Abstract
In a search for Polo-like kinase 1 (Plk1) interaction proteins, we have identified TRF1 (telomeric repeat binding factor 1) as a potential Plk1 target. In this communication we report further characterization of the interaction. We show that Plk1 associates with TRF1, and Plk1 phosphorylates TRF1 at Ser-435 in vivo. Moreover, Cdk1, serving as a priming kinase, phosphorylates TRF1 to generate a docking site for Plk1 toward TRF1. In the presence of nocodazole, ectopic expression of wild type TRF1 but not TRF1 with alanine mutation in the Plk1 phosphorylation site induces apoptosis in cells containing short telomeres but not in cells containing long telomeres. Unexpectedly, down-regulation of TRF1 by RNA interference affects cell proliferation and results in obvious apoptosis in cells with short telomeres but not in cells with long telomeres. Importantly, we observe that telomeric DNA binding ability of TRF1 is cell cycle-regulated and reaches a peak during mitosis. Upon phosphorylation by Plk1 in vivo and in vitro, the ability of TRF1 to bind telomeric DNA is dramatically increased. These results demonstrate that Plk1 interacts with and phosphorylates TRF1 and suggest that Plk1-mediated phosphorylation is involved in both TRF1 overexpression-induced apoptosis and its telomeric DNA binding ability.
Composed of repetitive DNA sequences of TTAGGG arrays (in vertebrates) and telomere-binding proteins (1), telomeres are specialized DNA structures positioned at the termini of eukaryotic chromosomes (2). Telomeric DNA together with specific and nonspecific telomere-binding proteins participates in forming highly ordered structures that protect the ends of chromosomes from exonucleolytic attack, end-to-end fusion, and degradation leading to cell death, possible genetic recombination, or survival selection (1, 3, 4). Most human normal somatic cells show a progressive loss of telomeric DNA during successive rounds of cell division due to incomplete DNA replication at the most terminal lagging-strand synthesis (4, 5). Thus, telomere shortening functions as a control mechanism that regulates the replicative capacity of cells and cellular senescence (6). Telomeres are regulated by a homeostatic mechanism that includes telomerase, a reverse transcriptase that adds telomeric TTAGGG repeats onto the 3′ end of chromosomes (7, 8), and telomeric repeat-binding proteins, TRF12 and TRF2, both of which share the highly conserved Myb-like telomeric DNA binding domain. Although the TRF1 complex contains TRF1, TRF1 interacting partners, TIN2, TPP1, and POT1, the TRF2 complex contains TRF2 and its interacting partner RAP1 (1, 4). TRF1 has been shown to negatively regulate telomere length. Overexpression of TRF1 accelerates telomere shortening, whereas dominant-negative inhibition of TRF1 leads to telomere elongation (9). TRF2, a distant homologue of TRF1, also binds to telomeric DNA as a homodimer through the Myb-like domain (10). TRF2 is required to protect chromosomal ends (van Steensel et al. (12)) by stabilizing a terminal loop structure called the t-loop (11).
In addition to its well documented role in telomere length control, accumulating evidence suggests that TRF1 is involved in mitosis as well. It has been reported that overexpression of TRF1 induces premature mitotic entry and subsequent apoptosis in cells with short telomeres (13). Furthermore, before entering apoptosis, TRF1 expression results in accumulation of cells in G2 or M phase of the cell cycle (13, 14). In Xenopus, TRF1 associates with telomere chromatin specifically in mitotic egg extracts and dissociates from it upon mitotic exit (15).
The polo-like kinases (Plks) are a conserved subfamily of serine/threonine protein kinases that play pivotal roles during cell cycle and proliferation (16). In mammalian cells, four Plks (Plk1–4) exist, but their expression patterns and functions appear to be distinct from each other (17). Among these, Plk1 has been the focus of extensive studies because of its association with neoplastic transformation of human cells (18). Studies show that Plk1, which is expressed and active in mitosis, plays a critical role in various aspects of mitotic events such as mitotic entry, centrosome maturation, spindle pole assembly, chromosome segregation, and cytokinesis (16, 19). In a search for Plk1 interacting proteins using a yeast two-hybrid system, we have identified TRF1 as a Plk1 interacting partner. In this study we further characterize the interaction between Plk1 and TRF1 and show that TRF1 is a Plk1 substrate in vitro and in vivo. Significantly, overexpression of wild type TRF1, but not TRF1, with an alanine mutation in the Plk1 phosphorylation site (TRF1-S435A) induces apoptosis in cells containing short telomeres, suggesting an essential role of Plk1 in TRF1-induced apoptosis in these cells. Down-regulation of TRF1 by RNA interference affects cell proliferation and also results in obvious apoptosis in cells with short telomeres but not in cells with long telomeres. Importantly, we observe that telomeric DNA binding ability of TRF1 is cell cycle-regulated, reaching a peak during mitosis. Upon phosphorylation by Plk1 in vivo and in vitro, the DNA binding ability of TRF1 is dramatically increased, suggesting that Plk1-mediated phosphorylation positively regulates the ability of TRF1 to bind telomeres.
EXPERIMENTAL PROCEDURES
Generation of Plasmid Constructs—Full-length human TRF1 (NM_017489), kindly provided by de Lange (Rockefeller University, New York, NY), was amplified by PCR and subcloned into pGEX-KG vector (GST fusion vector, Amersham Biosciences) and pEGFP-C1 (GFP fusion vector, Clontech). Human TRF1 mutants were made with site-directed mutagenesis by using the QuikChange kit (catalog number 200523) from Stratagene according to the manufacturer's instructions. pLKO.1-TRF1 RNA interference vector was constructed as described previously (20), and the targeting sequence of TRF1 is 5′-GGAACATGACAAACTTCATGA-3′, corresponding to 489–509 of the coding region relative to the first nucleotide of the start codon. All of the mutageneses were confirmed by sequencing.
Cell Culture, Synchronization, and DNA Transfections—HeLa, HEK293T, HT1080, and A-T22IJE-T (ATM-/-) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 100 units ml-1 penicillin, and 100 units ml-1 streptomycin at 37 °C in 8% CO2. For synchronization, cells were treated with mimosine (0.3 mm) for 16 h, hydroxyurea (4 mm) for 24 h, or nocodazole (100 ng ml-1) for 14 h to arrest cells at G1, S, or M phase, respectively. Alternatively, cells were treated with 2.5 mm thymidine for 16 h, released for 8 h, and then treated with thymidine a second time for 16 h. After two washes with phosphate-buffered saline (PBS), cells were cultured for different times as indicated in each experiment and harvested. Cells were transfected with mammalian expression constructs by GenePORTER transfection reagent from Genlantis (catalog number T201075).
Recombinant Protein Purification—Full-length and various domains of TRF1 were subcloned into pGEX-KG vector and expressed in Escherichia coli. Expression was induced by 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C for 5 h after the cell density had reached 0.5 at 600 nm. To express GST-Plk1, Hi5 insect cells were infected with baculovirus-encoding GST-Plk1 and harvested 36 h after infection. Recombinant GST fusion proteins were affinity-purified by incubation with glutathione-agarose beads from Sigma (catalog number G4510) followed by extensive washes with STE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 150 mm NaCl) and elution with glutathione elution buffer (40 mm glutathione, 50 mm Tris-HCl, 10 mm dithiothreitol, 200 mm NaCl, pH 8.0).
Cell Extract Preparations—Total nuclear extracts were prepared by using a kit from ActiveMotif (catalog number 40410) according to the manufacturer's instructions with a slight modification. Briefly, cells were lysed in the hypotonic buffer for 15 min on ice. After the lysates were vortexed for 10 s in the presence of detergent and centrifuged, the pellets were collected. The pellets were then resuspended in the complete lysis buffer containing Benzonase nuclease (Sigma, catalog number E1014), a nuclease that digests genomic DNA and releases nuclear proteins intimately associated with DNA. The suspensions were incubated for 1 h with gentle agitation at 4 °C. The lysates were collected by centrifugation at 12,000 × g for 10 min as nuclear extracts.
Immunoprecipitation (IP), Immunoblot, Far Western Blot, and GST Pulldown Assays—Cell lysates were incubated with GFP (Invitrogen, catalog number A11122), TRF1 (Sigma, catalog number T1948), or Plk1 (Zymed Laboratories Inc., catalog number 33-1700) antibodies overnight at 4 °C followed by a 2-h incubation with protein A/G PLUS-agarose beads (Santa Cruz Biotechnology.). Immunocomplexes were resolved by SDS-PAGE and transfer to Immobilon-P membranes (Millipore). Immunoblot analyses were performed using antibodies against TRF1 (Sigma or Abcam, catalog number ab1423), GFP, Erk2, Plk1 (Santa Cruz Biotechnology, catalog number sc-17783), TopoIIβ (TopGEN, catalog number 2010-3), and FLAG (Sigma, catalog number F3165) followed by anti-mouse or anti-rabbit horseradish peroxidase-linked secondary antibodies (Amersham Biosciences) and detection using ECL reagents (Amersham Biosciences). Far Western blots were performed as described previously (21). Briefly, membranes containing the different TRF1 proteins were incubated in blocking buffer (PBST (PBS, pH 7.4, and 0.1% Tween 20) and 3% w/v nonfat milk powder) for 2 h at room temperature followed by a 2-h incubation in PBS containing 2 μg/ml of recombinant GST-Plk1-PBD (polo-box domain). The membrane was extensively washed with PBST and probed with an anti-GST antibody (Santa Cruz Biotechnology, catalog number sc-138). The membrane was then stripped and probed with an anti-GFP antibody. For GST pulldown assays, the lysates of nocodazole-treated HeLa cells in TBSN buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 0.5% Nonidet P-40, 5 mm EGTA, 1.5 mm EDTA, 0.5 mm Na3VO4, 20 mm p-nitrophenyl phosphate) supplemented with protease inhibitors were precleared with glutathione-agarose beads at 4 °C for 2 h and incubated with glutathione-agarose beads containing GST or GST-TRF1 at 4 °C for 2.5 h. The beads were harvested, washed extensively with TBSN buffer, and subjected to Western blot analysis using an anti-Plk1 antibody.
Kinase Assays—In vitro kinase assays were performed in TBMD buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm dithiothreitol, 2 mm EGTA, 0.5 mm Na3VO4, 20 mm p-nitrophenyl phosphate) supplemented with 125 μm ATP and 10 μCi of [γ-32P]ATP (3000 Ci mmol-1, PerkinElmer Life Sciences) for 30 min at 30 °C in the presence of purified GST-TRF1 proteins. For sequential kinase assays, purified GST-TRF1-WT or GST-TRF1-T344A/T371A proteins were preincubated for 1 h at 30 °C in a total volume of 40 μl with or without Cdk1/cyclin B (New England Biolabs, catalog number P6020) in the presence of unlabeled ATP (1 mm). All samples were then incubated for 30 min at 30 °C in the presence of [γ-32P]ATP and GST-Plk1-WT. Samples were then resolved by SDS-PAGE and subjected to autoradiography.
Immunofluorescence Staining and Fluorescence-activated Cell Sorter (FACS) Analysis—Cells growing on coverslips were incubated with 0.1% Triton X-100 in PEM buffer (20 mm PIPES, pH 6.8, 0.2% Triton X-100, 1 mm MgCl2, 10 mm EGTA) for 1 min, fixed in 3.7% formaldehyde in PEM for 5 min, and subsequently permeabilized with 0.3% Triton X-100 in PEM buffer for 30 min. After blocking in PBS with 3% bovine serum albumin for 1 h, cells were incubated with an anti-TRF1 antibody at room temperature overnight followed by incubation with a secondary antibody for 2 h at room temperature. Finally, DNA was stained with 4′,6′-diamidino-2-phenylindole. For FACS analysis, cells were harvested and fixed for 30 min in ice-cold 70% ethanol. The fixed cells were resuspended in PBS containing RNase A (200 μgml-1) and propidium iodide (50 μgml-1) and incubated in the dark for 30 min at room temperature.
Metabolic Labeling—Metabolic labeling was performed essentially as described (22). Briefly, HEK293T cells were labeled for 3.5 h with [32P]orthophosphate (PerkinElmer Life Sciences) at 1 mCi ml-1 in phosphate-free Dulbecco's modified Eagle's medium followed by incubation with okadaic acid (1.0 μm) for 30 min. Nuclear extracts were prepared and incubated with an anti-GFP antibody overnight at 4 °C followed by incubation with protein A PLUS-agarose beads for additional 3 h. The beads were washed with TBSN buffer containing 500 mm NaCl for 5 times and with TBSN buffer containing 150 mm NaCl for 10 times. Samples were then resolved by SDS-PAGE and subjected to autoradiography and Western blot analysis.
In Vitro Gel-shift Assays—In vitro gel-shift assays were performed essentially as described (23) with an end-labeled 142-bp HindIII-Asp-718 fragment from plasmid pTH12 containing 12 tandem TTAGGG repeats (24). The bacterially expressed TRF1 proteins were incubated with the end-labeled DNA at room temperature for 30 min in a 40-μl reaction containing 20 mm HEPES-KOH, pH 7.9, 150 mm KCl, 5% (v/v) glycerol, 4% (w/v) Ficoll, 1 mm EDTA, 0.1 mm MgCl2, 0.5 mm dithiothreitol, 500 ng of sheared E. coli DNA, and 100 ng of casein. The DNA-protein complexes were fractionated on a 5% non-denaturing polyacrylamide gel with 1× TBE (90 mm Tris base, 90 mm boric acid, 2 mm EDTA) as a running buffer. Gels were dried onto Whatman DE81 paper and autoradiographed.
Chromatin Immunoprecipitation (ChIP)—ChIP assays were performed as described (25) with slight modifications. In brief, after digestion with trypsin and washing with PBS, cells were fixed in 1% (w/v) formaldehyde in PBS for 30 min at room temperature, quenched with 125 mm glycine, washed with PBS, and lysed in the lysis buffer containing 50 mm Tris-HCl, pH 8.0, 1% Triton X-100, 1% Nonidet P-40, 0.1% SDS, 0.1% deoxycholate, 150 mm NaCl, 5 mm EDTA, protease inhibitors mixture (BD Biosciences). Lysates were sonicated to obtain chromatin fragments <1 kilobase and centrifuged for 30 min at 4 °C. Two hundred microliters of lysates were diluted with 1 ml of 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, and 150 mm NaCl and incubated with antibodies overnight at 4 °C followed by incubation with 40 μl of protein A/G PLUS-agarose beads (pre-blocked with 30 μg of bovine serum albumin and 5 μg of sheared E. coli DNA) for 2 h at 4 °C. Immunoprecipitated pellets were washed with 0.1% SDS, 1% Triton X-100, 2 mm EDTA, pH 8.0, 20 mm Tris-HCl, pH 8.0, containing 150 mm NaCl in the first wash and 500 mm NaCl in the second wash. Further washes were with 0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, pH 8.0, 10 mm Tris-HCl, pH 8.0, and with 10 mm Tris-HCl, pH 8.0, 1 mm EDTA. Chromatin was eluted from the beads with 500 μl of 1% SDS, 0.1 m Na2CO3. After the addition of 20 μl of 5 m NaCl, cross links were reversed overnight at 65 °C. Samples were supplemented with 20 μl of 1 m Tris-HCl, pH 6.5, 10 μl of 0.5 m EDTA, and 20 μg of DNase-free RNase A and incubated at 37 °C for 30 min. After samples were digested with 50 μg of proteinase K (Calbiochem) for 60 min at 42 °C and phenol-extracted, the DNA was precipitated overnight at -20 °C with 2.5 volumes ethanol and 0.1 volume sodium acetate (3 m, pH 5.2). The precipitate was dissolved in water, denatured at 95 °C for 5 min, and blotted onto Hybond membranes in 20× SSC. Membranes were treated with 1.5 m NaCl, 0.5 n NaOH for 10 min, neutralized with 1 m NaCl, 0.5 m Tris-HCl, pH 7.0 for 10 min, dried for 1 h at 80 °C, rinsed with 6× SSC for 5 min, prehybridized with 5× Denhardt's solution, 6× SSC, 0.5% SDS, 100 μgml-1 denatured sperm DNA (Sigma) overnight at 68 °C, and hybridized with a 800-bp Klenow-labeled TTAGGG probe overnight at 68 °C (Addgene plasmid 12401) (26). Membranes were washed for 15 min at room temperature with 2× SSC, 0.5% SDS in the first wash and 2× SSC, 0.1% SDS in the second wash. Further washes were with 0.1× SSC, 0.5% SDS for 1 h at 37 °C and subsequent wash at 68 °C. Finally, the membrane was rinsed with 0.1× SSC and exposed to a PhosphorImager screen.
RESULTS
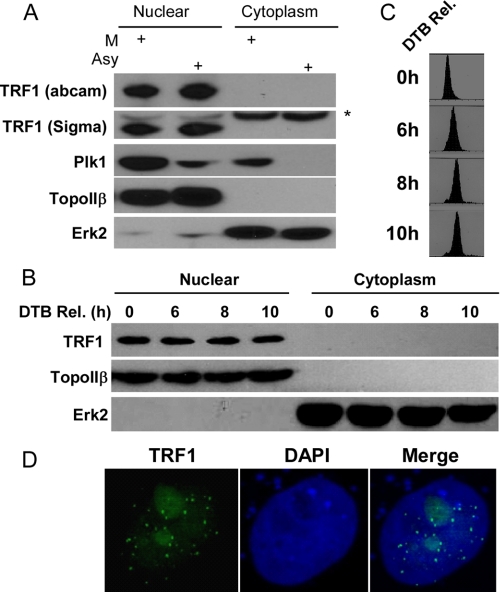
TRF1 Localizes In Nucleus, and Its Protein Expression Is Not Cell Cycle-regulated—It was reported that overexpressed HA-PIN2, a splicing variant of TRF1, is regulated during the cell cycle progression, with the highest expression level at mitosis (14). To examine whether the protein level of endogenous TRF1 is also cell cycle-regulated, HeLa cells were treated with nocodazole for 16 h and harvested by mitotic shake-off. Extracts of asynchronous or mitotic cells were fractioned into cytoplasmic and nuclear fractions and subjected to Western blot analysis using two different commercial antibodies against TRF1. We found that TRF1 is localized in the nuclear fraction, and its protein level in asynchronous cells is similar to that in mitotic cells, suggesting that TRF1 protein level is not cell cycle-regulated in HeLa cells (Fig. 1A). Next, we used double thymidine block to arrest HeLa cells at G1/S boundary, and then the cells were released to different cell cycle stages as monitored by flow cytometry analysis (Fig. 1C). Again, TRF1 protein level remains the same during cell cycle progression (Fig. 1B). Furthermore, we asked whether this is also the case in other cell lines. Our data showed that TRF1 protein level is not cell cycle-regulated in HEK293T and HT1080 cells (data not shown). Consistent with the data from Western blot analysis, anti-TRF1 immunofluorescence staining showed a punctate pattern in detergent-extracted nuclei (Fig. 1D).
FIGURE 1.
TRF1 localizes in nucleus and its protein level is not cell cycle-regulated in HeLa cells. A, cells were treated with nocodazole for 16 h and harvested by mitotic shake-off. Asynchronous (Asy) and mitotic (M) cells were fractioned into cytoplasmic and nuclear fractions and subjected to Western blot analysis. TopoIIβ and Erk2 are used as a marker for nuclear and cytoplasmic fraction, respectively. B and C, cells were synchronized using the double thymidine block (DTB) and harvested at different times after release from the block. Cells were fractionated into cytoplasmic and nuclear fractions and subjected to Western blot analysis using antibodies indicated on the left (B)or fixed and subjected to FACS analysis (C). D, cells growing on the coverslips were fixed and stained with an anti-TRF1 antibody and 4′,6′-diamidino-2-phenylindole (DAPI).
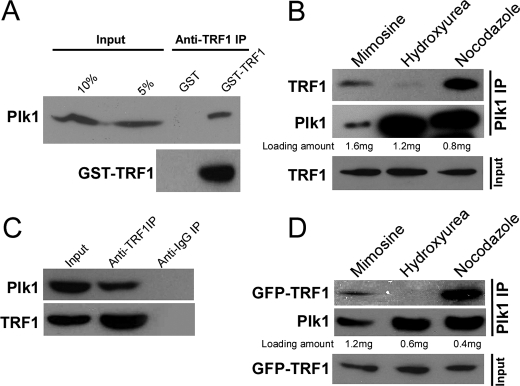
Interaction between Plk1 and TRF1 in Vitro and in Vivo—In a search for Plk1 interacting proteins using a yeast two-hybrid system as a candidate approach, we have identified TRF1 as a potential Plk1 target. To further confirm the association between Plk1 and TRF1, a GST pulldown assay was performed. Purified GST or GST-TRF1 protein (bound to glutathione-agarose beads) was incubated with precleared lysates of nocodazole-treated HeLa cells. As shown in Fig. 2A, Plk1 protein associated with GST-TRF1 protein, but not with GST protein, suggesting that Plk1 interacts with TRF1 in vitro. To confirm the association between TRF1 and Plk1 in vivo, HeLa cells were treated with mimosine, hydroxyurea, or nocodazole to arrest cells at G1, S, or M phase, respectively. Total nuclear extracts were prepared and subjected to Plk1 or TRF1 antibody IP. As indicated, endogenous or overexpressed TRF1 was coimmunoprecipitated with Plk1 in nocodazole- or mimosine-treated cells but not in hydroxyurea-treated cells, suggesting that the binding occurs in both mitosis and G1 phase (Fig. 2, B and D). Moreover, we found that Plk1 was also coimmunoprecipitated with TRF1 in nocodazole-treated cells, further confirming the binding of Plk1 with TRF1 during mitosis (Fig. 2C).
FIGURE 2.
Interaction between Plk1 and TRF1 in vitro and in vivo. A, GST-TRF1 binds to Plk1 protein in vitro. Bacterially expressed GST-TRF1 was incubated with lysates of nocodazole-treated cells. The Plk1 protein that associates with GST-TRF1 was detected by Western blot analysis with an anti-Plk1 antibody. B–D, coimmunoprecipitation of endogenous Plk1 and endogenous or overexpressed TRF1. HeLa cells were transfected (D) or not (B and C) with GFP-TRF1. At 36 h after transfection, cells were treated with mimosine, hydroxyurea, or nocodazole to arrest cells at G1, S, or M phase, respectively. Nuclear extracts were prepared, subjected to anti-Plk1 IP (B and D) or anti-TRF1 IP (C), and analyzed by Western blot.
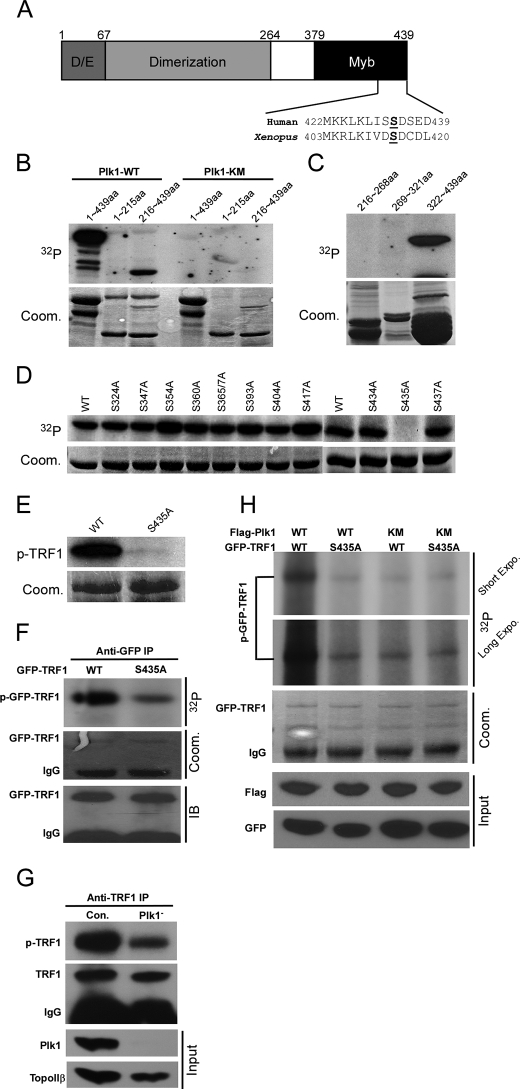
Plk1 Phosphorylates TRF1 in Vitro and in Vivo—Considering a strong binding between TRF1 and Plk1 in vivo, we asked whether TRF1 is a substrate of Plk1. Toward this end, purified full-length (aa 1–439), N-terminal (aa 1–215), or C-terminal (aa 216–439) TRF1 was incubated with purified Plk1-WT or -K82M (kinase dead mutant) in the presence of [γ-32P]ATP. As shown in Fig. 3B, both full-length and the C terminus of TRF1 yielded a strong phosphorylation signal. To further narrow down the site, three non-overlapping TRF1 fragments (aa 216–268, 269–321, and 322–439) were subjected to a kinase reaction, and only the domain containing amino acids 322–439 was a robust substrate for Plk1 (Fig. 3C). Next, virtually every serine in the amino acids 322–439 domain was mutated into alanine to map the potential phosphorylation site for Plk1. Compared with the phosphorylation level of TRF1-WT (aa 322–439), the phosphorylation level of TRF1-S435A was completely abolished, indicating that Ser-435 is the phosphorylation site for Plk1 in vitro (Fig. 3D). We further introduced S435A mutation in the context of full-length of TRF1 and showed that full-length TRF1-S435A was not detectably phosphorylated by Plk1 in vitro (Fig. 3E). As shown in Fig. 3A, Ser-435 localizes in Myb-type DNA binding domain, which is highly conserved from Xenopus to human.
FIGURE 3.
Plk1 phosphorylates TRF1-Ser-435 in vivo. A, schematic representation of TRF1 modular organization along with its major domains. The Myb type DNA binding domain is conserved in human and Xenopus TRF1 proteins. B, purified GST-Plk1-WT (wild type) or -KM (kinase-defective mutant K82M) was incubated with purified GST-TRF1 (aa 1–439), TRF1-N (aa 1–215), or TRF1-C (aa 216–439) for 30 min at 30 °C in the presence of [γ-32P]ATP. The reaction mixtures were resolved by SDS-PAGE and subjected to autoradiography. Arrowheads indicate the positions of TRF1 proteins. Coom., Coomassie Brilliant Blue. C, purified Plk1 was incubated with three C-terminal TRF1 fragments (aa 216–268, 269–361, and 362–439). D, purified Plk1 was incubated with various GST-TRF1 serine to alanine mutants (aa 362–439). p-TRF, phosphorylated TRF. E, purified Plk1 was incubated with GST-TRF1-WT or -S435A protein. F, mutation of Ser-435 to alanine significantly reduced phosphorylation of TRF1 in vivo. HeLa cells were transfected with GFP-TRF1-WT or -S435A. At 24 h after transfection, cells were treated with nocodazole for 8 h and labeled with [32P]orthophosphate in the presence of nocodazole for 3.5 h and further incubated with okadaic acid (1.0 μm) for 0.5 h. Phosphoproteins were immunoprecipitated with anti-GFP antibodies, resolved by SDS-PAGE, and subjected to autoradiography. G, Plk1 is a TRF1 kinase in vivo. HeLa cells were transfected with pBS/U6-Plk1 to deplete Plk1, metabolically labeled as in F, and harvested. Lysates were processed with anti-TRF1 antibodies. H, Plk1 is responsible for TRF1-Ser-435 phosphorylation in vivo. HeLa cells were cotransfected with FLAG-Plk1-WT or -KM and GFP-TRF1-WT or -S435A construct at a 3:1 ratio. 36 h post-transfection, and the cells were labeled with [32P]orthophosphate and processed as in F.
To determine whether this site influences TRF1 phosphorylation in vivo, HEK293T cells were transfected with GFP-TRF1-WT or -S435A. At 24 h post-transfection, cells were treated with nocodazole for 8 h and labeled for 3.5 h with [32P]orthophosphate in the presence of nocodazole followed by incubation with okadaic acid for 0.5 h. TRF1 was immunoprecipitated with an anti-GFP antibody, and the level of TRF1 phosphorylation was determined (Fig. 3F). The mutation S435A significantly reduced but not completely abolished TRF1 phosphorylation in vivo, suggesting that Ser-435 is one of major sites phosphorylated in vivo (Fig. 3F). To provide direct evidence that TRF1 is an in vivo substrate of Plk1, HeLa cells were transfected with pBS/U6-Plk1 to deplete Plk1, metabolically labeled, and harvested. Lysates were subjected to anti-TRF1 IP followed by autoradiography. As shown in Fig. 3G, depletion of Plk1 significantly reduced the phosphorylation of endogenous TRF1, suggesting that Plk1 is a TRF1 kinase in vivo. To determine whether Plk1 is responsible for TRF1-Ser-435 phosphorylation in vivo, 293T cells were cotransfected with GFP-TRF1 (WT or S435A) and FLAG-Plk1 (WT or KM). At 36 h post-transfection, cells were labeled with [32P]orthophosphate. Again, the mutation S435A significantly reduced, but not completely eliminated, TRF1 phosphorylation (Fig. 3H, upper panel, first versus second lanes). There was also detectable phosphorylation of both GFP-TRF1-WT and -S435A when cells were cotransfected with FLAG-Plk1-KM (Fig. 3G, upper panel, third and fourth lanes), suggesting that additional protein kinases may be involved in TRF1 phosphorylation in vivo. Taken collectively, these data strongly indicate that Plk1 phosphorylates TRF1 at Ser-435 in vitro and in vivo.
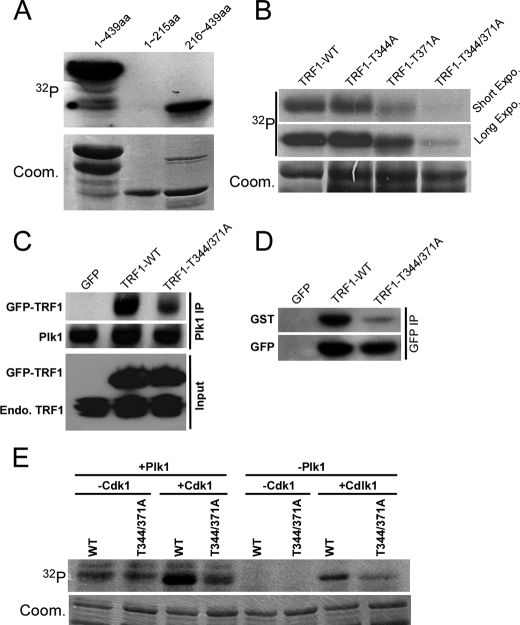
Priming Phosphorylation of TRF1 by Cdk1 Enhances Recruitment of Plk1 to TRF1—Cdk1-associated phosphorylation has been shown to generate a docking site to recruit Plk1 toward its substrates, such as Cdc25C (27, 28). We asked whether the TRF1-Plk1 interaction is regulated by a similar mechanism. Accordingly, recombinant Cdk1/cyclin B was incubated with purified TRF1-WT or various TRF1 threonine to alanine mutants in the presence of [λ-32P]ATP. We showed that TRF1 is a robust substrate of Cdk1 and that Thr-344 and Thr-371 are the two sites phosphorylated in vitro (Fig. 4, A and B). Next, we tested whether the phosphorylation state of Cdk1 sites would affect Plk1 binding to TRF1 in vivo. For that purpose, HeLa cells were transfected with GFP-TRF1-WT or -T344A/T371A constructs and then treated with nocodazole. Nuclear extracts were prepared, subjected to anti-Plk1 IP, and analyzed by Western blot. As shown in Fig. 4C, introduction of the T344A/T371A mutations strongly reduced the binding affinity between Plk1 and TRF1, suggesting that Cdk1-associated priming phosphorylation might generate a docking site for Plk1. Furthermore, Far Western blots were used to test the direct binding between Plk1-PBD and TRF1. After cells were transfected with GFP-TRF1 and treated with nocodazole, and nuclear extracts were prepared and subjected to anti-GFP IP. The IP pellets were resolved by SDS/PAGE and transferred to a membrane, and the membrane was incubated with GST-Plk1-PBD. After extensive washes, the membrane was probed with a GST antibody. As shown in Fig. 4D, T344A/T371A dramatically reduced the binding of Plk1-PBD with TRF1, indicating that Cdk1-associated priming phosphorylation enhances the recruitment of Plk1-PBD to TRF1. To determine whether the Cdk1-induced recruitment of Plk1 to TRF1 converts TRF1 into a more efficient Plk1 substrate, sequential kinase assays were performed (Fig. 4E). For that purpose, recombinant WT and T344A/T371A TRF1 proteins were incubated with Cdk1/cyclin B in the presence of unlabeled ATP followed by incubation with or without Plk1 in the presence of [λ-32P]ATP. Our results showed that the sequential exposure of TRF1 to Cdk1 and Plk1 resulted in strong phosphorylation of WT TRF1 but only weak phosphorylation of the T344A/T371A mutant, suggesting that Cdk1 serves as a priming kinase for Plk1 toward TRF1.
FIGURE 4.
Prior phosphorylation of TRF1 by Cdk1 enhances recruitment of Plk1 to TRF1. A, purified Cdk1/cyclin B was incubated with GST-TRF1 (aa 1–439), TRF1-N (aa 1–215), or TRF1-C (aa 216–439) for 30 min at 30 °C in the presence of [γ-32P]ATP. The reaction mixtures were resolved by SDS-PAGE followed by autoradiography. Arrowheads indicate the positions of TRF1 proteins. Coom., Coomassie Brilliant Blue. B, purified Cdk1/cyclin B was incubated with GST-TRF1 or various GST-TRF1 threonine to alanine mutants. C, HeLa cells were transfected with GFP-TRF1-WT or -T344A/T371A mutant. At 2 days after transfection, cells were treated with nocodazole for 14 h. Nuclear extracts were subjected to anti-Plk1 antibody IP and analyzed by Western blot. D, direct binding of TRF1 to Plk1-PBD. After cells were transfected as in C, nuclear extracts were subjected to anti-GFP IP. Upper, the IP pellets were resolved by SDS-PAGE, transferred to a membrane, and probed with a recombinant GST-Plk1-PBD protein followed by anti-GST Western blot analysis. Lower, the same membrane was stripped and reprobed with an anti-GFP antibody to assess the expression level of TRF1. E, purified GST-TRF1-WT or -T344A/T371A proteins were preincubated with or without Cdk1/cyclin B in the presence of unlabeled ATP. All samples were then incubated with or without Plk1 in the presence of [γ-32P]ATP.
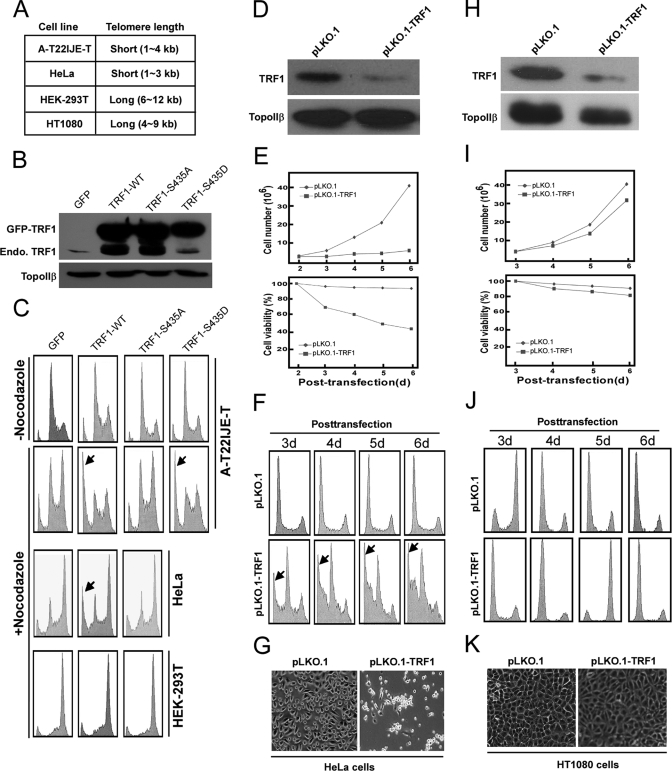
Overexpression of TRF1-WT but Not -S435A Leads to Apoptosis in Cells with Short Telomeres—It has been reported that overexpression of TRF1 induces mitotic entry and apoptosis in cells with short telomeres such as A-T22IJE-T and HeLa cells but not in cells with long telomeres such as 293T and HT1080 cells (13). We tried to test whether Plk1 is involved in this process. For that purpose, cells with different telomere lengths were transfected with GFP vector, GFP-TRF1-WT, Plk1 unphosphorylatable mutant S435A, or phosphomimetic mutant S435D, treated with or without nocodazole, and subjected to FACS analysis. In A-T22IJE-T cells, overexpression of various TRF1 constructs (WT or Ser-435 mutants) could not induce obvious apoptosis without treatment of nocodazole (Fig. 5C). In the presence of nocodazole, overexpression of TRF1-WT or -S435D could induce significant apoptosis in A-T22IJE-T and HeLa cells, whereas GFP vector or TRF1-S435A mutant could not induce apoptosis at all, indicating that Plk1-mediated phosphorylation is involved in TRF1 expression-induced apoptosis in cells with short telomeres. Moreover, we found that overexpression of TRF1 did not induce obvious apoptosis in 293T cells even in the presence of nocodazole, indicating that cells with long telomeres are resistant to changes of TRF1 protein level, consistent with the previous finding (13).
FIGURE 5.
Overexpression of TRF1-WT but not -S435A or depletion of TRF1 leads to apoptosis in cells with short telomeres. A, telomere lengths of different cell lines. B and C, at 24 h after transfection with GFP, GFP-TRF1-WT, -S435A, or -S435D, cells were treated with or without nocodazole for different times (8 h for A-T22IJE-T cells and 14 h for HeLa and 293T cells) and harvested for Western blot (B) or FACS (C) analysis. D–K, HeLa cells (D–G), and HT1080 cells (H–K) were transfected with pLKO.1 or pLKO.1-TRF1. At 1 day post-transfection, puromycin was added for additional 1 day to select transfection-positive cells. After floating cells were removed, cells were further incubated up to day 6 in the presence of puromycin. D and H, at 3 days post-transfection, cells were harvested for Western blot to follow the TRF1 depletion efficiency. E and I, cells were harvested at different times to monitor cell proliferation (upper panel) and viability (lower panel). F and J, FACS profiles of TRF1-depleted cells. G and K, morphology of TRF1-depleted cells at 5 days post-transfection.
Depletion of TRF1 Leads to Apoptosis in Cells with Short Telomeres but Not in Cells with Long Telomeres—It was reported that targeted deletion of exon 1 of the mouse gene encoding Trf1 causes early embryonic lethality (29). In conditional mouse Trf1 null mutant embryonic stem cells, Trf1 deletion induced growth defect and chromosomal instability (30). However, it remains unclear if TRF1 depletion could affect cell growth in mammalian somatic cells. Here, we used vector-based RNA interference to specifically deplete TRF1 in HeLa cells (with short telomeres) and HT1080 cells (with long telomeres). The depletion efficiency of pLKO.1-TRF1 in both cell lines was first determined. Accordingly, cells were transfected with pLKO.1-TRF1 or pLKO.1 (as a control vector). At 1 day of post-transfection, puromycin was added to select transfection-positive cells for 2 days. After floating cells were removed, attached cells were harvested for phenotype analysis. As shown in Fig. 5, D and H, endogenous TRF1 was significantly reduced at 3 days of post-transfection, suggesting that pLKO.1-TRF1 can efficiently deplete endogenous TRF1 in both cell lines. The requirement of TRF1 for cell proliferation and viability was then determined. Although cells transfected with a control vector grew at a normal rate, the proliferation and viability of TRF1-depleted HeLa cells were strongly reduced (Fig. 5E). In striking contrast, TRF1 depletion did not affect the proliferation and viability in HT1080 cells (Fig. 5I), indicating that cells with long telomeres are insensitive to changes of TRF1 protein level. To further characterize the inhibition of cell growth by TRF1 depletion, cell cycle progression was analyzed by FACS. As indicated in Fig. 5, F and J, no obvious cell cycle arrest occurred in TRF1-depleted or control cells, and apoptotic sub-G1 populations in FACS profile indicate that TRF1 depletion induces serious apoptosis in HeLa cells but not in HT1080 cells. Finally, TRF1 depletion leads to a rounded morphology in HeLa cells, whereas TRF1-depleted HT1080 cells remain flat (Fig. 5, G and K).
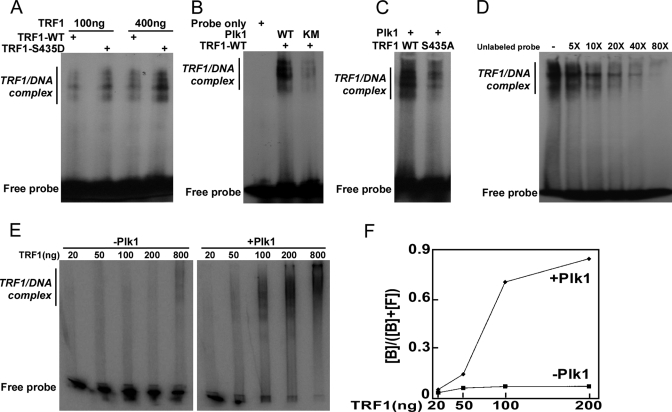
Plk1 Phosphorylation of TRF1 Enhances Its Binding to Telomeric DNA in Vitro and in Vivo—Considering that TRF1 is a telomeric DNA-binding protein and Plk1 targeting site is localized in its DNA binding domain (aa 379–439), we asked whether Plk1 phosphorylation affects TRF1 binding to telomeres. For that purpose, TRF1 proteins purified from bacteria were incubated with end-labeled telomeric repeats probe, and then the mixtures were resolved by non-denaturing PAGE followed by autoradiography. TRF1 formed three complexes with the telomeric repeat containing DNA (Fig. 6) corresponding to binding of one TRF1 dimer (complex I), two TRF1 dimers (complex II), and three TRF1 dimers (complex III) to DNA (31). We first directly compared the telomeric DNA binding ability of TRF1-WT versus phosphomimetic mutant S435D. Our data showed that the S435D mutant binds to telomeric DNA with much higher affinity than did the WT protein (Fig. 6A). The enhanced ability of TRF1-S435D to bind to telomeric DNA is not likely to be due to a difference in protein expression, as the expression of S435D is indistinguishable from that of TRF1-WT and S435A (data not shown). We then observed that upon preincubation with Plk1-WT, but not -KM, the telomeric DNA showed significantly increased binding ability to TRF1 (Fig. 6B). Moreover, after incubation with Plk1, the binding of TRF1-WT protein with telomeric DNA was much stronger than that of Plk1 unphosphorylatable mutant TRF1-S435A protein (Fig. 6C). The addition of excess amounts of unlabeled probe abolished the formation of TRF1-DNA complex, indicating the specificity of DNA-protein interaction (Fig. 6D). Finally, different amounts of Trf1 were incubated with or without Plk1 followed by band-shift analysis. Again, preincubation of Plk1 strongly enhanced telomere binding ability of TRF1 (Fig. 6, E and F).
FIGURE 6.
Phosphorylation by Plk1 promotes TRF1 binding to telomeric DNA in vitro. A, TRF1-S435D has higher binding affinity to telomeric DNA than TRF1-WT. End-labeled telomeric DNA was incubated with purified TRF1 (WT or S435D) at room temperature for 20 min. The DNA-protein complexes were fractionated on a native 5% acrylamide gel. Positions of the three TRF1-containing complexes (I, II, and III) are indicated on the right. B, Plk1 phosphorylation of TRF1 increases its binding affinity to telomeric DNA. Purified TRF1 was incubated with different forms of Plk1 (WT or KM mutant) in kinase assay conditions in the presence of cold ATP followed by incubation with the end-labeled probe as in A. C, alanine mutation at TRF1-Ser-435 decreases its binding affinity to telomeric DNA. Purified Plk1 was incubated with different forms of TRF1 (WT or S435A mutant) in kinase assay conditions in the presence of cold ATP followed by incubation with the end-labeled probe as in A. D, end-labeled telomeric DNA was incubated with purified TRF1-S435D in the presence of increasing concentrations of unlabeled probe. E and F, increasing amounts of TRF1 were incubated with Plk1 in kinase assay conditions in the presence of cold ATP followed by incubation with the end-labeled probe as in A. F, percentages of bound probe ([bound]/([bound] + [free])) were plotted versus TRF1 amounts.
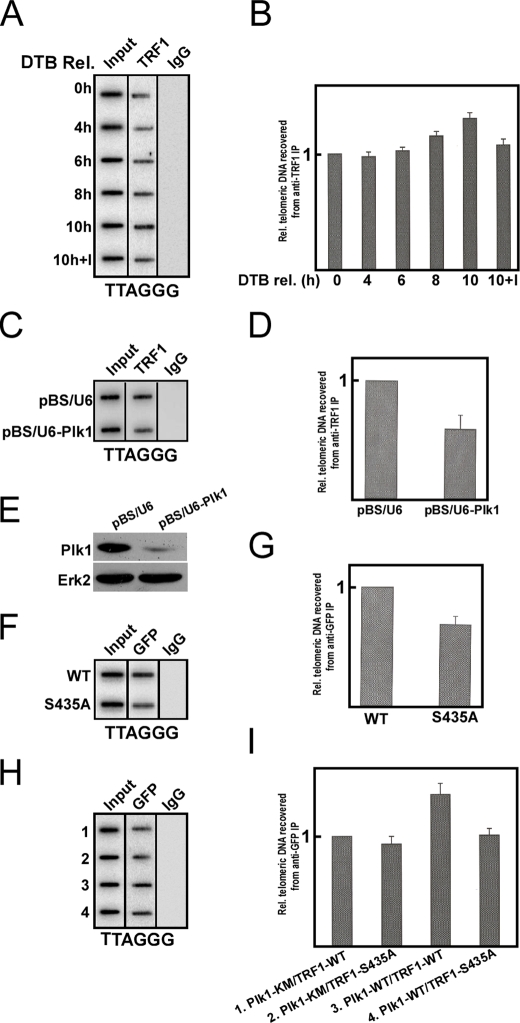
To further test whether Plk1-mediated phosphorylation of TRF1 enhances its in vivo telomeric DNA binding ability, the ChIP experiments were performed (Fig. 7). We observed that the binding ability of TRF1 significantly increased in nocodazole-treated cells compared with that in asynchronous cells (data not shown). Furthermore, HeLa cells were synchronized by the double thymidine block and released for different times. Our data showed that the in vivo telomeric DNA binding ability of TRF1 gradually increased as cells progressing from G1 (0-h point) and S (4-h and 6-points) to G2 (8-h point) and M phase (10-h point). When cells were treated with a Plk1 inhibitor BTO1 (32), the binding ability of TRF1 to telomeric DNA was significantly reduced, indicating that Plk1 might contribute to TRF1 DNA binding ability (Fig. 7, A and B). To provide direct in vivo evidence that TRF1 binding to telomeres depends on Plk1, HeLa cells were transfected with pBS/U6-Plk1 to deplete Plk1 and subjected to anti-TRF1 ChIP analysis. As shown in Fig. 7, C–E, Plk1 depletion strongly reduced TRF1 binding to telomeres, confirming that TRF1 telomere binding ability is regulated by Plk1. To test whether introduction of S435A mutation affects TRF1 binding ability to telomeric DNA in vivo, cells were transfected with GFP-TRF1-WT or -S435A, treated with nocodazole, and subjected to ChIP analysis. As shown in Fig. 7, F and G, the telomeric DNA binding ability of TRF1-S435A was decreased when compared with that of WT in nocodazole-treated cells. To further determine whether Plk1-associated kinase activity regulates the telomeric binding ability of TRF1, cells were cotransfected with GFP-TRF1 (WT or S435A) and FLAG-Plk1 (WT or KM). At 48 h post-transfection, cells were harvested and subjected to ChIP analysis. The mutation S435A significantly reduced the telomeric binding ability of TRF1 (Fig. 7H, lanes 3 versus 4). Furthermore, the telomeric binding ability of TRF1-WT was similar to that of the S435A mutant when cells were cotransfected with FLAG-Plk1-KM (Fig. 7H, lanes 1 versus 2). Together with the in vitro gel-shift analysis data, these results support the conclusion that Plk1-mediated phosphorylation is essential for the telomeric DNA binding ability of TRF1.
FIGURE 7.
Plk1 phosphorylation of TRF1 promotes its binding to telomeric DNA in vivo. A and B, TRF1 binding to telomeric DNA increases in mitosis. HeLa cells were synchronized by the double thymidine block (DTB) and released for different times as indicated. For the last sample (10 h + I), cells were treated with BTO1 (I, 50 μm) for the last 3 h before harvest. Precipitated DNA was probed for the presence of TTAGGG repeats. Although A is a representative slot blot of anti-TRF1 ChIP, B is the quantification results of A. C–E, Plk1 depletion reduces TRF1 binding to telomeres. HeLa cells were transfected with pBS/U6-Plk1 to deplete Plk1 and subjected to anti-TRF1 ChIP analysis. F–I, mutation of Ser-435 to alanine impairs TRF1 binding to telomeric DNA in vivo. F and G, cells were transfected with GFP-TRF1-WT or -S435A, treated with nocodazole for 8 h, and subjected to anti-GFP ChIP analysis. H and I, cells were cotransfected with FLAG-Plk1 (WT or KM) and GFP-TRF1 (WT or S435A) at a 3:1 ratio and subjected to anti-GFP ChIP analysis.
DISCUSSION
In a search for Plk1-interacting proteins, we observed the interaction between Plk1-PBD and TRF1. In the present study, using GST pulldown and coimmunoprecipitation approaches, we further show that TRF1 binds to Plk1 in vitro and in vivo (Fig. 2). The in vivo binding between TRF1 and Plk1 mainly occurs during mitosis, although the binding is also detected during G1 phase but not S phase. A possible explanation for this cell cycle-specific interaction is that Plk1 protein level peaks in mitosis (33).
It has been reported that TRF1 binds to ATM and its phosphorylation level is increased in an ATM-dependent manner upon DNA damage. Furthermore, activated ATM directly phosphorylates TRF1 preferentially on the conserved Ser219 site in vitro and in vivo (34). Although the TRF1/Polo kinase connection was first reported in Xenopus system (15), our work not only validates those initial reports, but more significantly, our work provides crucial insights into the mechanism of TRF1 regulation, linking TRF1 to Plk1 activity. 1) We have analyzed Plk1/TRF1 regulation in mammalian cells, in contrast to the Xenopus system in the previous study. 2) We have mapped Ser-435 of TRF1 as a Plk1 phosphorylation site in vivo; however, no Plx1 phosphorylation sites were identified in xTRF1. 3) Using the TRF1-S435A and -S435D mutants, we were able to directly demonstrate the functional significance of Plk1 phosphorylation of TRF1 in both telomere binding and TRF1 expression-induced mitotic catastrophe. The only experiment to show that xTRF1 function might be regulated by Plx1 is that xTRF1 binds inefficiently to telomeric chromatin in extracts immunodepleted of Plx1. No add-back rescue experiments could be performed due to the lack of information on phosphorylation sites. 4) Accordingly, our studies further advance the understanding of TRF1 function and regulation.
The sequence context of Ser-435 in TRF1, the major phosphorylation site for Plk1, is ISSD436 and highly conserved from human to Xenopus. Although the phosphorylation site in TRF1 identified here fails to exactly resemble the Plk1 consensus phosphorylation sequence (D/E)X(S/T)Φ (X, any amino acid; Φ, a hydrophobic amino acid) (21, 35), the sequence context of TRF1-Ser-435 is similar to that identified in other substrates of Plk1 in our laboratory: ESSY720 in Topors3 and TASE195 in CLIP-170.4 Thus, an aspartic acid or glutamic acid at the +1 or -1 position appears to be important for phosphorylation of substrates by Plk1.
During the preparation of this manuscript, it was reported that casein kinase 2 phosphorylates TRF1 in vitro (36). We also observed that casein kinase 2 is a TRF1 kinase in vitro, and our mapping experiments indicated that Ser-435 is not the phosphorylation site targeted by casein kinase 2 (data not shown). Here, we provide evidence that TRF1 could be phosphorylated by Plk1 at Ser-435 in vivo. Based on our in vitro and in vivo data, we propose that Plk1 is the major kinase that contributes phosphorylation of TRF1 during M phase.
Additionally, we observe that TRF1 could be phosphorylated by Cdk1 at Thr-344 and -371 (Fig. 4). The consensus phosphorylation motif for Cdk1 is (S/T)PX(R/K), in which the Pro at the +1 position is absolutely required, and a basic residue at the +3 position is preferred but not essential for kinase recognition (37). The sequence contexts of both Thr-344 (VGTPQS347) and Thr-371 (PVTPEK374) match this consensus phosphorylation motif, indicating that the phosphorylation of TRF1 by Cdk1 is specific. Thus far, several physiological substrates that bind to the PBD of Plk1 in a Cdk1 phosphorylation-dependent manner have been identified, such as Cdc25C (27, 28), the Plk1-interacting checkpoint “helicase” (38), and histone acetyltransferase binding to Orc1 (Hbo1) (21). Here, we provide another example that this type of interaction between TRF1 and Plk1 is physiologically relevant by demonstrating that Plk1-PBD binding to TRF1 is Cdk1 phosphorylation-dependent.
In the present study we observe that the telomeric DNA binding ability of TRF1 is cell cycle-regulated and reaches a peak during mitosis (Fig. 7A), which apparently contradicts the general belief in previous literature that TRF1 interacts with telomere throughout the cell cycle including M phase. It has been reported that TRF1 is detected at both interphase and metaphase telomeres in human cells (39). The possible explanation for the contradictory observations is that ChIP assays used in this study are more sensitive than the conventional immunofluorescence and Western blot assays and may be able to detect the cell cycle-dependent changes of TRF1 binding ability that was not observed previously (15). To support this explanation, we fail to detect cell-cycle dependent changes of TRF1 protein level using Western blot assay (Fig. 1). Our results are consistent with a previous report that xTRF1 dynamically associates with telomere chromatin specifically in mitotic Xenopus egg extracts and dissociates from it upon mitotic exit, and xTRF1 binds inefficiently to telomeric chromatin in the Plx1-depleted mitotic extracts (15). Furthermore, TRF1 negatively regulates the telomere length by inhibiting access of telomerase at telomere termini (9, 40), suggesting that the ability of TRF1 to interact with telomeric DNA is tightly regulated. Our data further reveal that the ability of TRF1 to bind telomeric DNA during mitosis could be reversed by pretreatment with a Plk1 inhibitor, suggesting that Plk1 is involved in the regulation of telomere binding of TRF1. The most significant finding of our study is that the S435A mutation results in an unphosphorylatable form of TRF1, which in turn leads to diminished DNA binding ability in vitro and in vivo (Figs. 6 and 7). The alanine substitution of Ser-435 significantly reduces the ability of TRF1 to bind telomeric DNA in cells cotransfected with Plk1-WT, but not with Plk1-KM, directly supporting the notion that Plk1-mediated phosphorylation is required for the ability of TRF1 to interact with telomeric DNA.
It has been reported that overexpression of TRF1 induces mitotic entry and apoptosis in cells containing short telomeres, and TRF1-induced apoptosis is further potentiated by arresting cells in mitosis (13). In the present study we observe that overexpression of TRF1-WT or TRF1-S435D but not TRF1-S435A induces obvious apoptosis in mitosis-arrested cells containing short telomeres, indicating that Plk1-mediated phosphorylation is involved in TRF1 expression-induced apoptosis and that Plk1 probably serves as a positive regulator in this process. Moreover, we fail to observe apoptosis in cells containing long telomeres even in the presence of nocodazole, suggesting that Plk1/TRF1-induced apoptosis may be dependent on telomere length. This finding is consistent with previous report that TRF1 has not been shown to induce apoptosis in some cells with long telomere (9, 41). It has been documented that apoptosis induced by inhibition of TRF2 is ATM- and p53-dependent (41). Considering that TRF1 also induces apoptosis in ATM-defective A-T22IJE-T cells and p53 is functionally absent in HeLa cells, it is conceivable to predict that Plk1/TRF1-induced apoptosis is ATM- and p53-independent. Interestingly, we also observe that down-regulation of TRF1 induces apoptosis and cell growth defects in mammalian somatic cells, which is consistent with previous finding that targeted deletion of Trf1 in the mouse causes embryonic lethality, and ES cells deprived of Trf1 function die rapidly (29, 30). Depletion of TRF1 reduces the presence of TRF2 on telomeres in both mammalian somatic cells and mouse ES cells (30, 42), indicating that TRF1 may be important for the binding of TRF2 to telomeres. This may provide an explanation for why depletion of TRF1 could induce apoptosis and growth defects.
In short, we show that Plk1 interacts with and phosphorylates TRF1 in vivo. Our data suggest that Plk1 phosphorylation of TRF1 is involved in TRF1 expression-induced apoptosis and TRF1 association to telomeric DNA.
Acknowledgments
We gratefully acknowledge Dr. Raymond L. Erikson (Harvard University, Cambridge, MA), in whose laboratory the preliminary experiments were performed, and for kindly providing the cell lines and plasmids, Dr. Titia de Lange (Rockefeller University, New York, NY) for pcDNA-Myc-TRF1 expression plasmid and pTH12 TTAGGG plasmid, Dr. Kun Ping Lu (Harvard Medical School) for HT1080 cells, and Drs. Ming Lei (University of Michigan), Jiabin Tang, and Hongchang Li for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant K01 CA114401 (a Howard Temin Award (to X. L.)). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRF1, telomeric repeat binding factor 1; Plk1, Polo-like kinase 1; GST, glutathione S-transferase; PBS, phosphate-buffered saline; IP, immunoprecipitation; GFP, green fluorescent protein; WT, wild type; PIPES, 1,4-piperazinediethanesulfonic acid; ChIP, chromatin immunoprecipitation; aa, amino acids; PBD, polo-box domain; FACS, fluorescence-activated cell sorter; KM, kinase-defective mutant.
X. Yang and X. Liu, unpublished data.
H. Li and X. Liu, unpublished data.
References
- 1.de Lange, T. (2005) Genes Dev. 19 2100-2110 [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. (2000) Nature 408 53-56 [DOI] [PubMed] [Google Scholar]
- 3.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A., and Greider, C. W. (1997) Cell 91 25-34 [DOI] [PubMed] [Google Scholar]
- 4.O'Connor, M. S., Safari, A., Xin, H., Liu, D., and Songyang, Z. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11874-11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingner, J., Cooper, J. P., and Cech, T. R. (1995) Science 269 1533-1534 [DOI] [PubMed] [Google Scholar]
- 6.Stewart, S. A., and Weinberg, R. A. (2006) Annu. Rev. Cell Dev. Biol. 22 531-557 [DOI] [PubMed] [Google Scholar]
- 7.Cech, T. R. (2004) Cell 116 273-279 [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. R., and Blackburn, E. H. (2004) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 359 109-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Steensel, B., and de Lange, T. (1997) Nature 385 740-743 [DOI] [PubMed] [Google Scholar]
- 10.Bilaud, T., Brun, C., Ancelin, K., Koering, C. E., Laroche, T., and Gilson, E. (1997) Nat. Genet. 17 236-239 [DOI] [PubMed] [Google Scholar]
- 11.Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., and de Lange, T. (1999) Cell 97 503-514 [DOI] [PubMed] [Google Scholar]
- 12.van Steensel, B., Smogorzewska, A., and de Lange, T. (1998) Cell 92 401-413 [DOI] [PubMed] [Google Scholar]
- 13.Kishi, S., Wulf, G., Nakamura, M., and Lu, K. P. (2001) Oncogene 20 1497-1508 [DOI] [PubMed] [Google Scholar]
- 14.Shen, M., Haggblom, C., Vogt, M., Hunter, T., and Lu, K. P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13618-13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiyama, A., Muraki, K., Saito, M., Ohsumi, K., Kishimoto, T., and Ishikawa, F. (2006) EMBO J. 25 575-584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Vugt, M. A., and Medema, R. H. (2005) Oncogene 24 2844-2859 [DOI] [PubMed] [Google Scholar]
- 17.Winkles, J. A., and Alberts, G. F. (2005) Oncogene 24 260-266 [DOI] [PubMed] [Google Scholar]
- 18.Eckerdt, F., Yuan, J., and Strebhardt, K. (2005) Oncogene 24 267-276 [DOI] [PubMed] [Google Scholar]
- 19.Barr, F. A., Sillje, H. H., and Nigg, E. A. (2004) Nat. Rev. Mol. Cell. Biol. 5 429-440 [DOI] [PubMed] [Google Scholar]
- 20.Liu, X., Lei, M., and Erikson, R. L. (2006) Mol. Cell. Biol. 26 2093-2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, Z. Q., and Liu, X. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1919-1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, C. Y., Madsen, M. L., Yarm, F. R., Jang, Y. J., Liu, X., and Erikson, R. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12589-12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, S., Giriat, I., Schmitt, A., and de Lange, T. (1998) Science 282 1484-1487 [DOI] [PubMed] [Google Scholar]
- 24.Zhong, Z., Shiue, L., Kaplan, S., and de Lange, T. (1992) Mol. Cell. Biol. 12 4834-4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loayza, D., and De Lange, T. (2003) Nature 423 1013-1018 [DOI] [PubMed] [Google Scholar]
- 26.de Lange, T. (1992) EMBO J. 11 717-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elia, A. E., Rellos, P., Haire, L. F., Chao, J. W., Ivins, F. J., Hoepker, K., Mohammad, D., Cantley, L. C., Smerdon, S. J., and Yaffe, M. B. (2003) Cell 115 83-95 [DOI] [PubMed] [Google Scholar]
- 28.Elia, A. E., Cantley, L. C., and Yaffe, M. B. (2003) Science 299 1228-1231 [DOI] [PubMed] [Google Scholar]
- 29.Karlseder, J., Kachatrian, L., Takai, H., Mercer, K., Hingorani, S., Jacks, T., and de Lange, T. (2003) Mol. Cell. Biol. 23 6533-6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwano, T., Tachibana, M., Reth, M., and Shinkai, Y. (2004) J. Biol. Chem. 279 1442-1448 [DOI] [PubMed] [Google Scholar]
- 31.Bianchi, A., Smith, S., Chong, L., Elias, P., and de Lange, T. (1997) EMBO J. 16 1785-1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan, I. M., Peters, U., Kapoor, T. M., and Straight, A. F. (2007) PLoS ONE 2 e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golsteyn, R. M., Schultz, S. J., Bartek, J., Ziemiecki, A., Ried, T., and Nigg, E. A. (1994) J. Cell Sci. 107 1509-1517 [DOI] [PubMed] [Google Scholar]
- 34.Kishi, S., Zhou, X. Z., Ziv, Y., Khoo, C., Hill, D. E., Shiloh, Y., and Lu, K. P. (2001) J. Biol. Chem. 276 29282-29291 [DOI] [PubMed] [Google Scholar]
- 35.Nakajima, H., Toyoshima-Morimoto, F., Taniguchi, E., and Nishida, E. (2003) J. Biol. Chem. 278 25277-25280 [DOI] [PubMed] [Google Scholar]
- 36.Kim, M. K., Kang, M. R., Nam, H. W., Bae, Y. S., Kim, Y. S., and Chung, I. K. (2008) J. Biol. Chem. 283 14144-14152 [DOI] [PubMed] [Google Scholar]
- 37.Songyang, Z., Blechner, S., Hoagland, N., Hoekstra, M. F., Piwnica-Worms, H., and Cantley, L. C. (1994) Curr. Biol. 4 973-982 [DOI] [PubMed] [Google Scholar]
- 38.Baumann, C., Korner, R., Hofmann, K., and Nigg, E. A. (2007) Cell 128 101-114 [DOI] [PubMed] [Google Scholar]
- 39.Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. (1995) Science 270 1663-1667 [DOI] [PubMed] [Google Scholar]
- 40.Smogorzewska, A., and de Lange, T. (2004) Annu. Rev. Biochem. 73 177-208 [DOI] [PubMed] [Google Scholar]
- 41.Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. (1999) Science 283 1321-1325 [DOI] [PubMed] [Google Scholar]
- 42.Ye, J. Z., Donigian, J. R., van Overbeek, M., Loayza, D., Luo, Y., Krutchinsky, A. N., Chait, B. T., and de Lange, T. (2004) J. Biol. Chem. 279 47264-47271 [DOI] [PubMed] [Google Scholar]