Abstract
Exported mRNAs are targeted for translation or can undergo degradation by several decay mechanisms. The 5′→3′ degradation machinery localizes to cytoplasmic P bodies (PBs). We followed the dynamic properties of PBs in vivo and investigated the mechanism by which PBs scan the cytoplasm. Using proteins of the decapping machinery, we asked whether PBs actively scan the cytoplasm or whether a diffusion-based mechanism is sufficient. Live-cell imaging showed that PBs were anchored mainly to microtubules. Quantitative single-particle tracking demonstrated that most PBs exhibited spatially confined motion dependent on microtubule motion, whereas stationary PB pairs were identified at the centrosome. Some PBs translocated in long-range movements on microtubules. PB mobility was compared with mitochondria, endoplasmic reticulum, peroxisomes, SMN bodies, and stress granules, and diffusion coefficients were calculated. Disruption of the microtubule network caused a significant reduction in PB mobility together with an induction of PB assembly. However, FRAP measurements showed that the dynamic flux of assembled PB components was not affected by such treatments. FRAP analysis showed that the decapping enzyme Dcp2 is a nondynamic PB core protein, whereas Dcp1 proteins continuously exchanged with the cytoplasm. This study reveals the mechanism of PB transport, and it demonstrates how PB assembly and disassembly integrate with the presence of an intact cytoskeleton.
INTRODUCTION
Gene expression begins with the synthesis of mRNA molecules in the nucleus. After processing events, transcripts are exported to the cytoplasm where they can face several posttranscriptional fates, elicited by a balance between cytoplasmic translation and mRNA degradation pathways. Quality control pathways regulate the degradation of mRNAs and facilitate their sequestration or translational repression (Meyer et al., 2004). In eukaryotes, mRNA degradation typically begins with deadenylation, and then either of two major pathways is used. The exosome protein complex degrades mRNAs in the 3′→5′ direction, whereas the 5′→3′ direction involves other factors, including a decapping enzyme followed by the Xrn1 exonuclease (Parker and Song, 2004).
The removal of the cap structure irreversibly marks the mRNA for degradation. The decapping process is tightly regulated biochemically and spatially. It was first discovered that the Xrn1 nuclease localizes in discrete cytoplasmic foci in eukaryotic cells (Bashkirov et al., 1997). Some years later, a decapping protein termed Dcp2 was found to colocalize in Xrn1-foci (Ingelfinger et al., 2002; Lykke-Andersen, 2002; van Dijk et al., 2002), finally leading to the understanding that both yeast (Sheth and Parker, 2003) and mammalian cells (Cougot et al., 2004) contain discrete areas in which mRNA decapping and 5′→3′ degradation can occur (Sheth and Parker, 2006). These cytoplasmic foci are now widely known as P bodies (PBs), and they have been referred to as Dcp-bodies, processing bodies, mRNA-decay foci, and GW182 bodies. Since these initial discoveries, a plethora of proteins have been found to reside within P bodies, linking them to processes of mRNA degradation, storage, translational repression, or silencing (Eulalio et al., 2007a; Jakymiw et al., 2007; Parker and Sheth, 2007).
Studies in yeast and mammalian cells provide a detailed, yet probably partial inventory of the conserved components that are found within PBs, and they demonstrate that factors of the 5′→3′ degradation pathway reside within PBs, whereas the 3′→5′ exosomal proteins portray a diffuse cytoplasmic distribution. This suggests a level of regulation based on cytosolic spatial compartmentalization of the two processes (Eulalio et al., 2007a). Several findings suggest that PBs are dynamic structures. Different-sized PBs are observed in fixed mammalian cells, although the significance of this remains unknown. Changes in PB number and size per cell correlate to the cell cycle, to the proliferation status of the cell, and to the availability of nutrients (Yang et al., 2004). In Saccharomyces cerevisiae, stresses such as glucose deprivation, osmotic stress, UV irradiation, or inhibition of translation initiation lead to PB assembly, whereas translation elongation inhibition dissociates PBs (Sheth and Parker, 2003; Brengues et al., 2005; Teixeira et al., 2005). In mammalian cells, cycloheximide and actinomycin D (ActD) treatments result in a gradual reduction in PBs (Cougot et al., 2004; Andrei et al., 2005; Ferraiuolo et al., 2005). Small interfering RNA (siRNA) to PB components GW182, Ge-1 (Hedls), LSm1, RCK/p54, eIF4E-T, Drosha, and other microRNA processing factors, cause the disassembly of PBs (Yang et al., 2004; Andrei et al., 2005; Ferraiuolo et al., 2005; Yu et al., 2005; Chu and Rana, 2006; Pauley et al., 2006). During stress, PBs can transiently associate with stress granules (SGs), structures that accumulate stalled translational preinitiation complexes (Kedersha et al., 2005). Photobleaching experiments of several PB components showed that the proteins dynamically exchange with the cyotsolic pool (Andrei et al., 2005; Kedersha et al., 2005; Leung et al., 2006). The significance of the changes in PB size and number are unknown as yet, and they may be an indication of different types of protein–protein and/or protein–mRNA interactions that comprise a functional PB structure.
The Dcp proteins Dcp1 and Dcp2 are conserved PB components in yeast, Caenorhabditis elegans, Drosophila, and mammalian cells (Andrei et al., 2005; Lall et al., 2005; Behm-Ansmant et al., 2006; Squirrell et al., 2006). Dcp2 is the decapping enzyme that cleaves the cap structure at the 5′ end of mRNA, releasing 7-methyl-GDP. Studies in S. cerevisiae deciphered the crystal structure of the protein (She et al., 2006, 2008) showing that Dcp1p can interact with Dcp2p and stimulate its decapping activity (Beelman et al., 1996; Dunckley and Parker, 1999; Steiger et al., 2003). The human homologues of Dcp1 (hDcp1a and hDcp1b) and Dcp2 (hDcp2) are also involved in decapping (Lykke-Andersen, 2002; van Dijk et al., 2002; Wang et al., 2002; Piccirillo et al., 2003). Interestingly, small interfering RNA (siRNA) to hDcp2 does not abolish PB structures, but has a stimulatory effect instead (Andrei et al., 2005). hDcp1a and hDcp1b proteins are encoded by separate genes located on different chromosomes. They share ∼70% homology in their N termini and ∼30% homology in their entire length (Lykke-Andersen, 2002). Most studies use Dcp1a as a PB marker and expect Dcp1b to behave as Dcp1a (Fenger-Gron et al., 2005).
Although several studies have indicated that PBs are dynamic structures, a quantitative analysis of PB mobility in living mammalian cells is lacking. This study examines the underlying mechanisms that PBs use for scanning the cytoplasm, asking whether PBs actively roam through the cytoplasm in pursuit of targets, or whether these associations depend on random interactions. We also show how the assembly and disassembly of PBs is coordinated with the presence of an intact microtubule network.
MATERIALS AND METHODS
Plasmids
Plasmids encoding green fluorescent protein (GFP)-Dcp1b, RFP-Dcp1b, GFP-Dcp2, RFP-Dcp2 were obtained from the Sonenberg laboratory. The human Dcp1a open reading frame was amplified by reverse transcription-polymerase chain reaction from U2OS total RNA by using primers ATACTCGAGAGATGGAGGCGCTGAGTCGAGCT and ATAGAATTCGTCAAAA CAAGGATCTGCTGGTA, and subcloned into XhoI and EcoRI restriction sites of the peGFP-C1 vector (Clontech, Mountain View, CA). GFP-TIAR was a gift from S. Hüttelmaier (University of Halle-Wittenberg, Germany; Stohr et al., 2006), GFP-centrin was from B. Motro (Bar-Ilan University), and GFP-ER was from R. Sarid (Bar-Ilan University).
Cell Culture and Transfections
Human U2OS cells were maintained in low glucose DMEM (Biological Industries, Kibbutz Beit Haemek, Israel) containing 10% fetal bovine serum (HyClone Laboratories, Logan, UT). The following U2OS stable lines were generated: GFP-Dcp1a, GFP-Dcp1b, RFP-Dcp1b, and GFP-Dcp2. Cyan fluorescent protein (CFP)-peroxisome-expressing cells (Darzacq et al., 2006) were induced with 1 μg/ml doxycycline (Sigma-Aldrich, St. Louis, MO). Mitochondria were labeled with MitoTracker Red CMXRos (Invitrogen, Carlsbad, CA).
Immunofluorescence
Immunofluorescence was performed as described previously (Shav-Tal et al., 2005). Primary antibodies were rabbit anti-hDcp1a, rabbit anti-hDcp1b (J. Lykke-Andersen, University of Colorado, Boulder, CO), rabbit anti–hDcp2 (S. Hüttelmaier), mouse anti-Dcp1a (Abnova, Taipei City, Taiwan), anti-GW182, anti-Xrn1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-eIF4E-T (N. Sonenberg), mouse anti-γ-tubulin, mouse anti-α-tubulin (Sigma-Aldrich), rabbit anti-α tubulin (Abcam, Cambridge, MA). Secondary antibodies included Cy5-labeled goat anti-mouse immunoglobulin G (IgG), Cy3-labeled goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), and Alexa488-labeled goat anti-rabbit IgG (Invitrogen). Nuclei were counterstained with Hoechst 33342, and coverslips were mounted in mounting medium.
Microtubule Destabilization and Cell Synchronization
For the various treatments, cells were treated with 600 nM nocodazole, 300 nM vinblastine, 5 μg/ml cycloheximide, 10 μg/ml puromycin, 5 μg/ml actinomycin D, and 500 μM arsenite (Sigma-Aldrich). Because some of the above-mentioned materials are solubilized in dimethyl sulfoxide (DMSO), we verified that DMSO alone had no effect on PB assembly, numbers, movements, and dynamics.
Western Blotting
SDS-polyacrylamide gel electrophoresis and Western blotting were performed as described previously (Shav-Tal et al., 2001). Primary antibodies used were anti-Dcp1a and anti-α-tubulin, and the secondary antibody was a horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (Sigma-Aldrich). Immunoreactive bands were detected by the enhanced chemiluminescence kit (ECL; Pierce Chemical, Rockford, IL).
Fluorescence Microscopy, Live-Cell Imaging, and Data Analysis
Wide-field fluorescence images were obtained using the cellR system based on an Olympus IX81 fully motorized inverted microscope (60× PlanApo objective, 1.42 numerical aperture [NA]) fitted with an Orca-AG charge-coupled device camera (Hamamatsu, Bridgewater, NJ), rapid wavelength switching, and driven by the cellR software. For time-lapse imaging cells were plated on glass-bottomed tissue culture plates (MatTek, Ashland, MA) in medium containing 10% fetal calf serum (FCS) at 37°C. The microscope is equipped with an on-scope incubator which includes temperature and CO2 control (Life Imaging Services, Reinach, Switzerland). For long-term imaging, several cell positions were chosen and recorded by a motorized stage (Scan IM; Märzhäuser, Wetzlar-Steindorf, Germany). In these experiments, cells were typically imaged in three dimensions (3D) (4 Z planes per time point) every 7 min, at 3.33-μm steps. For presentation of the movies, the four-dimensional (4D) image sequences were transformed into a time sequence by using the maximum projection option in the cellR software. Single particle tracking and track annotation were performed by the Manual Tracking tracking plug-in of ImageJ (National Institutes of Health, Bethesda, MD). For area calculations of PB motion, PBs were first tracked with a MatLab tracking program written in the laboratory (MathWorks, Natick, MA). To segment an object for tracking in a defined region of interest (ROI), the software takes the 25% of the highest values pixels as a threshold for creating a binary mask of the ROI that contains the object. The mask was eroded twice and dilated twice, creating a binary image where the largest contiguous object in the binary mask was the PB. It then traces the PB boundaries and its center of mass, and the program outputs a vector of the center of mass over time. PB position was measured per frame, and the data were imported into MatLab for calculations of total distance, instantaneous velocity, and plotting of mean square displacement versus time, as described previously (Shav-Tal et al., 2004). When necessary, movie sequences were deconvolved using the Huygens Essential software (Scientific Volume Imaging, Hilversum, The Netherlands). 3D analysis was performed with the Imaris software (Bitplane, Saint Paul, MN). For quantification of the proportion of PB markers assembled in PBs versus their diffuse cytoplasmic pool, we performed 3D signal intensity analysis from endogenous Dcp1a and Dcp1b-immunolabeled cells. A 3D volume of the cell containing PBs (7 z-slices of 0.25 μm; total height, 1.75 μm) was imaged and reconstructed using Imaris software. Borders of the cytoplasm, PBs, and nuclear volumes were thresholded and assigned, and signal intensities in the voxels of PBs only and cytoplasm only were calculated by the removal of the nuclear signal in addition to the removal of background levels, as measured in the same experiments by immunostaining with the secondary antibody only.
Fluorescence Recovery after Photobleaching (FRAP)
For FRAP, cells were maintained in Leibovitz's L-15 phenol red-free (Invitrogen) containing 10% FCS at 37°C. FRAP image sequences were obtained on a Zeiss LSM 510 META inverted scanning confocal microscope equipped with a heated chamber and objective heater, and a Plan-Apochromat 63×, 1.4 NA oil objective (Carl Zeiss, Jena, Germany). Cells were scanned using a 488 laser for detection of GFP-labeled PBs. For analysis of the fluorescence recovery, the moving PBs were tracked using the above-described MatLab tracking module. When the module could not identify the PB immediately after the bleach, the last mean area was kept until the signal was recognized. FRAP data were normalized and calculated as described previously (Shav-Tal et al., 2005).
Fluorescence In Situ Hybridization
Cells expressing GFP-labeled PBs were fixed and fluorescence hybridization was performed as described previously (Chartrand et al., 2000). 50-mer fluorescently labeled DNA probes were used: Cy3-oligo(dT), 18S: ACGCCCAGTATTCGAACGCAACTAATTCAGGGACGGGAAACATGTGTGGC, and mixtures of tumor necrosis factor (TNF)-α: 1) TGAGCCAGAAGAGGTTGAGGGTGTCTGAAGGAGGGGGTAATAAAGGGATT, 2) TTCTAGGTGAGGTCTTCTCAAGTCCTGCAGCATTCTGGCCAGAACCAAAG, and 3) TGTCTCAAGGAAGTCTGGAAACATCTGGAGAGAGGAAGGCCTAAGGTCCA; interferon (IFN)-γ: 1) GGCAGTAACAGCCAAGAGAACCCAAAACGATGCAGAGCTGAAAAGCCAAG, 2) ATTGCAGGCAGGACAACCATTACTGGGATGCTCTTCGACCTTGAAACAGC, and 3) CTGCCTAGTTGGCCCCTGAGATAAAGCCTTGTAATCACATAGCCTTGCCT; and c-jun: 1) GGCGTTGAGGGCATCGTCATAGAAGGTCGTTTCCATCTTTGCAGTCATAG, 2) TCGGCCAGGTTCAGGGTCATGCTCTGTTTCAGGATCTTGGGGTTACTGTA; 3) GTTTTCACTTTTTCCTCCAGCCGGGCGATTCTCTCCAGCTTCCTTTTTCG, and 4) CGTGGTTCATGACTTTCTGTTTAAGCTGTGCCACCTGTTCCCTGAGCATG.
RESULTS
P Body Movements Are Mostly Confined
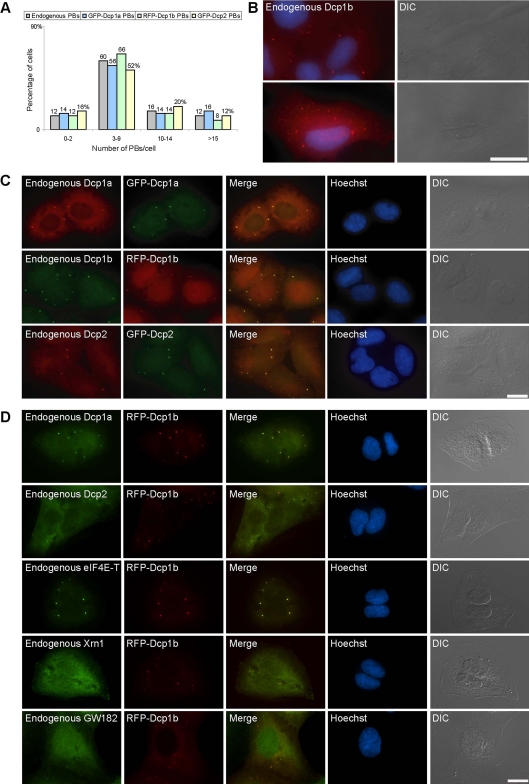
To follow the mechanism of PB transport dynamics in single living cells, we used fusion proteins of known components that are part of the PB multimeric complex. We used either GFP or red fluorescent protein (RFP) fusions of hDcp1a, hDcp1b, and hDcp2, stably integrated into the human U2OS osteosarcoma cell line. We selected clones that exhibited low expression levels of fluorescent PBs. To verify that exogenous Dcp proteins did not induce the formation of high numbers of PBs (high expressing cells showed many and very large PBs; data not shown), we first quantified the endogenous number of PBs in the parental U2OS cells by using specific antibodies to the endogenous Dcp proteins. The majority of cells contained between three and nine PBs (Figure 1A), although the numbers of PBs per cell varied up to ∼20 within the population (Figure 1, A and B), which may be due to cell cycle-related differences (Cougot et al., 2004; Yang et al., 2004). Besides the PB structures, PB components also show a diffuse distribution in the cytoplasm (for quantification, see Supplemental Figure 1A). We selected the stably expressing cell lines that showed moderate distributions of PBs (Figure 1, A and C). XFP-tagged PBs in these cells harbored other endogenous PB components (Figure 1D). Endogenous and fluorescent PBs were heterogenous in size (Yang et al., 2004) and distributed throughout the cytoplasm. However, there was preferential localization of PBs to the inner cytoplasmic region proximal to the nuclear envelope, and in some cells PBs were observed in proximity to the nuclear envelope. PBs were also found in areas overlapping with the nuclear DNA staining, but confocal 3D imaging showed that they were situated in the thin cytoplasmic volume above the nucleus and not within the nuclear volume (Supplemental Figure 1B).
Figure 1.
Quantification and distribution of endogenous and XFP-Dcp PBs. (A) Counting of endogenous PBs in U2OS cells and XFP-Dcp PBs in stable cell lines (n = 50 cells; numbers above bars represent percentages). (B) Although most cells have between three and nine PBs (endogenous hDcp1a; red) of varying sizes, some cells can have less (top) or many more PBs (bottom). Hoechst DNA counterstain (blue) and DIC. (C) PBs in the different stable cell lines expressing XFP-Dcp proteins contained both the endogenous and exogenous proteins and (D) other endogenous PB proteins. Bar, 20 μm.
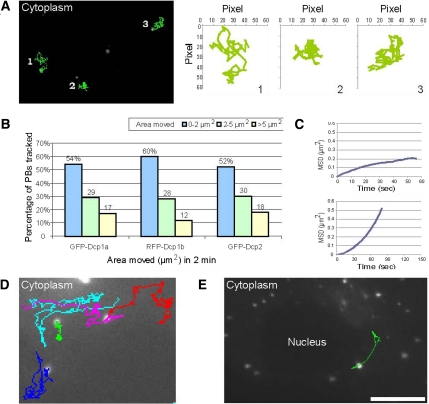
The travels of PBs in the cytoplasm of living cells were then followed using these stable cells. We conducted both long time-lapse live-cell imaging that spanned many hours (see below) and shorter measurements on the scale of minutes or seconds to obtain high-resolution data of PB movement. In general, long-range movies showed that PBs tended to remain in the same spatial neighborhood throughout the cell cycle, although some PBs did move extensively (Supplemental Video 1). Short-range movies showed that PBs were not static and exhibited saltatory movements (Supplemental Video 2), as detected previously (Yang et al., 2004; Kedersha et al., 2005). To quantitatively describe these movements, we performed rapid imaging followed by single particle tracking. This analysis showed that although dynamic, most PB movements were confined to a certain cytoplasmic region giving an impression of constrained movement (Figure 2A and Supplemental Video 2). Single particle tracking and mean square displacement (MSD) analysis (Saxton and Jacobson, 1997) were applied for the analysis of PB movement. Images were acquired every 2 s for a period of 2 min, and the cytoplasmic areas visited by PBs were measured. Measurements from the various stable cell lines showed similar movement dynamics (Figure 2B and Supplemental Figure 2A). For example, for RFP-Dcp1b PBs, 60% of the tracked PBs showed very confined movement and probed areas ranging up to 2 μm2, whereas 28% of the tracks spanned areas of up to 5 μm2, and 12% moved in larger areas. MSD analysis that models the type of movement measured, showed that the movement of the PBs fit a model that describes diffusion (x2 = 4Dt) (Figure 2C, top). The plots obtained from the MSD analysis could be divided into two groups (Saxton and Jacobson, 1997): tracks correlating to diffusion (MSD plotted over time Δt showing a linear correlation) and tracks that portrayed restricted diffusion (in these cases the plots began linearly but reached a plateau, characteristic of constrained diffusion). The measured diffusion coefficients, calculated over both long and short time periods, were mostly in the range of 10−3 to 10−2 μm2/s. Higher and lower diffusion coefficients values were also measured (see analysis below). Only few PBs per cell were relatively stationary.
Figure 2.
Live-cell imaging and single particle tracking of PBs. (A) RFP-Dcp1b PBs were imaged in living cells (60 frames; total 2 min). The first acquired frame is presented and the subsequent tracks from 60 frames of three PBs are annotated (green). The tracks and relative areas (in pixels) in which the PBs moved are plotted. (B) Summary of the distribution of the confined areas of movement for GFP-Dcp1a (n = 211 PBs), RFP-Dcp1b (n = 199), GFP-Dcp2 (n = 186) PBs imaged as described above. (C) Two examples of MSD analysis, depicting plots of a RFP-Dcp1b PB that exhibited confined motion (top) or moved in a directional manner (bottom). (D) Cytoplasmic tracks of RFP-Dcp1b PBs exhibiting directional movement (pink, red, and cyan tracks) or restricted movement (green) (Supplemental Video 3). The blue track shows a mixture of the two types of movement, with a back and forth directed movement on the same track. (E) A frame from a time-lapse movie showing the track of a PB moving along the nuclear periphery (Supplemental Video 4). Bar, 10 μm.
Although confined movements were the majority of movements observed, we could also detect less frequent directional movements of PBs. Supplemental Video 3 shows a cytoplasmic area containing several highly mobile PBs showing directional motility in part of their tracks and that seem to be using a similar portion of the same track (pink, red, and cyan tracks) (Figure 2D). Two of the PBs travel back and forth on the same track (blue and cyan tracks), and a PB with restricted movement is also observed (green track). MSD analysis performed on trajectories of directed PBs in time-lapse movies showed that indeed these movements exhibited directional properties (Figure 2C, bottom). Velocity analysis of the directed PBs demonstrated that they moved at speeds ranging from 0.5 to 1.1 μm/s and could be tracked for distances of 2–10 μm. In several imaged cells, we could detect PBs traveling along the rim of the nuclear envelope or above the nucleus (Figure 2E and Supplemental Video 4), providing a dynamic view of the same observations made with the immunofluorescent staining of endogenous PBs in fixed cells (Supplemental Figure 1B). PBs were also found to fuse to form larger PBs (Supplemental Video 5).
P Bodies Are Anchored to the Cytoskeleton
The directed movement of PBs suggested that PBs might be associated with cytoplasmic filamentous networks. Also, the confined movements of PBs indicated the possibility of anchoring to filaments, although another explanation could be slow diffusive movement limited by cytoplasmic organelles, because the cytoplasm is a crowded solution in which movement is restricted (Luby-Phelps, 2000). We tested these possibilities in living cells. Real-time tracking of PB movements, of which the majority were confined, showed track patterns with an oriented distribution running vectorially from the cell periphery toward the nucleus (Figure 3A and Supplemental Video 6). This implied that PBs exhibited confined movement due to anchoring to a cytoplasmic structure. To examine which cytoskeletal component the bodies associated with, we cotransfected RFP-Dcp1b and GFP-actin, which integrates into the actin cytoskeleton. Dual-color imaging showed that stationary PBs were associated with actin bundles, whereas other nonassociated PBs continued to move rapidly (Figure 3B and Supplemental Video 7). We could follow the rapid movements of a PB in the area of an actin bundle, and their termination once the PB attached (data not shown).
Figure 3.
PBs associate with the cytoskeleton. (A) The tracks of nine PBs show restricted movement with occasional directed motion occurring in the direction of the nucleus. Bar, 10 μm. (See Supplemental Video 6.) (B) RFP-Dcp1b–labeled PBs did not show any movement when associated with GFP-actin bundles (Supplemental Video 7). Two regions are enlarged and PBs tracked: 1) mobile PBs not connected to actin (red track; 30 frames) and 2) stationary PBs associated with an actin bundle (blue track; 30 frames). (C) The movement of PBs is due to their association with microtubules (Supplemental Video 8). 1 and 2, two subsequent frames showing the downward movement of a microtubule (yellow line) and an associated PB (pink). The other two PBs are colored yellow and cyan to ease their identification in the images. 3 and 4, two examples of multiple PBs associated with microtubules. (D) Directed movement of a PB on a microtubule toward the nucleus, and a change in direction when reaching the region of the nuclear membrane. Bar, 2 μm. (E) Directed movement of a PB on a microtubule above the nucleus showing track portions in which the PB is stationary and portions when the PB is moving on the microtubule. During the second stretch the PB changes the microtubule track it is moving on (Supplemental Video 9).
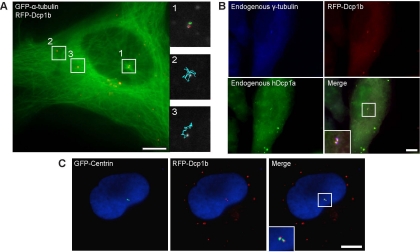
When GFP-α-tubulin was cotransfected into RFP-Dcp1b cells, we observed that PBs were associated with the microtubule network (Supplemental Video 8). In fact, the saltatory movements of PBs were due to the swaying motion of microtubules in the cytoplasm (Figure 3C and Supplemental Video 8). This explained the confined motion detected by particle tracking, resulting from the anchoring of PBs to the motile microtubules. It also explained the fact that PBs attached to actin bundles did not move, because the actin bundles do not have the flexibility of microtubules. In addition to the confined movements, directed movements on microtubules were detected (Figure 3D). Directed movements could also be followed as PBs traveled on microtubules crossing the thin cytoplasmic volume above the nucleus (Figure 3E and Supplemental Video 9). This also demonstrated the ability of PBs to arrive at microtubule intersections and to change direction by traveling from one microtubule to another.
Interestingly, in some live-cell experiments we could observe PBs that were stationary for long periods and that were situated directly above the nucleus at a major microtubule intersection (Figure 4A and Supplemental Video 10). Usually, these stationary PBs were found in pairs. To check whether these PBs might be localized at the main microtubule-organizing center of the cell, cells were fixed and stained with an antibody against endogenous γ-tubulin, marking the position of the centrosome. Figure 4B shows the localization of the PBs marked with an antibody against endogenous Dcp1a and RFP-Dcp1b in proximity to the centrosomal region. Cells transfected with GFP-centrin (White et al., 2000), another component of the centrosome, also showed that PBs and centrosomes were spatially associated (Figure 4C). However, the two signals did not colocalize, meaning that PBs and the centrosome are separated but that PBs can anchor in proximity to the centrosomal structure. Because it was recently shown that certain RNAs might be associated with the centrosome, in particular 18S rRNA that was associated with centrosomes isolated from oocytes of the surf clam (Alliegro et al., 2006), we performed fluorescent in situ hybridization (FISH) with probes against 18S rRNA and poly(A) mRNA. No accumulation of 18S rRNA or poly(A) mRNA was detected in PBs located at the centrosome or in the centrosomal domain (data not shown). Because not all PBs detected above the nucleus were stationary and some could be observed moving through noncentrosomal microtubule junctions above the nucleus, we suggest that some PBs traveling along microtubules might reach a dead-end at the centrosomal region, where the minus ends of microtubules terminate.
Figure 4.
PBs can associate with the microtubule organizing center. (A) RFP-Dcp1b cells were transfected with GFP-tubulin. The tracks of four PBs are shown. Although the two PBs in the peripheral cytoplasm were dynamic (regions 2 and 3; cyan tracks), the two PBs at the microtubule junction above the nucleus were stationary (region 1; red and green tracks) (Supplemental Video 10). (B) RFP-Dcp1b cells were coimmunostained with antibodies to endogenous γ-tubulin (blue) and Dcp1a (green). The Dcp1a and 1b signals colocalized and were adjacent to the centrosome. (C) RFP-Dcp1b cells transfected with GFP-centrin. The two PBs were adjacent to the centrosome. Bar, 10 μm.
P Body Dynamics Are Abrogated When the Microtubule Cytoskeleton Is Disrupted
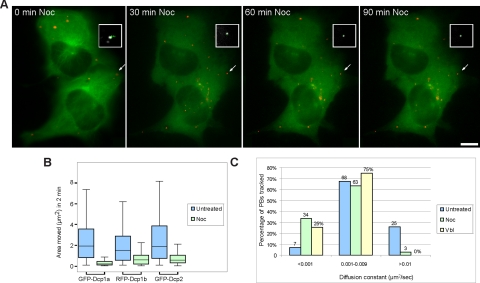
To further analyze the dynamic properties of PBs in vivo, cells were treated with different inhibitors of microtubule assembly. Cells were treated with either nocodazole or vinblastine that binds to tubulin monomers, leading to microtubule destabilization. We acquired rapid time-lapse movies of RFP-labeled PBs and GFP-tubulin in the same cells before and after microtubule disruption. Both treatments significantly reduced the mobility of PBs (Figure 5, A and B). For example, in nocodazole-treated cells the reduction in mobility could be observed 30 min after treatment and was enhanced at 60 and 90 min (Figure 5A and Supplemental Video 11). PB movements were significantly reduced, as was the area of confinement, in all XFP-Dcp cell lines (Figure 5B). For example, in RFP-Dcp1b cells, the area of PB movement was reduced from a median area of 1.5 μm2 in untreated cells to 0.6 μm2 in nocodazole-treated cells. A similar effect was obtained with vinblastine and the median area of movement was reduced to 0.5 μm2 (Supplemental Figure 2, C and D). The measured diffusion coefficients were reduced after treatment. Before treatment, 25% of PBs had diffusion coefficients >0.01 μm2/s, and only 7% were <0.001 μm2/s (Figure 5C). After treatment, the high diffusion coefficients were eliminated, whereas the lower diffusion coefficients increased to 25–34%. This analysis indicated that PB movement after microtubule-destabilizing treatment was highly reduced and more confined.
Figure 5.
Disruption of the microtubule network reduces PB mobility. (A) RFP-Dcp1b PBs (red) were imaged in cells expressing GFP-tubulin (green) before and after nocodazole treatment (30, 60, and 90 min after treatment). The white demarcations show a PB and the track of the same PB during this period. Before treatment the track (green) covers a confined but relatively large area. After destabilization of the microtubule network the tracks show a major reduction in PB motion (Supplemental Video 11). Bar, 10 μm. (B) Box-plot of measurements from tracked PBs: GFP-Dcp1a (n = 427; median untreated = 1.91, noc = 0.21 μm2), RFP-Dcp1b (n = 318; median untreated = 1.5, noc = 0.6 μm2), GFP-Dcp2 (n = 391; median untreated = 1.9, noc = 0.55 μm2), showing the distribution of tracked areas, and a reduction in the areas monitored by PBs after treatment with nocodazole (median, horizontal line). This type of single-cell analysis is probably the reason for the variations observed in between the cell lines. (C) Distribution of the calculated diffusion coefficients (μm2/sec) of the tracked RFP-Dcp1b PBs before (blue) and after treatments (green, nocodazole; yellow, vinblastine).
We assumed that the loss of PB anchoring to microtubules would increase PB mobility; however, the data showed the contrary. We first verified that the movements of PBs were reduced due to the absence of microtubules and not because of a change in the physical properties of the cytoplasm due to treatment, which might have caused hindering of their mobility. Using FRAP of polymerized or depolymerized GFP-tubulin before and after vinblastine treatment, we found that GFP-labeled microtubules in untreated cells recovered slowly as expected, whereas GFP-tubulin monomers in treated cells were free to rapidly diffuse in the cytoplasm (Supplemental Figure 3). Next, we tested whether cytoplasmic crowding (Luby-Phelps, 2000) might explain the reduction in PB diffusion rates. The microtubule network serves as a highway for trafficking of a variety of cytoplasmic vesicles, and so we compared the spatial distribution of PBs relative to other organelles. A cell line expressing CFP-labeled peroxisomes (Darzacq et al., 2006) was transfected with RFP-Dcp1b and showed that PBs and peroxisomes did not interact (Supplemental Video 12A). Peroxisomes exceeded PBs per cell, and many were found surrounding the nucleus. Even PBs in the crowded perinuclear region did not codistribute with peroxisomes. Fluorescently marked mitochondria together with GFP-Dcp1b showed that most mitochondria surrounded the nucleus. PBs in this region were found to move within channel-like domains formed in between the mass of mitochondria and were not spatially or dynamically associated with mitochondria (Supplemental Figure 4A and Supplemental Video 12B). Similar results were obtained with endoplasmic reticulum (ER) labeling (Supplemental Figure 4B and Supplemental Video 12C) or survival of motor neurons protein (SMN) body labeling (Supplemental Video 12D). We then examined whether microtubule disruption trapped PBs in between other vesicles. Depolymerizing microtubules in mitochondria-labeled cells showed that PB movement was very constrained (Supplemental Video 12E). Finally, we examined the distribution of PBs and stress granules (SGs) because these structures can associate during stress (Kedersha et al., 2005). In arsenite-treated cells, PBs and SGs (labeled with GFP-TIAR) were in motion and also associated (Supplemental Video 13, left). However, when microtubules were disrupted in SG-containing cells, both PBs and SGs ceased to move and connections were lost (Supplemental Video 13, right). We therefore conclude that PBs require an intact microtubule network for anchoring and moving within the cytoplasm. This increases the cytoplasmic volume that PBs can access, whereas nonanchored PBs can only diffuse over short distances. Therefore, microtubule anchoring might be a crucial component of the mechanism by which PBs interacts with RNA and SGs.
Microtubule Disruption Leads to Increased PB Assembly
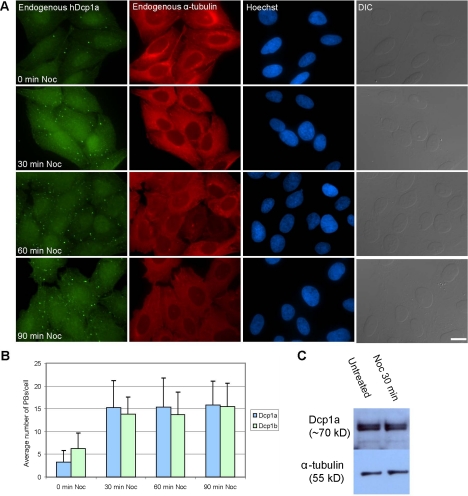
Nocodazole or vinblastine treatments caused an increase in PB numbers. We used this finding to probe the mechanism of PB assembly and disassembly in cells. We tested whether this phenomenon was observed also with endogenous PBs, and using antibodies to endogenous hDcp1a, hDcp1b, hDcp2, eIF4E-T, and GW182 we indeed found a marked increase in the number and size of PBs at 30 min after treatment with the inhibitors (Figure 6, A and B). Western blotting with an antibody against endogenous hDcp1a showed that there was no significant change in the protein levels during the time of nocodazole treatment (Figure 6C). We then checked whether these treatments increased the accumulation of endogenous RNAs within the PBs. We probed the PBs for RNAs by using either an oligo(dT) probe to poly(A) mRNAs, or several probes against endogenous RNAs that contain AU-rich elements (AREs) such as TNF-α, interleukin-2, IFNγ, and c-jun. However, no accumulation of RNA FISH signal was detected in these PBs after microtubule disassembly (data not shown).
Figure 6.
Disruption of the microtubule network causes an increase in PB numbers. (A) U2OS cells treated with nocodazole were fixed at different time points after treatment and costained with anti-hDcp1a (green) and anti-α-tubulin (red) antibodies. DNA was counterstained with Hoechst (blue) and cells were imaged also in differential interference contrast (DIC). Bar, 20 μm. (B) The increase in PB numbers per cell was quantified by counting 20 cells. Error bars indicate SD. (C) Western blot analysis of cell extracts from cells before and during nocodazole treatment, blotted with anti-hDcp1a and with anti-α-tubulin for quantification.
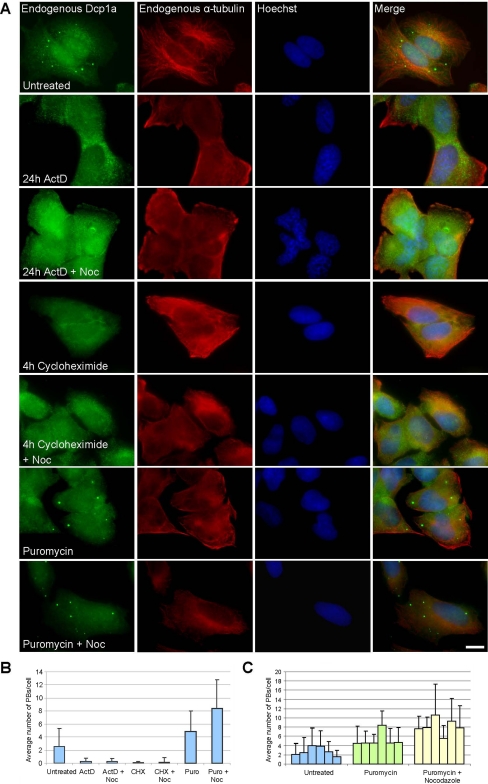
Inhibition of transcription or translation reduces PB numbers in cells (Cougot et al., 2004; Andrei et al., 2005; Ferraiuolo et al., 2005). We tested whether this reduction can be reversed when the microtubule network is disrupted. Inhibition of transcription with actinomycin D (4–24 h) caused a reduction in PB numbers (Figure 7, A and B). ActD treatment followed by nocodazole treatment did not lead to an increase in PB numbers (Figure 7, A and B). Inhibition of translation with cycloheximide (2–4 h) also caused a reduction in PB numbers (Figure 7, A and B). Cycloheximide treatment followed by nocodazole treatment did not lead to an increase in PB numbers either (Figure 7, A and B). When transcription is inhibited mRNA levels decrease, and when translation is inhibited with cycloheximide, mRNAs are trapped in polysomes, and therefore in both cases PBs cannot form. In contrast, puromycin inhibits translation by the release of mRNA from polyribosomes. Indeed, puromycin inhibition caused an increase in PB size and number (Figure 7, A and B) (Eulalio et al., 2007b). Moreover, although PB numbers varied between cells under untreated conditions, and some cells could contain either 0 or 1 PB (Figure 1A), in puromycin-treated cells this was not observed. Instead, a global increase in PB numbers was observed in all cells in the population (Figure 7C). When puromycin treatment was followed by nocodazole treatment, an additional increase in PB numbers was observed (Figure 7, A–C). Together, these data suggest that under conditions of microtubule disruption, the assembly of PBs may depend on the availability of free cytoplasmic RNA (see Discussion).
Figure 7.
PB assembly/disassembly after inhibition of transcription and translation. (A) Endogenous hDcp1b (green), α-tubulin (red), DNA (blue), and DIC images show that after inhibition of transcription (ActD) or translation (cycloheximide) PBs disassembled. No increase in PBs was found in conjunction with microtubule disruption by nocodazole. However, during puromycin treatment that releases mRNA from polysomes, PBs did not disassemble and increased in number and size together with nocodazole treatment. Bar, 10 μm. (B) Counting of PBs in 20 U2OS cells before and after the different treatments. (C) The average number of PBs in random populations of cells before and after puromycin (± nocodazole) treatment. Error bars indicate SD.
PB-assembled Dcp Proteins Exhibit Different Exchange Kinetics
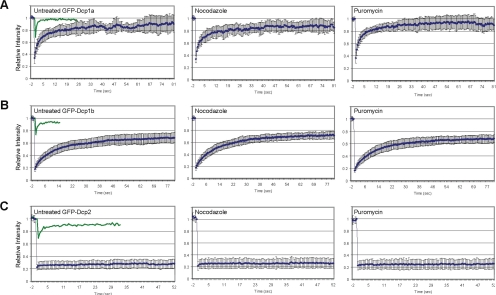
We then wanted to examine whether the internal dynamics of PB components might change during microtubule disruption and PB assembly. To determine the flux of Dcp proteins entering and exiting PBs, we performed photobleaching experiments. FRAP of GFP-Dcp1a PBs showed that this protein rapidly exchanges between PBs and the cytosol (t1/2 of ∼6 s) (Figure 8A), as described previously (Kedersha et al., 2005; Leung et al., 2006). GFP-Dcp1b showed slower recovery kinetics (t1/2 of ∼11 s) and recovered only to 67% (Figure 8B), whereas Dcp1a recovered to 86%. The presence of fixed/immobile fractions for both proteins suggests that the PB structure contains a core structure with components that do not exchange with the cytoplasmic pool. This finding was corroborated when GFP-Dcp2 labeled PBs were photobleached. The fluorescence of Dcp2 within PBs did not recover after the bleaching, whereas the cytoplasmic pool rapidly recovered (Figure 8C), suggesting that the decapping enzyme is a core fixed component of the PB. Furthermore, although the overexpression of Dcp1a or Dcp1b could result in large-sized PBs thereby indicating the multimerization of these proteins, overexpression of Dcp2 never formed large PBs and the Dcp2 signal was always tightly packed. In addition, a previous study has shown that the down-regulation of Dcp2 by siRNA leads to the hyperassembly of PBs (Andrei et al., 2005). Together, these data suggest that Dcp2 serves as a core component of the PB that can regulate the correct assembly and stoichiometry of protein components within the PB core. Finally, we found that the dynamic properties of the Dcp proteins did not change when the cells were treated with either nocodazole or puromycin as described above (Figure 8, A–C).
Figure 8.
PB-assembled Dcp proteins exhibit different exchange kinetics. FRAP recovery curves for GFP-Dcp1a (A), GFP-Dcp1b (B), and GFP-Dcp2 (C). Left, untreated control cells (green curves depict the fast recovery of the diffusing cytoplasmic pool). Middle, nocodazole treated. Right, puromycin treated. Error bars indicate SD.
DISCUSSION
The In Vivo Dynamics of PBs
In this study, we examined the dynamic properties of P body complexes in mammalian cells. We found that PBs are normally associated with the cytoskeleton. Although one might expect these bodies to roam through the cytoplasm in search for RNAs destined for degradation, the contrary is shown by the live-cell data. PB movements were rather spatially confined and PBs tended to remain within the same cytoplasmic region throughout the cell cycle, suggesting that their targets must find their own way to these sites of degradation/storage. Because cytoplasmic mRNA motion is mostly random (Fusco et al., 2003) and PB movement is typically passive, we conclude that the interactions of PBs with mRNA transcripts is based on random encounters rather than on an active or directed mechanism.
PBs were also detected moving in a directional manner on microtubules. Some localized RNAs use the microtubule and actin networks for their travels within cells (Shav-Tal and Singer, 2005). After nuclear export, nonlocalized mRNAs are distributed throughout the cytoplasm via several types of movement, including random or diffusive motion, which can stochastically change to fast and directional motion (Fusco et al., 2003). Furthermore, mRNAs were shown to be associated with mitotic microtubules (Blower et al., 2007). It therefore stands to reason that PBs can also encounter mRNAs during travels on microtubules. We suggest that PBs are anchored to microtubules to increase the probability of colliding with RNAs, and this conclusion is supported by the reduction in diffusion observed when the microtubule network was disrupted. Still, we do not know whether all mRNAs encountering PBs on microtubules functionally interact, because it is possible that some mRNAs are protected, for example by the granule complexes containing them. That some PBs were actively moving along the nuclear periphery might suggest a mechanism of PB patrolling of exported mRNAs. It has been suggested that NMD-targeted mRNAs are recognized by the NMD machinery as they emerge into the cytoplasm. Because PBs contain some NMD factors, it will be interesting to test this possibility.
PBs and other cytoplasmic organelles/structures examined are independent entities that do not associate (except for SGs under stress). PBs moved in channel-like areas in between mitochondria and when the microtubule network was disrupted, the areas of movement and diffusion were greatly reduced. Other studies have also found that depolymerization of actin or microtubules did not increase the diffusion of vesicles/granules (Felder and Kam, 1994; Jones et al., 1998). The calculated diffusion coefficients for PBs were in the range of 10−3–10−2 μm2/s, within the same range as other cytoplasmic granules and vesicles, such as mitochondria (5 × 10−4 μm2/s) (Salmeen et al., 1985), neutrophilic vesicles (2.5 × 10−2 μm2/s) (Felder and Kam, 1994), secretory vesicles (3.9 × 10−4–7.4 × 10−3 μm2/s) (Burke et al., 1997), and chromaffin granules (3 × 10−3 μm2/s) (Steyer et al., 1997). Slow diffusion and restricted movements of cytoplasmic organelles are attributed to the nonhomogeneity of the cytoplasm and to the effects that crowding, obstruction and exclusion have on transport (Luby-Phelps, 2000), meaning that long-range delivery within the cytoplasm has to rely on mechanisms of active transport. In contrast, cytoplasmic messenger ribonucleoproteins (mRNPs) diffuse a 100-fold faster than PBs and other cytoplasmic vesicles (0.1 μm2/s) (Fusco et al., 2003), and they are at least 1000 times more abundant (Femino et al., 1998), thus providing the probability of interactions between RNAs and PBs based on the random movement of both. When no microtubule network is available the diffusion of PBs is reduced, ensuring that when PBs detach from microtubules, they remain in the vicinity to reattach. We conclude that the relative mobility of the microtubule network is necessary for increasing the volumes that PBs probe and the targets that they encounter.
The Assembly and Disassembly of PBs
PBs can assemble or disassemble in response to various cellular cues. Unlike most structures that function in degradation processes in the cytosol, PBs are not membrane enclosed (Yang et al., 2004). Under stress conditions PBs and stress granules associate (Kedersha et al., 2005). In Drosophila, PBs can interact with U bodies, cytoplasmic structures that contain U small nuclear ribonucleoproteins (snRNPs) and that might function in the assembly and storage of U snRNPs before their import into the nucleus (Liu and Gall, 2007). An “open” nonmembrane-bound PB complex could facilitate interactions with the RNAs and the PB enzymes. Indeed, RNA FISH has shown RNA encircling the PB, indicating that interactions are occurring at the PB periphery (Durand et al., 2007). Yeast and mammalian studies have shown that PBs have a role in the storage of mRNAs in an untranslated state (Brengues et al., 2005; Bhattacharyya et al., 2006; Parker and Sheth, 2007). For example, mRNAs can be stored during the stationary phase and then when growth is induced they exit and become translated (Brengues et al., 2005). Thus, a nonmembranous structure allows the robust exchange of mRNPs in and out of the PBs in response to cellular cues.
Our findings show that when the microtubule network is disassembled, PBs grow in number. Likewise, microtubule disruption increases PB formation in yeast cells (Sweet et al., 2007). Most striking is the fact that endogenously only few PBs assemble per cell, and that this takes place in a particular cytoplasmic region in which the PB remains for most of the cell cycle. These findings suggest that PB nucleation and formation are tightly controlled under normal conditions and that a significant portion of the PB proteins are diffusely distributed and are unable to assemble into PBs. When there is no microtubule network, this control is overrun and more of the Dcp proteins can now enter PBs. Sweet et al. (2007) suggest that the microtubules themselves act in the restriction of PB size under normal conditions. We showed that drug-induced microtubule disruption caused an increase in PB numbers, which was dependent on ongoing transcription and translation. If mRNAs were unavailable, PBs did not form or increase in number. This could suggest that mRNA is important for the assembly of PBs under such conditions. Sweet et al. (2007) showed that mRNA metabolism was not modified during microtubule disassembly and concluded that PB aggregation was uncoupled from changes in mRNA. Indeed, an additional scenario should be considered, in which the reduction of mRNA levels leads to the degradation of an unidentified protein factor normally required for PB assembly. In such a case the inhibition of transcription or protein synthesis would not allow PB assembly even when the microtubule network is disassembled. Future efforts in identifying PB components will assist in determining the minimal required factors for the assembly of a functional PB.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to colleagues who contributed reagents: J. Lykke-Andersen, S. Hüttelmaier, B. Motro, and R. Sarid. We thank A. Faragy for assistance. Y.S.-T. thanks the Israel Science Foundation for the fluorescence live-cell imaging microscope. Y.S.-T. is the Jane Stern Lebell Family Fellow in Life Sciences at Bar-Ilan University. This work was supported by grants to Y.S.-T. by the Israel Science Foundation (grant 250/06), France-Israel Biomedical Imaging project, the Israel Cancer Research Fund, the Israel Ministry of Health, and the Alon Fellowship. R.H.S. is supported by National Institutes of Health grants.
Abbreviations used:
- ActD
actinomycin D
- FISH
fluorescence in situ hybridization
- MSD
mean square displacement
- PB
P body
- SG
stress granule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0513) on July 23, 2008.
Author contributions: A. A. and Y.S.-T. designed and performed the experiments. Y. B. programmed the tracking module and performed the FRAPs. L.L.W., N. S., and R.H.S. contributed reagents/materials/ analysis tools. Y.S.-T. wrote the paper.
REFERENCES
- Alliegro M. C., Alliegro M. A., Palazzo R. E. Centrosome-associated RNA in surf clam oocytes. Proc. Natl. Acad. Sci. USA. 2006;103:9034–9038. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei M. A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V. I., Scherthan H., Solinger J. A., Buerstedde J. M., Heyer W. D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C. A., Stevens A., Caponigro G., LaGrandeur T. E., Hatfield L., Fortner D. M., Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Feric E., Weis K., Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke N. V., Han W., Li D., Takimoto K., Watkins S. C., Levitan E. S. Neuronal peptide release is limited by secretory granule mobility. Neuron. 1997;19:1095–1102. doi: 10.1016/s0896-6273(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Bertrand E., Singer R. H., Long R. M. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- Chu C. Y., Rana T. M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R. H., Meier U. T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S., Cougot N., Mahuteau-Betzer F., Nguyen C. H., Grierson D. S., Bertrand E., Tazi J., Lejeune F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Kam Z. Human neutrophil motility: time-dependent three-dimensional shape and granule diffusion. Cell Motil. Cytoskeleton. 1994;28:285–302. doi: 10.1002/cm.970280403. [DOI] [PubMed] [Google Scholar]
- Femino A. M., Fay F. S., Fogarty K., Singer R. H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo M. A., Basak S., Dostie J., Murray E. L., Schoenberg D. R., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S. M., Blanchard J. M., Singer R. H., Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D. J., Luhrmann R., Achsel T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Pauley K. M., Li S., Ikeda K., Lian S., Eystathioy T., Satoh M., Fritzler M. J., Chan E. K. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Jones J. D., Ragsdale G. K., Rozelle A., Yin H. L., Luby-Phelps K. Diffusion of vesicle-sized particles in living cells is restricted by intermediate filaments. Mol. Biol. Cell. 1998;8:174a. [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S., Piano F., Davis R. E. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Calabrese J. M., Sharp P. A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Gall J. G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Temme C., Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pauley K. M., Eystathioy T., Jakymiw A., Hamel J. C., Fritzler M. J., Chan E. K. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo C., Khanna R., Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen I., Zacmanidis P., Jesion G., Feldkamp L. A. Motion of mitochondria in cultured cells quantified by analysis of digitized images. Biophys. J. 1985;48:681–686. doi: 10.1016/S0006-3495(85)83825-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M. J., Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Cohen M., Lapter S., Dye B., Patton J. G., Vandekerckhove J., Zipori D. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol. Biol. Cell. 2001;12:2328–2340. doi: 10.1091/mbc.12.8.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S. M., Fusco D., Janicki S. M., Spector D. L., Singer R. H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Singer R. H. RNA localization. J. Cell Sci. 2005;118:4077–4081. doi: 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Chen N., Tumati S., Parker R., Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Svergun D. I., Round A., Chen N., Muhlrad D., Parker R., Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell J. M., Eggers Z. T., Luedke N., Saari B., Grimson A., Lyons G. E., Anderson P., White J. G. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol. Biol. Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M., Carr-Schmid A., Schwartz D. C., Kiledjian M., Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer J. A., Horstmann H., Almers W. Transport, docking and exocytosis of single secretory granules in live chromaffin cells. Nature. 1997;388:474–478. doi: 10.1038/41329. [DOI] [PubMed] [Google Scholar]
- Stohr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R. H., Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T. J., Boyer B., Hu W., Baker K. E., Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B. Human Dcp 2, a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Jiao X., Carr-Schmid A., Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. A., Pan Z., Salisbury J. L. GFP-centrin as a marker for centriole dynamics in living cells. Microsc. Res. Tech. 2000;49:451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Yang Z., Jakymiw A., Wood M. R., Eystathioy T., Rubin R. L., Fritzler M. J., Chan E. K. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Yu J. H., Yang W. H., Gulick T., Bloch K. D., Bloch D. B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.