR3-N-CKA1 | Contains the prey CKA1 found from a cDNA NubG-X screen. Contains a TRP1 marker. | This study |
| pOST1-NubI | The entire ORF of the OST1 gene is fused to the NubI-CYT tag.b | Iyer et al. (36) |
| pOST1-NubG | The entire ORF of the OST1 gene is fused to the NubG-CYT tag.b | Iyer et al. (36) |
| pCT-L2 | A cassette containing the C-terminal end of ubiquitin (Cub) and transcription factor followed by KanMX marker. | Paumi et al. (34) |
| pCYT-L3 | A cassette containing the C-terminal end of ubiquitin (Cub), yellow fluorescent protein, and transcription factor followed by KanMX marker. | Paumi et al. (34) |
Construction of a NubG-X Yeast cDNA Library and iMYTH Screen with YCF1-CT as Bait—An oligo(dT)-primed, size-selected (0.8–5 kb; average insert size, 1.6 kb) yeast cDNA library (S. cerevisiae, strain Jel1) with 2 × 106 independent clones was custom constructed in the prey vector, pDL2Nx, by Dualsystems Biotech Inc. This cDNA library was transformed into the yeast reporter strain THY AP4 expressing the chromosomally tagged YCF1-CT bait (34). Approximately 8 × 106 transformants were screened for colonies that would grow on media lacking tryptophan (W), adenine (A), and histidine (H) supplemented with 25 mm 3-aminotriazole. Library plasmids were isolated from 102 positive clones, amplified in Escherichia coli, and analyzed by restriction analysis for insert sizes. The plasmids that contained an insert were further processed by a bait dependence test. For this purpose, the individual prey plasmids were retransformed into THY AP4 yeast expressing YCF1-CT and THY AP4 expressing a yeast plasma membrane protein, SHO1-CT, after which 77 YCF1-CT-dependent clones were identified, four of which encoded Cka1p and five of which encoded Hal5p, respectively. The nature of the remaining 68 YCF1-CT-dependent clones will be described elsewhere. Lastly, the remaining 25 clones interacted with both YCF1-CT and SHO1-CT and were therefore considered false positives.
Growth Inhibition by Cadmium—Growth inhibition by CdCl2 was monitored by spot tests on plates essentially as described previously (9, 23). Briefly, cells were grown overnight to saturation in SC-dropout medium, subcultured at a 1:5000 dilution in the same medium, and grown overnight to an A600 of ∼1.0. The overnight culture was diluted to an A600 of 0.1, which in turn was diluted in 10-fold increments. Aliquots (4 μl) of each 10-fold dilution were spotted onto SC-dropout plates containing no drug (vehicle alone), 50 μm, or 200 μm CdSO4 and incubated for 2 days (no drug) or 4 days (50 and 200 μm drug), respectively.
To examine growth inhibition by CdCl2 in liquid media, cells were grown overnight to saturation in SC medium, subcultured at a 1:5000 dilution in SC medium, and grown overnight to an A600 of ∼1.0. The overnight culture was diluted to an A600 of 0.1 in YPD medium (yeast/peptone/dextrose) containing 400 μm CdCl2, or to an A600 of 0.02 in YPD alone, and incubated at 25 °C for 20 h. A600 was measured and plotted in a bar graph as percent of control ((A600 of deletion strain/A600 of WT) × 100%).
Immunoblotting Analysis of GFP-tagged Ycf1p—Cell extracts and immunoblots to examine Western blots of WT and mutant Ycf1p were prepared essentially as described previously (22), with modifications as follows. Fifty ml of cells grown in log phase to an A600 of 0.8–1.0 units were harvested, resuspended in 100 μl of radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, and 0.5% deoxycholate) containing 1% SDS, fresh protease inhibitors (Roche Complete protease inhibitor mixture), phenylmethylsulfonyl fluoride (1 μm), and pepstatin A (5 μg/ml)), and broken with glass beads (1 min of vortexing and 1 min on ice for 10 times). Following bead beating, 900 μl of radioimmune precipitation assay buffer plus protease inhibitors without SDS was added, and the broken cells were microfuged at 2000 rpm for 5 min at 4 °C to remove debris. The protein concentration in the supernatant was measured by the Bradford assay. For standard Western blot analysis, 12.5 μg of total protein was solubilized in an equal volume of 2× SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 8 m urea, 0.1 mm EDTA, 15% SDS, 0.01% bromphenol blue, and 150 mm dithiothreitol added fresh), frozen overnight, and subsequently incubated at 37 °C for 1 h prior to SDS-PAGE and transfer to nitrocellulose. For Western blot analysis of GFP-tagged Ycf1p from total protein extracts, blots were incubated with the primary monoclonal mouse anti-GFP (1:1000) (Roche Applied Science) or polyclonal rabbit anti-hexokinase (1:200,000) (courtesy of Dr. Rob Jensen) and the respective secondary antibodies, sheep anti-mouse (1:2000) and sheep anti-rabbit (1:5000). Protein was visualized and quantitated using a chemiluminescent reagent (Roche Applied Science) on a Bio-Rad Versadoc imaging system.
In Vitro Transport of Radiolabeled Substrate by Ycf1p into Vesiculated Vacuoles—To assay transport activity of Ycf1p in vitro, vacuoles were isolated and purified essentially as described previously (34, 42). Ycf1p-dependent transport into the purified vacuoles was assayed by measuring the amount of radiolabeled Ycf1p substrate, [3H]estradiol-β-17-glucuronide ([3H]E2β17G), transported from outside to inside the vesiculated vacuoles. Transport assays were conducted as described previously (34) with minor modification as follows. 100-μl reaction volumes contained NTPs, NaATP, or NaAMP-PCP (at 4 mm); 10 mm MgCl2;5 μm gramicidin-D; 10 mm creatine phosphate and 16 units/ml creatine kinase; 50 mm KCl; 400 mm sorbitol; 25 mm Tris-Mes (pH 8.0); and a Ycf1p substrate, [3H]E2β17G (Amersham Biosciences), at concentrations of 50, 100, 200, 400, 800, 1600, and 2400 μm. Reactions were initiated by the addition of 1–2 μg of purified vacuoles and incubated at 25 °C for 5 or 10 min; they were stopped by the addition of 1 ml of ice-cold stop buffer (400 mm sorbitol, and 3 mm Tris/Mes (pH 8.0)). Vacuoles were collected on Millipore 0.22-μm GV filters via vacuum manifold and washed. Transport was measured as total dpm, normalized to Ycf1p protein levels as determined by Western blot (described above), and reported as nmol of substrate ([3H]E217G) sequestered within the vacuoles/mg of total protein over 10 min (34).
The efficiency of transport for Ycf1p, Ycf1p-S251A, Ycf1p-S251E, and Ycf1p-S908A,T911A was analyzed by standard Michaelis-Menten kinetics in Kaleidagraph (Synergy Software). Vmax and Kmapp values of E2β17G transport for each protein were extrapolated from the curve. Transport efficiency (see Table 3) is reported as Vmax/Kmapp, and relative transport efficiency of mutants is expressed as (Vmax/Kmapp of mutant/Vmax/Kmapp of WT).
TABLE 3.
Michaelis-Menten kinetics for Ycf1p transport of [3H]E2β17G
| Vacuole type | Km,appa | Vmaxa | Vmax/Km,app | Relative efficiencyb |
|---|---|---|---|---|
| μm | nmol/10 min/mg | |||
| Ycf1p | 394 | 260 | 0.66 | 1.0 |
| Ycf1p-S251A | 187 | 246 | 1.32 | 2.0 |
| Ycf1p-S251E | 481 | 276 | 0.57 | 0.86 |
| Ycf1p-S908A, T911A | 548 | 154 | 0.28 | 0.42 |
Values are extrapolated from Fig. 3B
Relative efficiency is calculated as (Vmax/Km,app of mutant)/(Vmax/Km,app of WT)
Fluorescence Microscopy—To examine the localization of Ycf1p-GFP fusion proteins, cells were grown overnight to saturation in minimal medium and then subcultured at a 1:1000 dilution in minimal medium and grown overnight in log phase to an A600 of ∼0.7. Cells were examined at ×100 magnification on polylysine-coated slides using an Axioskop microscope equipped with fluorescence and Nomarski optics (Carl Zeiss, Thornwood, NY). Images were captured with a Cooke charge-coupled device camera and IP Lab Spectrum Software (Biovision Technologies, Exton, PA) (9, 23, 34).
RESULTS
Phosphorylation of Serine 251 Negatively Regulates Ycf1p Function in Vivo—Ycf1p function is known to be positively regulated by phosphorylation at Ser-908 and Thr-911 within the ABC core (Fig. 1A) (35). Among phosphopeptides reported in recent high throughput yeast screens were two that contained a novel phosphorylation site at Ser-251 within the NTE of Ycf1p (Fig. 1A) (39, 40). To determine if phosphorylation at Ser-251 regulates Ycf1p function, we generated a Ycf1p-S251A mutation to inhibit phosphorylation and a Ycf1p-S251E mutation to mimic phosphorylation.
C-terminally GFP-tagged versions of these mutants were expressed in a Δycf1 strain background along with WT Ycf1p and the previously described phosphorylation mutant Ycf1p-S908A,T911A (35). All of these proteins are expressed roughly the same expression level and correctly localized to the vacuole membrane (Fig. 1, B and C, respectively). The somewhat diminished expression of Ycf1p-S908A,T911A seen here (∼50% of WT; Fig. 1B, compare lanes 2 and 5) can also be inferred from a study published by others (35). A slight mobility shift is also evident for the Ycf1p-S908A,T911A mutant protein, as was observed previously. The mutation of S251A did not result in a detectable SDS-PAGE mobility shift for full-length Ycf1p or the N-terminal fragment4; however, a mobility shift due to phosphorylation is often not detectable (43–45).
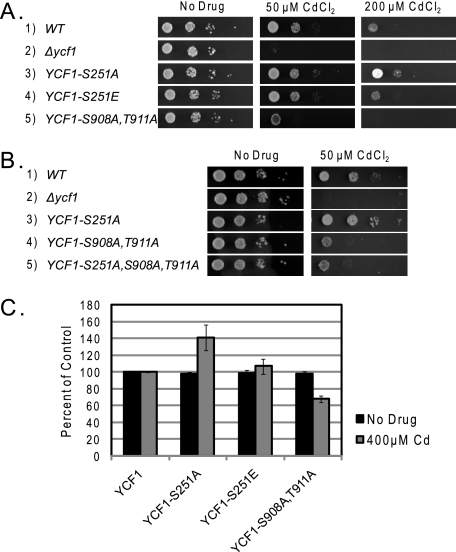
We examined the ability of strains expressing GFP-tagged WT and mutant Ycf1p to grown on plates containing cadmium. Surprisingly, the Ycf1p-S251A mutant is more resistant to cadmium than the strain expressing WT Ycf1p (Fig. 2A, compare rows 1 and 3). This result is the opposite of that from the Ycf1p-S908A,T911A mutant (Fig. 2A, compare rows 1 and 5), which is more sensitive to cadmium than the WT but not as sensitive as the Δycf1 strain (row 2). The resistance to cadmium associated with the expression of Ycf1p-S251A is reversed to approximately that of the WT (Ser-251) level in a strain expressing Ycf1p-S251E (Fig. 2A, compare rows 1 and 4). The increased cadmium resistance observed for the Ycf1p-S908A,T911A mutant is not altered by the addition of the S251A mutation (Fig. 2B, compare rows 4 and 5), suggesting that regulation of Ycf1p activity via phosphorylation of Ser-251 is dependent upon the functionality imparted by phosphorylation of Ser-908 and Thr-911.
FIGURE 2.
Ycf1p-S251A exhibits increased resistance to cadmium as compared with WT Ycf1p. The ability of strains expressing WT and mutant forms of Ycf1p to grow on CdCl2-containing SC plates was examined by spot dilution (A and B) or by growth in liquid media containing CdCl2 (C), as described under “Experimental Procedures.” Tests were performed in triplicate, and error bars are indicated. Yeast strains used were SM4460, SM5270, SM5280, SM5506, SM5507, SM5508, and SM5546.
We also carried out these studies in liquid media in which cadmium was present and observed an analogous relationship in the growth of the single and double mutant strains (Fig. 2C). These results suggest that phosphorylation of Ser-251 negatively regulates Ycf1p function in vivo.
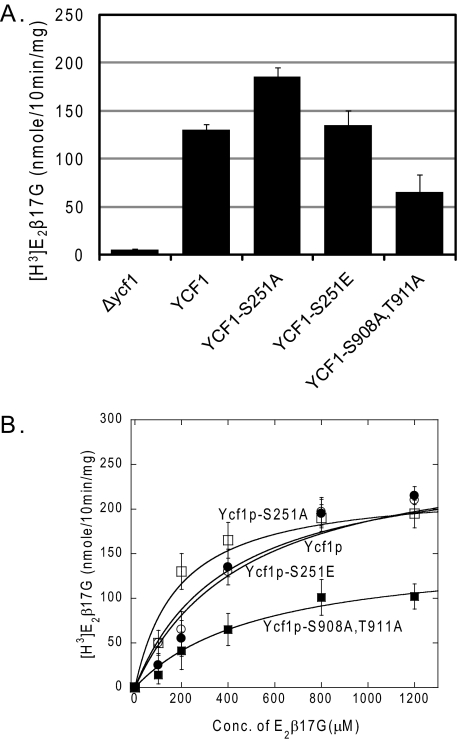
Phosphorylation of Ycf1p at Residue Ser-251 Negatively Regulates Transport Activity in Vitro—To determine whether the increased cadmium resistance of the Ycf1p-S251A mutant is directly due to increased Ycf1p function, we measured Ycf1p transport activity using an in vitro transport assay and a well characterized Ycf1p substrate, E2β17G (34). Assays were performed with vesiculated vacuoles derived from strains expressing no Ycf1p, WT Ycf1p, and various Ycf1p phosphorylation site mutants. The results shown in Fig. 3 reflect the in vivo Cd2+ resistance findings. Vacuoles derived from Ycf1p-S251A strains have greater transport activity than WT, whereas Ycf1p-S251E-derived vacuoles have transport activity similar to that of WT (Fig. 3A). These findings further support the conclusion that phosphorylation at Ser-251 promotes negative regulation of Ycf1p function. In comparison, vacuoles derived from a Ycf1p-S908A,T911A strain have decreased transport activity, in agreement with what has been reported previously (35).
FIGURE 3.
Ycf1p-S251A exhibits enhanced transport activity in vitro as compared with WT Ycf1p. A, Ycf1p-dependent transport of [3H]E2β17G into vacuoles derived from strains expressing WT and mutant Ycf1p was measured as described under “Experimental Procedures.” Experiments were conducted in triplicate or quintuplet, and error bars represent mean ± S.D. B, Michaelis-Menten kinetics of Ycf1p-dependent transport of [3H]E2β17G by vacuoles derived from strains expressing WT Ycf1p (open circle), Ycf1p-S251A (open square), Ycf1p-S251E (closed circle), and Ycf1p-S908A,T911A (closed square) were measured as described under “Experimental Procedures.” Values for Kmapp, Vmax,app, and relative transport efficiency (Vmax,app/Kmapp) were extrapolated from plots of transport assays and are shown in Table 3. All data points were calculated from an n = 3 or 4, and error bars represent mean ± S.D. Yeast strains used were SM5270, SM5280, SM5506, SM5507, and SM5508.
To gain insight into the mechanism by which phosphorylation of Ser-251 regulates Ycf1p transporter activity, we determined the Michaelis-Menten transport kinetics of the WT and mutant Ycf1p proteins (Fig. 3B and Table 3). Ycf1p-S251A is ∼2 fold more efficient in its ability to transport E2β17G than WT (Table 3, last column), largely because of a change in the Kmapp for substrate binding. This trend is reversed by the Ycf1p-S251E mutation. Ycf1p-S908A,T911A kinetics have not been reported previously. Here we show this mutation is ∼2.5 fold less efficient for transport than that of WT (Fig. 3B and Table 3, last column). The decrease in transport efficiency for Ycf1p-S908A,T911A is the result of both an increase in Kmapp for the substrate (E2β17G) and a decrease in Vmax,app as compared with WT (Fig. 3B and Table 3). Together these experiments suggest that phosphorylation in different regions within Ycf1p has opposing effects: phosphorylation of Ser-251 in the NTE appears to negatively regulate Ycf1p function, whereas phosphorylation of Ser-908 and Thr-911 in the ABC core positively regulates Ycf1p function.
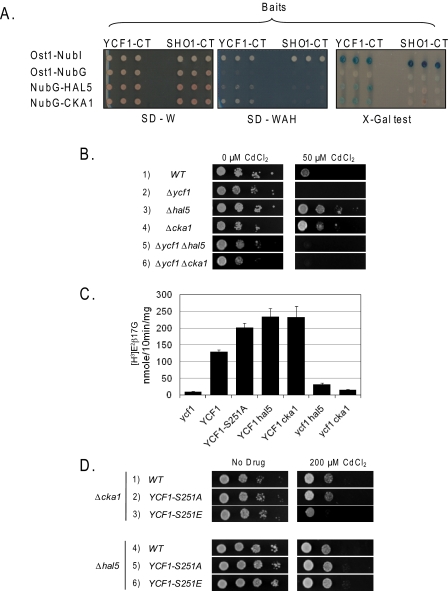
Identification of Hal5p and Cka1p as Ycf1p Protein Interactors by iMYTH—To seek a kinase(s) that could be involved in the negative regulation of Ycf1p, we used a modified yeast two-hybrid “split ubiquitin” screen called the integrated membrane yeast two-hybrid assay. iMYTH is specifically designed for detecting membrane protein interactors and has proven extremely useful in identifying the physiologically relevant Ycf1p interactor, Tus1p, which positively regulates Ycf1p transport activity (34). Complete details of the iMYTH screening procedure are provided under “Experimental Procedures” and elsewhere (34). In the library used here, and in contrast to our recently published work (34), cDNA fragments of ∼0.8 to 5 kb in length were inserted C-terminally to the NubG sequence, generating the library in NubG-X orientation (where X is a cDNA insert). This library was introduced by transformation into the yeast strain THY AP4-YCF1-CT expressing the endogenously tagged YCF1-CT “bait.” From an iMYTH screen of ∼8 × 106 yeast transformants, 18 novel Ycf1p interactors were identified, among which two new kinases were found (Fig. 4A). One of these is Cka1p, a catalytic component of the yeast multisubunit casein kinase II (CKII) (46), and the other is Hal5p, a Ser/Thr kinase associated with yeast halotolerance (Fig. 4A) (47). In this study, we focused our attention on these two new kinases; the analysis of the remaining 16 newly identified Ycf1p interactors will be described elsewhere.
FIGURE 4.
Identification of Ycf1p-protein interactors, Hal5p and Cka1p, by iMYTH. Deletion of Hal5p and Cka1p resulted in increases transport by Ycf1p, similar to Ycf1p-S251A. A, iMYTH identifies Hal5p and Cka1p kinases as novel Ycf1p-specific interactors. The yeast reporter strain THY AP4, expressing either YCF1-CT (left) or SHO1-CT (right) bait constructs from the chromosome, was transformed with the indicated (TRP1 marked) NubG prey plasmids. The growth of yeast cells expressing the YCF1-CT and SHO1-CT chimeras with the indicated NubG fusions was monitored on agar plates lacking tryptophan (SD-W, left) and tryptophan, adenine, and histidine (SD-WAH, middle). Three independent colonies were pinned onto SD-W- and SD-WAH-selective plates prior to assessment of β-galactosidase activity using an 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) plate test (right). Both Cka1p and Hal5p kinases show specific activation of the reporter gene system only in the presence of YCF1-CT bait but not in the presence of an unrelated yeast plasma membrane protein, Sho1p, fused to the C terminus of ubiquitin followed by an artificial transcription factor, LexA-VP16, collectively called SHO1-CT. Please note that the control construct (Ost1-NubI) is designed to activate the yeast reporter system independently of any protein-protein interaction because of the high affinity of NubI to associate with the C terminus of ubiquitin and form active ubiquitin irrespective of a protein-protein interaction. The OST1-Nub control indicates that the YCF1-CT and SHO1-CT chimeric bait proteins are expressed and the proteins are properly inserted into the membrane. B, growth of the indicated strains on plates containing CdCl2 was examined by spot dilution assay. Yeast strains used are SM5270, SM5509, SM5510, SM5515, and SM5516. C, in vitro transport activity of vacuoles derived from the indicated strains was measured as described for Fig. 3A and under “Experimental Procedures.” Experiments were conducted in triplicate or quintuplet, and error bars represent mean ± S.D. Yeast strains used in this experiment are SM5270, SM5511, SM5512, SM5513, and SM5514. D, growth of the indicated strains on plates containing CdCl2 was examined by spot dilution assay. Yeast strains used are SM5519, SM5520, SM5521, SM5522, SM5523, and SM5524.
Deletion of Hal5p or Cka1p Results in Increased Cadmium Resistance in Vivo and Increased Transport Activity in Vitro—To determine whether the deletion of the CKA1 and HAL5 genes affects cadmium resistance, we carried out growth tests by spot dilution on yeast agar plates containing 50 μm Cd2+ (Fig. 4B). We observed a significant increase in cadmium resistance for both the Δhal5 and the Δcka1 mutants (Fig. 4B, compare rows 3 and 4 with row 1). Increased resistance is dependent upon Ycf1p, as it does not occur in the Δycf1 Δhal5 and Δycf1 Δcka1double mutant strains (Fig. 4B, compare rows 5 and 6 with rows 3 and 4). These experiments indicate that Hal5p and Cka1p either directly or indirectly negatively regulate Ycf1p function.
To test whether the increased cadmium resistance observed in the Δhal5 and Δcka1 mutants in vivo is due to increased Ycf1p function, we conducted in vitro transport assays with the Ycf1p substrate [3H]E2β17G and vesiculated vacuoles derived from the Δhal5 and Δcka1 strains (Fig. 4C). If Hal5p and Cka1p kinases are negative regulators of Ycf1p function, then vacuoles derived from Δhal5 and Δcka1 deletion strains should exhibit increased Ycf1p-dependent transport. This is indeed the case, as shown in Fig. 4C. Notably, the increased activity observed for the kinase deletions is similar to the increased activity observed for strains expressing the Ycf1p-S251A mutant. Importantly when Ycf1p is absent in the Δhal5 and Δcka1 strains (Fig. 4C, ycf1 hal5 and ycf1 cka1), the kinase deletions show almost no transport activity, indicating that the increased activity observed for Δhal5 and Δcka1 is Ycf1p-dependent and not due to activation of another transporter. These experiments indicate that the Hal5p and Cka1p kinases negatively regulate Ycf1p activity.
To examine whether Hal5p and/or Cka1p regulate Ycf1p through phosphorylation of Ser-251, the phenotypes of the hal5Δ and cka1Δ strains expressing WT and mutant forms of Ycf1p were examined on cadmium-containing plates (Fig. 4D). Interestingly, in the cka1Δ strain, the cadmium resistance of WT Ycf1p is indistinguishable from that of Ycf1p-S251A (Fig. 4D, rows 1 and 2), possibly because WT Ycf1p cannot be phosphorylated at Ser-251 in the cka1Δ mutant, and thus negative regulation is lifted. In contrast, the kinase-independent mutant Ycf1p-S251E shows reduced cadmium resistance (Fig. 4D, row 3). These results are consistent with the possibility that Cka1p could negatively regulate Ycf1p function through phosphorylation of Ser-251, either directly or indirectly. In contrast, growth of the hal5Δ strains expressing either WT or mutant Ycf1p constructs grew similarly on cadmium, suggesting that Hal5p does not act through Ser-251 (Fig. 4D, lower panel, rows 4–6).
DISCUSSION
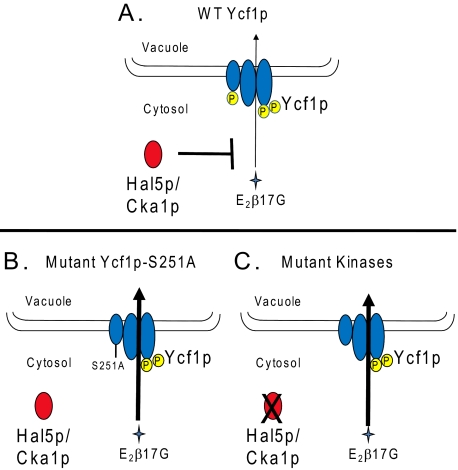
The ABCC transporter Ycf1p is critical for the detoxification of heavy metals in yeast. One of our long-term goals is to understand how Ycf1p activity is regulated. Like other members of the ABCC subfamily of transporters, including MRP1 and MRP2 in mammalian cells, Ycf1p contains an NTE with five membrane spans (MSD0) and a cytosolic loop (L0) in addition to its ABC core transporter domain (48, 49). Our studies and those of others have shown that L0 plays a key role in activity, because when L0 is deleted, Ycf1p, ABCC1 (MRP1), and ABCC2 (MRP2) are properly localized but exhibit no transport activity (8–10). This finding is somewhat surprising, as the ABC core suffices for the activity of most other ABC transporters, and it suggests a regulatory role for the L0 domain in the ABCC subfamily (4, 50). Two recent high throughput study identified residue Ser-251 in L0 of Ycf1p as a site of phosphorylation (39, 40). In the present study we have provided evidence that phosphorylation of Ser-251 negatively regulates Ycf1p activity. We show that cells expressing the mutant protein Ycf1p-S251A, in which phosphorylation should be abrogated, exhibited higher Ycf1p activity than cells expressing WT Ycf1p, both in vivo (cadmium resistance) and in vitro (E2β17G transport). On the other hand, cells expressing Ycf1-S251E, expected to mimic the phosphorylated form, exhibited wild-type activity. Thus, phosphorylation of L0 within the NTE appears to negatively regulate Ycf1p, as indicated in the model shown in Fig. 5A, and when phosphorylation is absent, the activity of Ycf1p increases (Fig. 5B).
FIGURE 5.
Hal5p, Cka1p, and phosphorylation at Ser-251 negatively regulate Ycf1p function. A, in a WT strain, Ycf1p is negatively regulated by phosphorylation at Ser-251 within the L0 domain of the NTE, which diminishes Ycf1p function (thin arrow, weak transport). In addition, Ycf1p interacts with the Cka1p subunit of CKII and the Hal5p kinase, which also negatively regulate Ycf1p function. These kinases could negatively regulate Ycf1p via phosphorylation at Ser-251; genetic evidence suggests that this may be the case for Cka1p (see Fig. 4D). Alternatively, they may act in an as yet undetermined way to decrease Ycf1p function, as discussed in the text. B and C, mutation of Ser-251 to alanine or deletion of the Ycf1p kinase interactor genes HAL5 and CKA1 results in increased Ycf1p function (thick arrow, strong transport). Apparently, in both cases, negative regulation is abolished.
Previous studies have shown that phosphorylation within the ABC core of Ycf1p at residues Ser-908 and Thr-911 positively regulates Ycf1 function (26, 35). Mutation of both sites to alanine decreases cadmium resistance and Ycf1p-dependent transport activity, a result recapitulated in this study. Thus, Ycf1p appears to be highly regulated by phosphorylation, both negatively, in the NTE, and positively, in the ABC core domain.
The L0 domain of the NTE is critical for ABCC function (7, 11, 51). MRP1 cross-linking studies suggest that L0 contributes to substrate binding, either directly or indirectly, by facilitating an interaction of substrate and the ABC core (10, 14). Phosphorylation of L0 at Ser-251 could inhibit substrate interaction with the Ycf1p core, accounting for the decrease in Km that we observed here for the S251A mutant. The ability of the L0 to interact with the membrane via two predicted amphipathic helices is required for MRP1 function. These amphipathic helices are conserved throughout the ABCC subfamily, including Ycf1p. Ser-251 of Ycf1p lies at the N-terminal proximal end of amphipathic helix 2 (11). Further, this region of L0 is required for homodimerization of MRP1. The inability of MRP1 to form a dimer appears to be required for function (15). Therefore, it is tempting to speculate that phosphorylation of Ser-251 could inhibit the interaction of amphipathic helix 2 with the membrane, thereby decreasing protein function. Alternatively, phosphorylation of Ser-251 may inhibit homodimerization, resulting in decreased Ycf1p function. In contrast, we observed an increased Km and decreased Vmax of transport for the S908A,T911A double mutant, suggesting that phosphorylation within the ABC core could negatively affect both substrate binding and transport.
We were interested in identifying the kinases that regulate Ycf1p activity. To do so, we used iMYTH technology, which we have previously shown to be an extremely effective method of identifying interactors (i.e. Tus1p) of membrane proteins such as Ycf1p (34). Here, we screened among kinase interactors for those that negatively regulate transporter function in a manner similar to phosphorylation of Ser-251. In this way, we identified genes encoding the kinase Hal5p and the catalytic subunit Cka1p of the serine/threonine kinase CKII (46). Hal5p plays a role in salt and pH tolerance in yeast through regulating the trafficking of the potassium transporters Trk1p and Trk2p (47, 52). CKII is involved in multiple processes, including the cell cycle, cell polarization, and transcription regulation (53). The residues flanking Ser-251 in Ycf1p conform to the CKII consensus sequence, which is conserved from yeast to humans (Fig. 6) Furthermore, the L0 region of the human ABCC6 (MRP6) shares strong homology with that of Ycf1p (11), including Ser-244 in ABCC6, which is positionally conserved with Ser-251 in Ycf1p. Both lie within a CKII consensus sequence, and Ser-251 of Ycf1p and Ser-244 in ABCC6 were predicted to be sites of CKII phosphorylation by the programs Scansite and Group-based Phosphorylation Scoring (GPS) (Fig. 6) (54–57). It will therefore be interesting to determine whether mutation of Ser-244 impacts ABCC6 function. Mutations in Ser-244 have not been associated with pseudoxanthoma elasticum (58); however, an interesting possibility is that mutations at this site could potentially modify the severity of pseudoxanthoma elasticum phenotypes.
FIGURE 6.
A CKII consensus that contains Ycf1p-Ser-251 is conserved from yeast to humans. The protein sequence of Ycf1p from S. cerevisiae, human ABCC6 (MRP6), and Ycf1p from Saccharomyces bayanus were aligned using ClustalW. Conserved residues have been highlighted using BOXSHADE. The arrow indicates Ser-251 in Ycf1p and the conserved homologous residues in the alignment. The residues surrounding Ser-251 fit a CKII consensus sequence, which is shown below the alignment.
The single deletion mutants Δhal5 and Δcka1 exhibited increased resistance to cadmium in vivo as compared with WT cells; vacuoles derived from these mutants have increased Ycf1p-dependent transport activity in vitro, a pattern similar to a strain expressing Ycf1p-S251A. These results suggest that the Hal5p and CKII kinases negatively regulate Ycf1p function, but they could do so either by similar or distinct mechanisms. Further genetic evidence (Fig. 4) provides support for the possibility that Cka1p may directly or indirectly phosphorylate Ser-251, as WT Ycf1p function is indistinguishable from Ycf1p-S251A in a cka1Δ strain as measured by cadmium resistance. However, because we currently cannot directly measure phosphorylation of Ser-251, it would be premature to draw the conclusion that Cka1p phosphorylates this residue. It is certainly possible that Cka1p does not phosphorylate Ycf1p directly but instead influences an as yet unknown factor that is the direct regulator of Ycf1p phosphorylation or in some other way negatively impacts Ycf1p function (but does not affect the expression level or vacuolar localization of Ycf1p). Taken together, the results presented here and elsewhere suggest an important mechanistic role of phosphorylation in negatively and positively regulating Ycf1p transporter function. Future studies will focus on directly establishing which kinases mediate phosphorylation at specific sites in Ycf1p and developing a complete phosphoprotein map of Ycf1p.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51508 (to S. M.) and National Institutes of Health Postdoctoral Award GM077024 (to C. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ABC, ATP-binding cassette; MRP, multidrug resistance-associated protein; CFTR, cystic fibrosis transmembrane conductance regulator; MSD, membrane-spanning domain; NBD, nucleotide-binding domain; NTE, N-terminal extension; WT, wild type; E2β17G, estradiol-β-17-glucuronide; AMP-PCP, adenosine 5′-(β,γ-methylenetriphosphate); Mes, 4-morpholineethanesulfonic acid; iMYTH, integrated membrane yeast two-hybrid; CKII, casein kinase II; GFP, green fluorescent protein.
C. M. Paumi and S. Michaelis, unpublished data.
References
- 1.Dean, M. (2005) Methods Enzymol. 400 409-429 [DOI] [PubMed] [Google Scholar]
- 2.Gottesman, M. M., and Ambudkar, S. V. (2001) J. Bioenerg. Biomembr. 33 453-458 [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. P., and Deeley, R. G. (2006) Trends Pharmacol. Sci. 27 438-446 [DOI] [PubMed] [Google Scholar]
- 4.Haimeur, A., Conseil, G., Deeley, R. G., and Cole, S. P. (2004) Curr. Drug Metab. 5 21-53 [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. P., Bhardwaj, G., Gerlach, J. H., Mackie, J. E., Grant, C. E., Almquist, K. C., Stewart, A. J., Kurz, E. U., Duncan, A. M., and Deeley, R. G. (1992) Science 258 1650-1654 [DOI] [PubMed] [Google Scholar]
- 6.Borst, P., and Elferink, R. O. (2002) Annu. Rev. Biochem. 71 537-592 [DOI] [PubMed] [Google Scholar]
- 7.Bakos, E., Evers, R., Calenda, G., Tusnady, G. E., Szakacs, G., Varadi, A., and Sarkadi, B. (2000) J. Cell Sci. 113 4451-4461 [DOI] [PubMed] [Google Scholar]
- 8.Fernandez, S. B., Hollo, Z., Kern, A., Bakos, E., Fischer, P. A., Borst, P., and Evers, R. (2002) J. Biol. Chem. 277 31048-31055 [DOI] [PubMed] [Google Scholar]
- 9.Mason, D. L., and Michaelis, S. (2002) Mol. Biol. Cell 13 4443-4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian, Y. M., Grant, C. E., Westlake, C. J., Zhang, D. W., Lander, P. A., Shepard, R. L., Dantzig, A. H., Cole, S. P., and Deeley, R. G. (2002) J. Biol. Chem. 277 35225-35231 [DOI] [PubMed] [Google Scholar]
- 11.Westlake, C. J., Qian, Y. M., Gao, M., Vasa, M., Cole, S. P., and Deeley, R. G. (2003) Biochemistry 42 14099-14113 [DOI] [PubMed] [Google Scholar]
- 12.Westlake, C. J., Cole, S. P., and Deeley, R. G. (2005) Mol. Biol. Cell 16 2483-2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren, X. Q., Furukawa, T., Yamamoto, M., Aoki, S., Kobayashi, M., Nakagawa, M., and Akiyama, S. (2006) J. Biochem. 140 313-318 [DOI] [PubMed] [Google Scholar]
- 14.Ren, X. Q., Furukawa, T., Aoki, S., Nakajima, T., Sumizawa, T., Haraguchi, M., Chen, Z. S., Kobayashi, M., and Akiyama, S. (2001) J. Biol. Chem. 276 23197-23206 [DOI] [PubMed] [Google Scholar]
- 15.Yang, Y., Liu, Y., Dong, Z., Xu, J., Peng, H., Liu, Z., and Zhang, J. T. (2007) J. Biol. Chem. 282 8821-8830 [DOI] [PubMed] [Google Scholar]
- 16.Sharma, K. G., Mason, D. L., Liu, G., Rea, P. A., Bachhawat, A. K., and Michaelis, S. (2002) Eukaryot. Cell 1 391-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wemmie, J. A., Szczypka, M. S., Thiele, D. J., and Moye-Rowley, W. S. (1994) J. Biol. Chem. 269 32592-32597 [PubMed] [Google Scholar]
- 18.Chappe, V., Hinkson, D. A., Zhu, T., Chang, X. B., Riordan, J. R., and Hanrahan, J. W. (2003) J. Physiol. 548 39-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chappe, V., Irvine, T., Liao, J., Evagelidis, A., and Hanrahan, J. W. (2005) EMBO J. 24 2730-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell, L. D., Borchardt, R., Kole, J., Kaz, A. M., Randak, C., and Cohn, J. A. (2004) Biochem. J. 378 151-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csanady, L., Seto-Young, D., Chan, K. W., Cenciarelli, C., Angel, B. B., Qin, J., McLachlin, D. T., Krutchinsky, A. N., Chait, B. T., Nairn, A. C., and Gadsby, D. C. (2005) J. Gen. Physiol. 125 171-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wemmie, J. A., and Moye-Rowley, W. S. (1997) Mol. Microbiol. 25 683-694 [DOI] [PubMed] [Google Scholar]
- 23.Mason, D. L., Mallampalli, M. P., Huyer, G., and Michaelis, S. (2003) Eukaryot. Cell 2 588-598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Z. S., Lu, Y. P., Zhen, R. G., Szczypka, M., Thiele, D. J., and Rea, P. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 42-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Z. S., Szczypka, M., Lu, Y. P., Thiele, D. J., and Rea, P. A. (1996) J. Biol. Chem. 271 6509-6517 [DOI] [PubMed] [Google Scholar]
- 26.Szczypka, M. S., Wemmie, J. A., Moye-Rowley, W. S., and Thiele, D. J. (1994) J. Biol. Chem. 269 22853-22857 [PubMed] [Google Scholar]
- 27.Tommasini, R., Evers, R., Vogt, E., Mornet, C., Zaman, G. J., Schinkel, A. H., Borst, P., and Martinoia, E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6743-6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri, B., Ingavale, S., and Bachhawat, A. K. (1997) Genetics 145 75-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascolo, L., Petrovic, S., Cupelli, F., Bruschi, C. V., Anelli, P. L., Lorusso, V., Visigalli, M., Uggeri, F., and Tiribelli, C. (2001) Biochem. Biophys. Res. Commun. 282 60-66 [DOI] [PubMed] [Google Scholar]
- 30.Ghosh, M., Shen, J., and Rosen, B. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5001-5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gueldry, O., Lazard, M., Delort, F., Dauplais, M., Grigoras, I., Blanquet, S., and Plateau, P. (2003) Eur. J. Biochem. 270 2486-2496 [DOI] [PubMed] [Google Scholar]
- 32.Jungwirth, H., and Kuchler, K. (2006) FEBS Lett. 580 1131-1138 [DOI] [PubMed] [Google Scholar]
- 33.Song, W. Y., Sohn, E. J., Martinoia, E., Lee, Y. J., Yang, Y. Y., Jasinski, M., Forestier, C., Hwang, I., and Lee, Y. (2003) Nat. Biotechnol. 21 914-919 [DOI] [PubMed] [Google Scholar]
- 34.Paumi, C. M., Menendez, J., Arnoldo, A., Engels, K., Iyer, K. R., Thaminy, S., Georgiev, O., Barral, Y., Michaelis, S., and Stagljar, I. (2007) Mol. Cell 26 15-25 [DOI] [PubMed] [Google Scholar]
- 35.Eraso, P., Martinez-Burgos, M., Falcon-Perez, J. M., Portillo, F., and Mazon, M. J. (2004) FEBS Lett. 577 322-326 [DOI] [PubMed] [Google Scholar]
- 36.Iyer, K., Burkle, L., Auerbach, D., Thaminy, S., Dinkel, M., Engels, K., and Stagljar, I. (2005) Sci. STKE 2005, p13. [DOI] [PubMed]
- 37.Stagljar, I., and Fields, S. (2002) Trends Biochem. Sci. 27 559-563 [DOI] [PubMed] [Google Scholar]
- 38.Li, X., Gerber, S. A., Rudner, A. D., Beausoleil, S. A., Haas, W., Villen, J., Elias, J. E., and Gygi, S. P. (2007) J. Proteome Res. 6 1190-1197 [DOI] [PubMed] [Google Scholar]
- 39.Smolka, M. B., Albuquerque, C. P., Chen, S. H., and Zhou, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10364-10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi, A., Huttenhower, C., Geer, L. Y., Coon, J. J., Syka, J. E., Bai, D. L., Shabanowitz, J., Burke, D. J., Troyanskaya, O. G., and Hunt, D. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2193-2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke, D., Dawson, D., and Stearns, T. (2000) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 42.Roberts, C. J., Raymond, C. K., Yamashiro, C. T., and Stevens, T. H. (1991) Methods Enzymol. 194 644-661 [DOI] [PubMed] [Google Scholar]
- 43.Cheng, S. H., Rich, D. P., Marshall, J., Gregory, R. J., Welsh, M. J., and Smith, A. E. (1991) Cell 66 1027-1036 [DOI] [PubMed] [Google Scholar]
- 44.Kelm, K. B., Huyer, G., Huang, J. C., and Michaelis, S. (2004) Traffic 5 165-180 [DOI] [PubMed] [Google Scholar]
- 45.King, S. A., and Sorscher, E. J. (2000) Biochemistry 39 9868-9875 [DOI] [PubMed] [Google Scholar]
- 46.Chen-Wu, J. L., Padmanabha, R., and Glover, C. V. (1988) Mol. Cell. Biol. 8 4981-4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulet, J. M., Leube, M. P., Kron, S. J., Rios, G., Fink, G. R., and Serrano, R. (1999) Mol. Cell. Biol. 19 3328-3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taglicht, D., and Michaelis, S. (1998) Methods Enzymol. 292 130-162 [DOI] [PubMed] [Google Scholar]
- 49.Deleted in proof
- 50.Higgins, C. F. (2001) Res. Microbiol. 152 205-210 [DOI] [PubMed] [Google Scholar]
- 51.Karwatsky, J. M., and Georges, E. (2004) Curr. Med. Chem. Anticancer Agents 4 19-30 [DOI] [PubMed] [Google Scholar]
- 52.Mulet, J. M., Alejandro, S., Romero, C., and Serrano, R. (2004) Yeast 21 569-582 [DOI] [PubMed] [Google Scholar]
- 53.Glover, C. V., III (1998) Prog. Nucleic Acids Res. Mol. Biol. 59 95-133 [DOI] [PubMed] [Google Scholar]
- 54.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635-3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obenauer, J. C., and Yaffe, M. B. (2004) Methods Mol. Biol. 261 445-468 [DOI] [PubMed] [Google Scholar]
- 56.Zhou, F. F., Xue, Y., Chen, G. L., and Yao, X. (2004) Biochem. Biophys. Res. Commun. 325 1443-1448 [DOI] [PubMed] [Google Scholar]
- 57.Xue, Y., Zhou, F., Zhu, M., Ahmed, K., Chen, G., and Yao, X. (2005) Nucleic Acids Res. 33 W184-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfendner, E. G., Vanakker, O. M., Terry, S. F., Vourthis, S., McAndrew, P. E., McClain, M. R., Fratta, S., Marais, A.-S., Hariri, S., Coucke, P. J., Ramsay, M., Viljoen, D., Terry, P. F., De Paepe, A., Uitto, J., and Bercovitch, L. G. (2007) J. Med. Genet. 44 621-628 [DOI] [PMC free article] [PubMed] [Google Scholar]