Abstract
Many protein regions have been shown to be intrinsically disordered, lacking unique structure under physiological conditions. These intrinsically disordered regions are not only very common in proteomes, they are also crucial to the function of many proteins, especially those involved in signaling, recognition, and regulation. The goal of this work was to identify the prevalence, characteristics, and functions of conserved disordered regions within protein domains and families.
A database was created to store the amino acid sequences of nearly one million proteins and their domain matches from the InterPro database, a resource integrating eight different protein family and domain databases. Disorder prediction was performed on these protein sequences. Regions of sequence corresponding to domains were aligned using a multiple sequence alignment tool. From this initial information, regions of conserved predicted disorder were found within the domains. The methodology for this search consisted of finding regions of consecutive positions in the multiple sequence alignments in which a 90% or more of the sequences were predicted to be disordered. This procedure was constrained to find such regions of conserved disorder prediction that were at least 20 amino acids in length.
The results of this work were 3,653 regions of conserved disorder prediction, found within 2,898 distinct InterPro entries. Most regions of conserved predicted disorder detected were short, with less than 10% of those found exceeding 30 residues in length.
Keywords: intrinsic disorder, protein structure-function, disorder prediction, PONDR
Introduction
Historically, a specific protein function has been ascribed to its unique 3-D structure. Emil Fischer's work on glycolytic enzymes, published in 1894, led him to the “lock-and-key” concept for enzymes and substrates, which postulated that it was the shape of these proteins that conferred their ability to bind with their substrates, and hence carry out their functions.1, 2 Within the next 40 years, scientists showed that it was possible for a protein to lose its native activity and to gain this activity back again. By the 1930's, papers had been published on protein denaturation, stating in essence that proteins have a structure which confers a specific function, and that this structure, and hence the function, is lost by denaturation.3, 4 Linderstrøm-Lang further refined the idea of protein structure, describing how proteins had a primary structure (the amino acid sequence), a secondary structure (such as helices, coils, and sheets), and a tertiary structure, formed by folding of the secondary structure.5, 6 This tertiary structure was presumed to be what determined the protein's function. The use of X-ray crystallography to determine the structure of proteins starting in 1960 further cemented the position of the “sequence-structure-function” paradigm as accepted truth.7, 8 However, there were signs indicating that not all parts of proteins were ordered within these structures determined by X-ray crystallography. Regions of some proteins had missing electron density, even in regions that were known to have function.9 Thus, although the function of a protein is generally thought to arise from its unique and rigid 3-D structure, in some cases, the lack of structure (also known as intrinsic disorder) might be crucial for the function of many proteins,10-23 especially proteins involved in signaling, recognition and regulation.22-25
Recent studies revealed that intrinsic disorder is an abundant phenomenon and many proteins lack rigid 3-D structure under physiological conditions in vitro, existing instead as dynamic ensembles of interconverting structures.26-30 In contrast to ordered proteins whose 3-D structures are relatively stable and whose Ramachandran angles vary slightly around their equilibrium positions with occasional cooperative conformational switches, intrinsically disordered proteins or regions exist as dynamic ensembles in which the atom positions and backbone Ramachandran angles vary significantly over time with no specific equilibrium values and typically undergo non-cooperative conformational changes.23
Statistical analysis shows that amino acid sequences encoding for the disordered proteins or regions are significantly different from those characteristic for the ordered proteins on the basis of local amino acid composition, flexibility, hydropathy, charge, coordination number and several other factors.10, 26, 31-33 A signature of a probable intrinsically disordered region is the presence of low sequence complexity coupled with amino-acid compositional bias, characterized by a low content of bulky hydrophobic amino acids (Val, Leu, Ile, Met, Phe, Trp and Tyr), which would normally form the core of a folded globular protein, and a high proportion of particular polar and charged amino acids (Gln, Ser, Pro, Glu, Lys and, on occasion, Gly and Ala).27, 34 Several predictors of intrinsic disorder have been elaborated based on these observations and on the assumption that the absence of rigid structure is encoded in the specific features of the amino acid sequence.35 The earliest of these predictors, PONDR® (predictor of naturally disordered regions), used neural networks to predict disorder on a residue-by-residue basis and achieved an accuracy of 73% against testing data 26. This early work was refined into the PONDR® VL-XT predictor, which was trained against long regions of disorder identified from regions missing in x-ray structures.27, 36 It has been shown that the PONDR® VL-XT predictor has an accuracy of about 73% when tested against a dataset from the fifth Critical Assessment of Structure Prediction.37 Its sensitivity was 58% for disordered regions of length 30 or less, and 79% for those longer than 30 residues. Numerous other predictors of intrinsic disorder were developed using various computational approaches, including analytical algebraic functions, linear least squares, logistic regression, neural networks, and support vector machines.14, 32, 33, 36, 38-49
There exist many methodologies for classifying proteins and protein regions into domains and families based on their amino acid sequences.50-54 With the advent of bioinformatics techniques, many databases of protein families have been developed. These databases consist of groupings of protein sequences or parts of protein sequences based on some criteria. Most protein family databases attempt to group together proteins that have a common function. Because of the large number of protein sequences now known, this process must be automated in order to cover any substantial fraction of the known proteins. Therefore most protein family databases group their members based on primary protein sequence. Some databases have human-curated patterns, models, or profiles which they then apply on a large scale while others use various algorithms to develop the patterns without the aid of humans. Curated databases tend to be more accurate but will miss classifying protein families or domains that exist in nature but are unknown to humans. Some databases combine the two approaches, having some curated entries and some computer-generated.
The European Bioinformatics Institute's InterPro database combines several protein family databases.55 Different techniques are used to identify family members in each database within InterPro. The largest of the members of InterPro is the curated Pfam database, which uses hidden Markov models and multiple sequence alignments to identify members of its families.50 Curated PIR SuperFamily database54 uses a hierarchical clustering method for identifying “homeomorphic” families, i.e., families “sharing full-length sequence similarity and a common domain architecture”. The PRINTS database56 is a database of protein fingerprints, which are built by an iterative process involving manual sequence alignment and identification of conserved motifs. These motifs (fingerprints) are then used to search the source database for more matches, which are used to adjust the frequency matrices of the motifs. The ProDom database57 is based partially on the Pfam database. In addition to the Pfam domains, which are curated, ProDom also contains domains built automatically using a position-specific iterative BLAST search. Thus, ProDom consists of curated and non-curated domains. PROSITE51 contains two kinds of family and domain signatures. Most of its signatures are in the form of patterns, which are short regular expressions used to match similar regions of sequences. The rest of the signatures are called profiles, which are position-specific scoring matrices built using hidden Markov models. The SMART database53 claims to represent genetically mobile domains. It contains mainly extracellular domains. These domain signatures are built, like Pfam and ProDom, using hidden Markov models and multiple sequence alignments. Unlike the other InterPro member databases, the SUPERFAMILY database58 is built from a source database of proteins of known structure. It uses hidden Markov models to build its families of structured proteins. Proteins within the same superfamily have “structural, functional and sequence evidence for a common evolutionary ancestor.” The last member database is TIGRFams,52 which groups related proteins into orthologous groups, termed “equivalogs”. These groups are sets of “homologous proteins that are conserved with respect to function since their last common ancestor.” TIGRFams also uses hidden Markov models and curated multiple sequence alignments to define their families.
The members of a given domain or protein family are generally assumed to share common functionality, which is derived from the specific structure. However, it is likely that there are many examples of domain or protein families which have a common disordered region or which are entirely disordered, and that this intrinsic disorder is the basis for the shared functionality. It is important to know which families and domains have functional disordered regions, because when novel proteins are identified, membership in a protein family is often used to assign the protein a potential function. Thus, if regions of conserved disorder were identified in protein families, it would enable the identification of new proteins that are likely to contain the same disordered region. The goal of this work was to identify the prevalence, characteristics, and functions of conserved disordered regions within protein domains and families.
Experimental Section
Software
Third party software used for this project is listed below. MySQL (v4.0.20a-nt; http://www.mysql.com) was used for a relational database to store and query data. XEmacs (v21.4; http://www.xemacs.org) was used to write Perl scripts to perform required tasks and calculations for the project. ActivePerl (v5.8.3; http://www.activestate.com) implementation of Perl was used to run these Perl scripts. PONDR® VL-XT software (http://www.pondr.com) was used to predict order and disorder of protein sequences. For generating multiple sequence alignments, CLUSTAL W (v1.83; http://www.ebi.ac.uk/clustalw) was used. For some file parsing and manipulation of multiple sequence alignments, BioPerl (http://www.bioperl.org), an open-source Perl module for bioinformatics, was used. Finally, BLAST59 queries were performed using the Blastall program (v2.2.10; http://www.ncbi.nlm.nih.gov/BLAST/).
In building the initial database for this work, data from a two public databases, UniProt (Release 1.9; http://www.uniprot.org) and InterPro (Release 7.2; http://www.ebi.ac.uk/interpro/), were downloaded and imported into a relational database. UniProt is a protein resource containing all of SwissProt, TrEMBL, and PIR protein information. InterPro is a domain database that integrates eight individual domain databases. Each InterPro entry consists of one or more signatures from one or more of the member databases representing a single domain concept. Because of different methods of detecting domains used in the different member databases, each match within an InterPro entry may span a different region of a protein. InterPro also divides its entries by “type”, which is the type of entity it represents. There are six types defined in the InterPro database: active site (26 entries), binding site (20 entries), domain (2411 entries), family (8035 entries), post-translational modification site (20 entries), and repeat (197 entries). Most InterPro entries are domains or families. For the purposes of this work, the word “domain” will be used to generically represent all of these types.
Initial Database Creation
A relational database was created using standard SQL syntax. Into this database, protein and domain information was imported by parsing downloaded data files using perl scripts. Protein information from UniProt added to the database included the protein accession number, name, amino acid sequence, and the kingdom and species the protein is from. Domain information, from InterPro, added to the database included the domain accession number, name, and type. In addition to this information, UniProt-to-InterPro mappings were imported from InterPro to the local database. Each mapping lists an InterPro entry, a member database accession number, a protein accession number, and start and end positions, which indicate the location of the domain match within the protein. Additional tables and attributes were added to the database for containing data which was created in later stages of the project. The final form of the relational database contained all the necessary information for location and analyzing regions of conserved predicted disorder. The tables of the database are described in Table 1. Specific attributes for each relational table are described in Tables S1 through S13 (see Supplementary Materials).
Table 1.
Description of tables in relational database
| Table Name | Description | Refers To |
|---|---|---|
| align_char | Represents characters of sequences within multiple sequence alignments | align_column, protein_domain |
| align_column | Represents columns of multiple sequence alignments of domains | domain_signature |
| cpd | Represents regions of conserved disorder within domain signatures | domain_signature |
| cpd_pdb_match | Represents overlaps in position between a CPD region and a region of sequence found in the PDB database | cpd, pdb_hit |
| domain_signature | Represents domains found in InterPro member databases | ipr_entry, ipr_db |
| gap | Represents horizontal gaps in the alignments of protein domains | protein_domain |
| ipr_db | Represents member databases of InterPro | |
| ipr_entry | Represents InterPro entries (which may consist of multiple domain signatures from member databases) | |
| pdb_hit | Represents BLAST hits on the PDB database for protein matches to a domain | protein_domain |
| protein | Represents proteins from UniProt | |
| protein_domain | Represents domain signature matches in proteins | domain_signature, protein |
| residue | Represents residues of proteins | protein |
| species | Represents species whose proteins are in the database |
Disorder Prediction
PONDR® VL-XT, software for prediction of disorder tendency of protein sequences, was run against all of the amino acid sequences of the proteins in the database. The resulting disorder score was saved in the database for each amino acid position. This raw disorder prediction information was used in later steps to find consecutive regions of conserved disorder prediction.
Discovery of Conserved Predicted Disorder
A methodology was developed to search for regions of conserved predicted disorder (CPD) in domains. Briefly, this methodology consists of finding regions of consecutive positions in a multiple sequence alignment of all domain matches in which a high percent of sequences are predicted to be disordered. This procedure, which is described below, was followed for each individual domain signature in the database.
First, all of the protein regions matching the domain signature were extracted from the database. If there were more than 100 such matches, then 100 were randomly selected. The amino acid sequences of these protein regions were then aligned using default settings with CLUSTAL W. The resulting text files, containing the multiple sequence alignment (MSA), were stored in a designated directory. Second, the character of the gaps in the MSA was analyzed. This was done to weed out any domains whose alignments were potentially incorrect. For this analysis, each gap length (a gap is a sequential row of positions within a single sequence in an alignment containing gap characters) was compared to the length of the domain for which the MSA was done. Any protein sequence within the alignment which contained a gap whose length was greater than a certain percentage of the domain length was removed from consideration in the alignment. This cutoff percentage was decided on by looking at the mode and standard deviation of the value for all gaps. Proteins containing gaps that were more than two standard deviations from the mode in size were eliminated. Third, the flanking residues for each gap were checked for their disorder prediction score. Each gap was characterized based on whether the amino acid before and the amino acid after the gap were both predicted to be ordered, both predicted to be disordered, or one of each (‘mixed’). This information was saved to be used in the next step. Fourth, the percent of included positions in each alignment column which were predicted to be disordered was calculated and stored in the database. For sequences in a column that were gapped at that position, the disorder/order prediction for the flanking amino acids was used, as described in the previous paragraph. If both were ordered or disordered, then the gap character was counted as ordered or disordered. If they were mixed, then the gap character order/disorder was decided by a weighted coin flip. The coin flip was weighted based on the proportion of ordered and disordered residues in the database. Lastly, the alignment columns were searched for regions where 90% or more of the sequences were predicted to be disordered. The smallest consecutive region that was considered was 20 columns in a row. Information about each of these regions (the domain accession number, and the start and end positions in the alignment) was stored in the database.
Once these initial CPD regions had been found, several types of statistical analysis were performed for each such region. The “gap area” was calculated as the percentage of characters in the alignment between the start and end points of the CPD that were gap characters. The “effective length” of the CPD was calculated by finding the average length of the protein sequences within the region, omitting gaps. The standard deviation for the effective length was also calculated. The two statistics, gap area and effective length, are related, as the higher the gap area, the smaller the effective length will be, relative to the actual length. Additionally, the overall percent of positions predicted to be disordered in the entire CPD was calculated. Although the lower limit was 90%, this was done to determine exactly how conserved the disorder prediction was.
Finally, the average position of the CPD within the domain was calculated for each CPD. While the start and end positions of the region found previously were alignment-based, these domain positions were domain-based. For example, if a CPD region was found by the methodology described above in a certain alignment of a domain from positions 1 to 40, it is likely that the actual ending position of this region were it mapped onto an actual protein sequence, would be less than 40. The domain-based start and end positions were calculated by finding the average length of the protein sequences, not counting gaps, up to the start or end position. From the initial set of CPD regions found as described in the preceding paragraphs, only those with 10 or more protein matches used in the alignment and an effective length of 20 or more were kept as true CPD regions. This eliminated those regions found from too small a set of protein matches, and those that, although having a raw length of at least 20, had too many gaps, shrinking the effective length below the acceptable threshold.
Ranking of CPD Regions
In order to focus on a smaller number of CPD regions, a scoring system was devised to indicate the priority level for each conserved disorder region. This score for each CPD was calculated as the effective length of the conserved region multiplied by the actual percent predicted disorder for that region of the alignment. The effective length, as explained previously, was calculated finding the average length of non-gapped sequences in the region. All CPD regions were ranked using this measure, from highest to lowest.
Sequence Conservation Analysis
Sequence conservation for each alignment column was calculated by applying Shannon's entropy formula:60
where H is the entropy value, X is the alignment column number, and pi is the frequency of the ith letter of the alphabet. For this project, the alphabet was all amino acids plus the gap character. The formula results in a number which represents the degree of variability in the amino acids represented in that column. This number was normalized by dividing by the maximum possible Shannon's entropy score. For each alignment column, the maximum possible entropy is calculated based on the number of sequences in that column. These normalized entropy values can be directly compared, with a range from 0 (all sequences the same) to 1 (all sequences different, as much as possible).
The average entropy for each CPD region was calculated by averaging the values for each column involved in the CPD region. For comparison purposes, Conserved Predicted Order (CPO) regions were found in the same way that CPDs were found, with the difference being that 90% or more of sequences had to be predicted to be ordered rather than disordered. These CPOs were subject to the same effective length and minimum protein sequence restrictions as CPDs. The average entropy was then calculated for all CPOs. Additionally, the average entropy for all “disordered” alignment columns (those with 90% or more sequences predicted to be disordered) as well as the average entropy for all “ordered” alignment columns (those with 90% or more sequences predicted to be ordered) was calculated.
Results
Database
The database constructed contained nearly one million proteins (961,216) from 62,305 different species. The most commonly represented kingdom was eukaryota, followed by bacteria. There were about 800 proteins whose classification was unknown. Table 2 shows some basic statistics on the proteins by Kingdom.
Table 2.
Proteins in the database by Kingdom
| Kingdom | # Proteins | # Species | Average Length |
|---|---|---|---|
| Archaea | 28,888 | 336 | 318.5 |
| Bacteria | 342,300 | 7,111 | 344.7 |
| Eukaryota | 405,146 | 47,660 | 398.1 |
| Viruses | 184,101 | 7,425 | 247.5 |
| Unclassified | 463 | 23 | 163.2 |
| Unknown | 316 | 1 | 837.6 |
| Plasmids | 1 | 1 | 260.0 |
| Transposons | 1 | 1 | 287.0 |
| Total | 961,216 | 62,305 | 347.8 |
The database included 15,498 distinct domain signatures from the eight member databases, representing 10,709 InterPro entries. Pfam accounted for nearly half of the domain signatures. There were over 4.5 million domain matches to proteins. Over 90% of the proteins in the database contained a match to a Pfam domain. A breakdown of the domains and protein matches by member database is shown in Table 3.
Table 3.
Domains and domain matches by database
| Member Database | Domains | Domain Matches | Proteins with Domain Matches | Average Matches per Domain | Average Domain Match Length | Domains with 10+ Protein Matches | |
|---|---|---|---|---|---|---|---|
| Number of Domains | % of Initial Domains | ||||||
| Pfam | 7,316 | 1,413,574 | 896,537 | 193 | 144 | 5,265 | 72.0% |
| PIR Superfamily | 406 | 8289 | 8,289 | 20 | 356 | 231 | 56.9% |
| PRINTS | 1,849 | 1,235,460 | 228,163 | 668 | 17 | 1012 | 54.7% |
| ProDom | 993 | 185,352 | 165,298 | 186 | 136 | 680 | 68.5% |
| PROSITE | 1,752 | 857,410 | 441,789 | 489 | 57 | 1,289 | 73.6% |
| SMART | 659 | 337,000 | 168,072 | 511 | 91 | 409 | 62.1% |
| SUPERFAMILY | 602 | 488,936 | 345,782 | 812 | 125 | 369 | 61.3% |
| TIGRFams | 1,921 | 171,877 | 140,690 | 89 | 295 | 1,547 | 80.5% |
| Total | 15,498 | 4,697,898 | 963,428 | 303 | 95 | 10,802 | 69.7% |
Conserved Disorder Prediction
Multiple Sequence Alignments
Of the 15,498 domains in the database, 13,824 were successfully used to generate multiple sequence alignments. The rest had too few protein matches or resulted in CLUSTAL W errors for various reasons and were discarded. These alignments contained 3,129,498 columns in total.
Gap Analysis
The gaps in the alignments were analyzed in order to eliminate protein sequences that were a poor match to the domain alignment, as described below. There were 5,018,083 gaps in all sequences of all alignments, with an average of 363 gaps per alignment. The average raw gap length was 10.3 positions with a standard deviation of 38.6. The length of the gaps when calculated as a percentage of the domain length was on average 5.9% with a standard deviation of 17.6%, with gaps whose lengths were between 0% and 2% of their domain length, accounting for 59% of the gaps.
The cutoff for gap length was set at 35%, which is approximately equivalent to two standard deviations from the mode. Any protein sequence containing a gap larger than 35% of the domain length was excluded from further analysis. There were 85,142 protein sequences that did not fit this criterion and were eliminated. Based on these eliminations, 10,802 domain signatures had 10 or more protein matches. Table 3 (see last two columns) shows the number and percent of domains for each member database that had 10 or more protein matches. Next, the disorder predictions for the residues before and after each gap were analyzed. Seventy-two percent of the gaps had ordered residues to each side, while almost twenty-four percent had disordered resides to each side. Only 4.4% of the gaps had mixed (one ordered and one disordered) flanking residues. ‘Mixed’ gaps were about three times longer, on average, than ordered or disordered-flanked gaps.
Disorder Prediction for Alignments
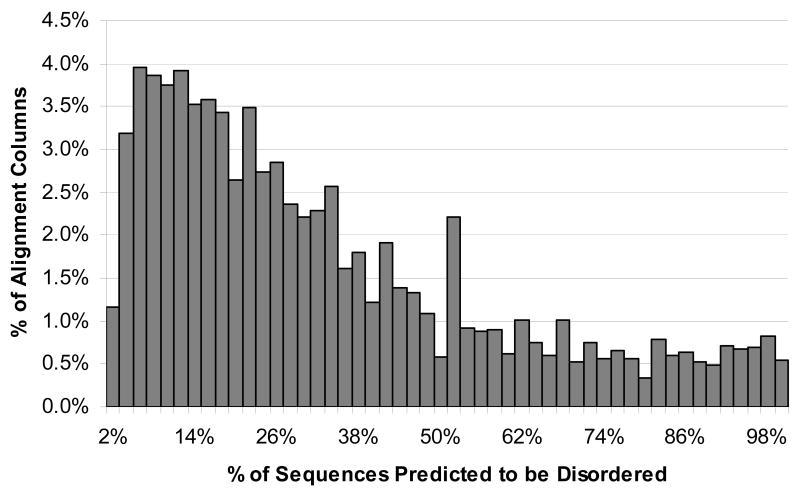
Part of the procedure for looking for the conserved disordered regions included finding the percent of disorder prediction for each column of each alignment, as explained below. The histogram for percent of sequences with predicted disorder is shown in Figure 1. This histogram does not include 542,355 columns (18.8% of columns) with no predicted disorder. The distribution is centered at around 10% with a long tail extending all the way to 100%.
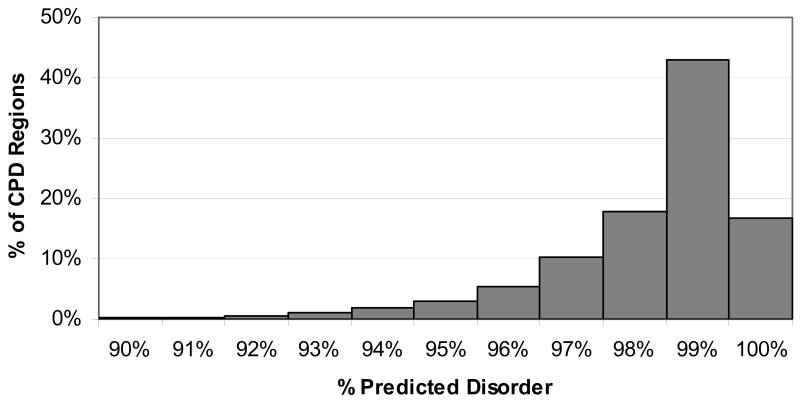
Figure 1.
Histogram of percent predicted disorder for alignment columns. Columns with exactly 0% disorder, accounting for 18.8% of columns, were not included in the histogram.
Conserved Predicted Disorder Regions
A total of 3,653 Conserved Predicted Disorder (CPD) regions in 3,392 domains, representing 2,898 distinct InterPro entries, were discovered in the database. As explained below, only those regions with an effective length of 20 amino acid residues or greater and which were based on alignments of 10 or more sequences were included in the final set of CPD regions.
CPD Regions by InterPro Member Database
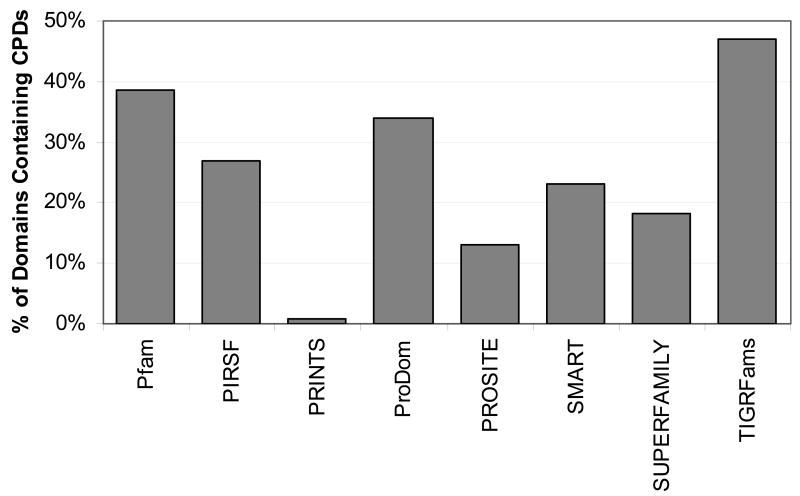
These CPDs were found in all eight member databases, although very few were found in the PRINTS database. The percent of domains containing CPDs for each database was calculated based on the number of domains that, after gap analysis and elimination of protein sequences, had at least 10 protein matches (see Table 3). Figure 2 compares the percent of domains containing CPDs for each database and shows that nearly half of the TIGRFams domains and almost 40% of Pfam domains contained at least one CPD region.
Figure 2.
Percent of domains for each member database containing one or more CPD.
CPDs by Kingdom
Each domain was assigned to a kingdom (archaea, bacteria, eukaryota, viruses) based on the proteins that matched it. Those domains for which over 90% of the proteins with matching regions belonged to the same kingdom were assigned to that kingdom, whereas domains for which no kingdom's proteins made up more than 90% of the matches were assigned to the kingdom ‘Multiple’, meaning the domain was present in proteins from more than one kingdom of life. The most common kingdom assignment was eukaryota, followed closely by Multiple. Archaea had the fewest domains assigned to it. Table 4 shows the number of domains assigned to each kingdom, as well as the number of domains assigned to each kingdom that had 10 or more protein matches.
Table 4.
Domain Kingdom Assignments and CPD regions by kingdom
| Kingdom | Domains Assigned | Domains with 10+ Protein Matches | Conserved Predicted Disorder (CDP) | ||
|---|---|---|---|---|---|
| Number of CPD Regions | Domains in Kingdom Containing CPDs (%) | Average CPD Effective Length | |||
| Archaea | 270 | 125 | 53 | 50 (40.0%) | 22.4 |
| Bacteria | 3757 | 2930 | 1174 | 1136 (38.8%) | 22.6 |
| Eukaryota | 5553 | 3329 | 910 | 795 (23.9%) | 25.9 |
| Viruses | 1062 | 800 | 540 | 446 (55.9%) | 27.2 |
| Multiple | 4856 | 3618 | 976 | 965 (26.7%) | 22.6 |
There were CPD regions found in domains assigned each kingdom in the database. Table 4 shows the number of distinct domains in each kingdom that contained CPDs (see last three rows). The percent of domains is calculated out of the total number of distinct domains containing at least 10 protein matches, since those with fewer matches were not considered when searching for CPD regions. More than half of the viral domains contained CPD regions. The CPD regions in viruses and eukaryotes were longer on average than other kingdoms.
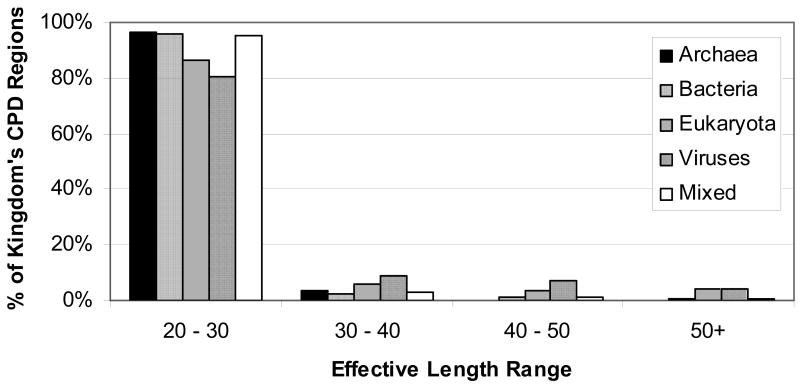
Figure 3 shows the percent of CPDs assigned to each kingdom by different length classes. Only domains assigned to eukaryota and viruses had a significant proportion of long CPD regions. In fact, these two kingdoms had ten times more CPDs of length 50 or more than bacteria and archeae. Another way of looking at the prevalence of CPD regions in different kingdoms is by calculating the percent of proteins in each kingdom which contains at least one CPD region. The percent of sequences containing a CPD of any length (noting that the minimum CPD length is 20) varies from about 19% for archaea to 37% for viruses, with eukaryota (21.5%) and bacteria (30%) falling in between.
Figure 3.
Histogram of CPD effective length classes by kingdom
Length of CPD Regions
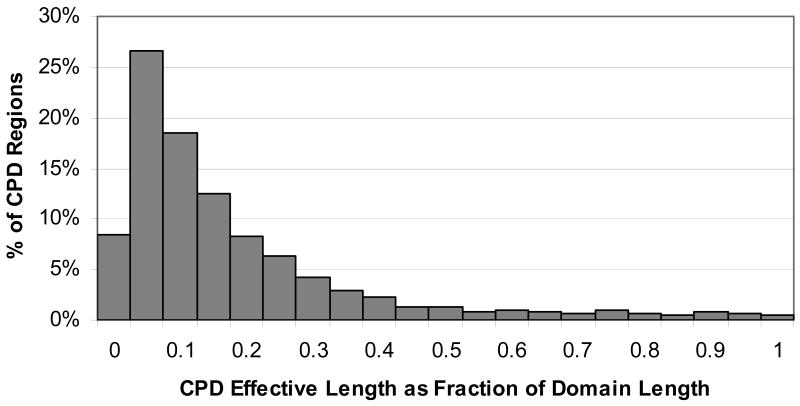
The minimum required effective length of CPD regions was 20; the vast majority of CPDs (91%) had an effective length between 20 and 30 (see Figure 3). The largest effective length was 171, within the Dentin matrix 1 domain. However, a relatively small number of CPD regions (less than 9%) exceeded 30 in length. Table 5 lists the number of regions at or above certain effective length thresholds. When the CPD effective length was taken as a fraction of the domain length, it showed that most CPD lengths were less than 15% of the domain. However, 316 (8.7%) of the CPDs covered more than half of the domain length, and 16 CPD regions covered the entire domain. Figure 4 shows a histogram of CPD effective length as a fraction of domain length.
Table 5.
Long CPD Regions
| Effective Length | Number of CPD regions | % of CPD regions |
|---|---|---|
| ≥30 | 326 | 8.9% |
| ≥40 | 168 | 4.6% |
| ≥50 | 77 | 2.1% |
| ≥60 | 39 | 1.1% |
Figure 4.
Histogram of CPD length as a fraction of domain length
Actual Percent Predicted Disorder
Although the minimum percent predicted disorder for a CPD region was 90%, most CPD regions had a much higher actual percent disorder. The percent of positions that were disordered within the region for all sequences was calculated as the actual percent disorder for the region. Almost 60% of the regions were 99% or more disordered, while only a few were actually 90% disordered. Figure 5 shows a histogram of the percent disorder for all CPD regions.
Figure 5.
Histogram of percent predicted disorder of CPD regions.
Sequence Conservation
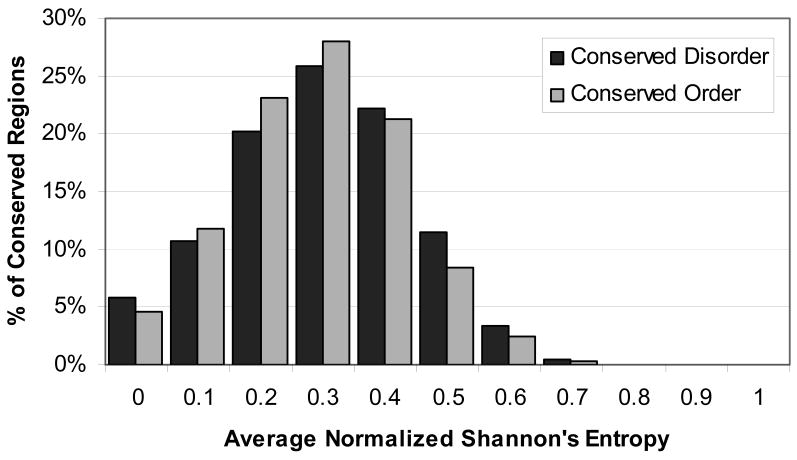
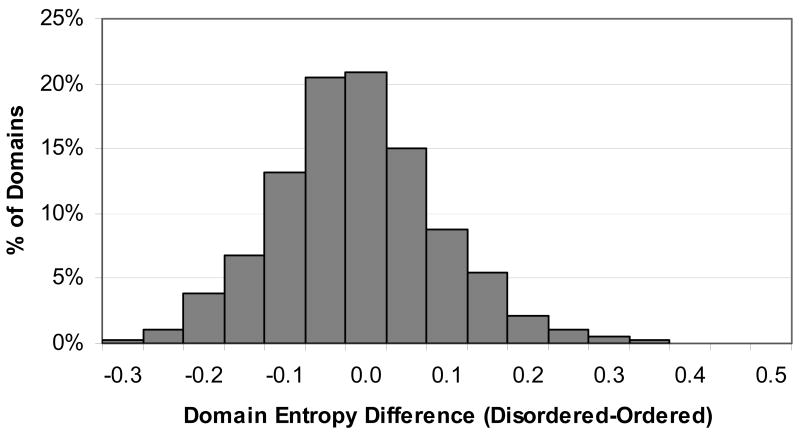
Shannon's entropy was calculated for all alignment columns as a measure of sequence conservation. On average, disordered alignment columns (those with 90% or more sequences disordered at that position) had a higher entropy value than ordered alignment columns (those with 90% or more sequences predicted to be ordered at that position) (0.38 vs. 0.33). However, when just the alignment columns that were part of either a CPD or CPO region were taken, the entropy values were roughly equal (0.34 vs. 0.33). Then the average Shannon's entropy value was calculated for all CPD and CPO regions. The average of these values for CPD regions was 0.35 with a standard deviation of 0.15, and for CPO regions was 0.34 with a standard deviation of 0.14. A histogram of the average Shannon's entropy values for CPD and CPO regions is shown in Figure 6. Altogether, there were 1,511 different domains containing both CPD regions and CPO regions. The average CPD entropy and the average CPO entropy were calculated for all of these domains. On average, the CPD entropy was slightly higher than the CPO entropy (with the average difference between CPD entropy and CPO entropy for domains being 0.015 with a standard deviation of 0.10). However, the histogram of the difference between the two entropy values (Figure 7) shows that for some domains, the disordered regions are more conserved in terms of amino acid sequence than the ordered regions.
Figure 6.
Histogram of average Shannon's Entropy for all CPD and CPO regions
Figure 7.
Histogram of difference between average CPD and CPO entropy for domains containing both types of conserved regions
Discussion
Prevalence and Characteristics of Conserved Disorder
Regions of conserved disorder prediction were found in protein domains from all available InterPro member databases
The percent of domains from each member database containing one or more CPDs varied from 0.8% to 47%. The PRINTS database had CPD regions in only 0.8% of its domains. This is due to the short length of most PRINTS region matches. With the average PRINTS domain match length of 17, it would be impossible for many regions to contain a CPD region of 20 or longer.
In the TIGRFams database, 47% of its domains contained CPD regions. The TIGRFams database is built by grouping proteins into clusters of orthologous groups, which implies a similar function across the members. This may be why such a high percentage of domains contained CPD regions: if the function is conserved across protein members, and if the disorder is necessary for the function, then the disorder will be conserved. In contrast, databases which group more distant family members together, which may have diverged in function, may be more likely to have domains in which disorder tendencies are not conserved across all members of the family.
The Pfam and ProDom databases also had fairly high percentages of domains containing CPDs, at 39% and 34%, respectively. These databases build families in different ways than TIGRFams, in that they do not cluster orthologues. Although these databases supposedly classify domains and families of similar function, it may be that they are including more distant relatives, leading to slightly less conservation of disorder. The fact that TIGRFams domains have on average less than half as many protein members as Pfam and ProDom, indicating a more exclusive family membership, lends support to this theory.
It is surprising that 18% of SUPERFAMILY's domains contained CPD regions, given that the database is derived from proteins of known structure. As expected, nearly all of its CPD regions had a known structure, and 50% had a known structure alone. However, only two CPDs (0.5%) derived from SUPERFAMILY had length 30 or greater and had a known 3D structure not in a complex. The implications of this for the accuracy of this work with respect to shorter regions of conserved disorder are discussed in the accompanying paper.61
Regions of conserved disorder prediction were found in all kingdoms of life, including viruses
When considering CPD regions of all lengths, viruses have the greatest proportion of proteins containing conserved disorder, and archaea have the least. However, when only long CPD regions are considered, viruses and eukaryota have far more conserved disorder (roughly 1% of proteins) than archaea and bacteria (0.1% of proteins). This finding is in line with previous work,10, 30 showing that eukaryotic proteomes have a higher long disorder (>50) content than bacterial and archaeal proteomes. In both this work and previous work, eukaryotes had on the order of ten times more proteins containing long disordered regions than did archaea and bacteria. The fact that viruses also have higher disorder content has not previously been reported, so cannot be verified by comparison to earlier work.
The difference between this and earlier work is that the percent of domains containing conserved disordered regions per kingdom is roughly a factor of ten less than the percent of proteins containing long disordered regions. One interpretation for this is that many disordered regions are not within regions of sequence conservation. A certain level of sequence conservation is required for membership in a protein domain or family, since the sequence is what is used to identify the domain signature. It is very likely that there are other regions of conserved disorder which are not in regions of sequence conservation, which would therefore not be detected by the methodology used for this project.
Most regions of conserved predicted disorder detected were short
The previous work on disorder prediction done using PONDR® VL-XT has focused on long (>30 residues) disordered regions. This was done because the PONDR® VL-XT predictor is more accurate over longer stretches of continuous disorder. Although short disordered regions are known to exist in nature, they have a different amino acid composition than long disordered regions, and so are difficult to predict with current long disorder predictors.62 This work focused on regions of disorder that were are least 20 residues long. Although the error rate for PONDR® VL-XT predictions at this length is higher than for longer stretches of disorder, it was thought that the methodology used to find conserved disorder, using multiple sequence alignments, might improve the accuracy compared to predicting short disorder regions in a single sequence. That is, a prediction of disorder might be more likely to be correct if that prediction was found in the same region in nearly all family members.
Although most regions of conserved disorder prediction were short, there were a significant number than exceeded 30 in length. Most of these were in domains from eukaryotic or viral proteins. As explained above, this is in line with previous work, which found that long regions of intrinsic disorder were much more prevalent in eukaryotes than in prokaryotes.
Sequence conservation in regions of conserved disorder varied, but was on average slightly lower than in regions of conserved order
Positions within CPD regions were just slightly less conserved based on Shannon's entropy than positions within regions of conserved order. When all alignment columns were considered, not just those in conserved order/disorder regions, regions of disorder were noticeably less conserved than regions of order. That is, among positions in the alignments that were 90% or more disordered those within CPD regions were more conserved than those that weren't. This indicates that these CPD regions are important for some function of the protein, since both the sequences and the disorder tendencies are conserved.
When the average Shannon's entropy was taken for regions of conserved order and disorder within a single domain, it was seen that for most domains, regions of conserved order and disorder were about equally conserved in sequence. However, 25% of domains had a higher degree of sequence conservation within conserved disorder as compared to conserved order. Roughly 18% of domains had a lower degree of sequence conservation within conserved disorder than within conserved order. In previous work studying rates of evolution in disordered regions, it was found that nearly 75% of disordered regions evolved faster, and therefore would have lower sequence conservation among family members, than ordered regions within the same protein.63 Only 9% of the disordered regions were more conserved than the ordered regions, and the remaining 18% were equally conserved. However, the sample size for the previous work was much smaller (26 families), and the families used were built using a BLAST search, rather than taken from protein family databases. Additionally, the previous work used only families with a region of experimentally characterized disorder, whereas this work used predicted disorder. Because of these differences in methodology, the results cannot be viewed as contradictory. In fact, both indicate that in some cases, disordered regions evolve faster, in others they evolve slower, and in the rest they evolve at roughly the same rate.
Conclusions
In this work, regions of conserved predicted disorder were identified in domains from all member databases of InterPro and in domains occurring in all kingdoms of life. Although most of these conserved disordered regions were relatively short, between 20 and 30 residues, some were long. These long regions of conserved disorder were much more common in protein families and domains occurring in eukaryotic organisms and viruses. However, conserved predicted disorder was much less common than predicted disorder in general.
This work has also shown that protein domains and families have regions of conserved disorder as well as conserved sequence. Most conserved disordered regions had sequence conservation greater than or equal to that in conserved ordered regions within the same protein. This indicates that disorder tendencies are kept in these proteins, indicating that their function depends on disorder.
Supplementary Material
Footnotes
This work was supported in part by the NIH grant R01 LM007688-01A1 (to A. K. D.) and the Programs of the Russian Academy of Sciences for the “Molecular and cellular biology” and “Fundamental science for medicine” (to V. N. U.)
References
- 1.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber Dt Chem Ges. 1894;27:2985–2993. [Google Scholar]
- 2.Lemieux RU, Spohr U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv Carbohydr Chem Biochem. 1994;50:1–20. [PubMed] [Google Scholar]
- 3.Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci U S A. 1936;22:439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edsall JT. Hsien Wu and the first theory of protein denaturation (1931) Adv Prot Chem. 1995;46 [PubMed] [Google Scholar]
- 5.Linderstrøm-Lang KU. The Lane Medical Lectures. Stanford University Press; Stanford, California: 1952. Proteins and Enzymes. III. The initial stages in the breakdown of proteins by enzymes; pp. 1–115. [Google Scholar]
- 6.Linderstrøm-Lang KU, Schellman JA. Protein structure and enzyme activity. In: Boyer PD, Lardy H, Myrback K, editors. The Enzymes. Academic Press; New York: 1959. pp. 443–510. [Google Scholar]
- 7.Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies RD, Phillips DC, Shore VC. Structure of myoglobin: a three-dimensional Fourier synthesis at 2.0Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 8.Perutz MF, Rossman MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5Å resolution, obtained by X-ray analysis. Nature. 1960;185 doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 9.Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of Protein Folding. Wiley-VCH, Verlag GmbH & Co KGaA; Weinheim, Germany: 2005. pp. 271–353. [Google Scholar]
- 10.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 11.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60:1852–71. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–56. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 14.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–27. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–6. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 16.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 17.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 18.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 19.Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 20.Demchenko AP. Recognition between flexible protein molecules: induced and assisted folding. J Mol Recognit. 2001;14:42–61. doi: 10.1002/1099-1352(200101/02)14:1<42::AID-JMR518>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272:5129–48. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–84. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 24.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 25.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 26.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 1998:437–48. [PubMed] [Google Scholar]
- 27.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–71. [PubMed] [Google Scholar]
- 29.Bax B, Carter PS, Lewis C, Guy AR, Bridges A, Tanner R, Pettman G, Mannix C, Culbert AA, Brown MJ, Smith DG, Reith AD. The structure of phosphorylated GSK-3beta complexed with a peptide, FRATtide, that inhibits beta-catenin phosphorylation. Structure (Camb) 2001;9:1143–52. doi: 10.1016/s0969-2126(01)00679-7. [DOI] [PubMed] [Google Scholar]
- 30.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998:473–84. [PubMed] [Google Scholar]
- 32.Romero P, Obradovic Z, Dunker AK. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Informatics. 1997;8:110–124. [PubMed] [Google Scholar]
- 33.Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. 1997; Proceedings of International Conference on Neural Networks; 1997. pp. 90–95. [Google Scholar]
- 34.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–84. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 35.Bracken C, Iakoucheva LM, Romero PR, Dunker AK. Combining prediction, computation and experiment for the characterization of protein disorder. Curr Opin Struct Biol. 2004;14:570–6. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 37.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53 6:566–72. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 38.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure (Camb) 2003;11:1453–9. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–8. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 2004;576:348–52. doi: 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 41.Coeytaux K, Poupon A. Prediction of unfolded segments in a protein sequence based on amino acid composition. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- 42.Pe'er I, Felder CE, Man O, Silman I, Sussman JL, Beckmann JS. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- 43.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 44.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53 6:573–8. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 45.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–9. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 46.Prilunski J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckman JS, Silman I, Sussman JL. FoldIndex© predicts whether a given protein is intrinsically disordered. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti537. In press. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Rost B. NORSp: Predictions of long regions without regular secondary structure. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson R, Esnouf R. Prediction of natively disordered regions in proteins using a bio-basis function neural network. In: Yang ZR, Yin H, Everson R, editors. ntelligent Data Engineering and Automated Learning - IDEAL 2004: 5th International Conference, Exeter, UK. August 25-27, 2004. Proceedings; Springer-Verlag GmbH; 2004. [Google Scholar]
- 49.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–85. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 50.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–8. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–3. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponting CP, Schultz J, Milpetz F, Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;27:229–32. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, Ledley RS, Suzek BE, Arminski L, Chen Y, Zhang J, Cardenas JL, Chung S, Castro-Alvear J, Dinkov G, Barker WC. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–4. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischmann W, Gouzy J, Griffith-Jones S, Haft D, Hermjakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJ. InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3:225–35. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- 56.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, Uddin A, Zygouri C. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–2. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Servant F, Bru C, Carrere S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–51. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 58.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–19. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. 623–656. [Google Scholar]
- 61.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. Functions of conserved disorder. J Proteome Res. 2006 doi: 10.1021/pr060049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13:71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–10. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.