Abstract
Insertions and precise eliminations of transposable elements generated numerous transposon insertion polymorphisms (TIPs) in rice (Oryza sativa). We observed that TIPs represent more than 50% of large insertions and deletions (>100 bp) in the rice genome. Using a comparative genomic approach, we identified 2,041 TIPs between the genomes of two cultivars, japonica Nipponbare and indica 93-11. We also identified 691 TIPs between Nipponbare and indica Guangluai 4 in the 23-Mb collinear regions of chromosome 4. Among them, retrotransposon-based insertion polymorphisms were used to reveal the evolutionary relationships of these three cultivars. Our conservative estimates suggest that the TIPs generated approximately 14% of the genomic DNA sequence differences between subspecies indica and japonica. It was also found that more than 10% of TIPs were located in expressed gene regions, representing an important source of genetic variation. Transcript evidence implies that these TIPs induced a series of genetic differences between two subspecies, including interrupting host genes, creating different expression forms, drastically changing intron length, and affecting expression levels of adjacent genes. These analyses provide genome-wide insights into evolutionary history and genetic variation of rice.
Transposons were first discovered and characterized in maize (Zea mays; McClintock, 1948). It was found that transposons have a great impact on genome structure and gene function in nearly all organisms (Kidwell and Lisch, 1997). Transposable elements (TEs) occupy a large proportion of nuclear genomes in many plants (Vicient et al., 1999; Meyers et al., 2001). Activities of TEs can affect individual genes, leading to the alteration of gene structure and expression (Bennetzen, 2000). Furthermore, TEs play an important role in unequal homologous recombination events (Kazazian, 2004). Recent insertion and excision of TEs have given rise to a series of transposon insertion polymorphisms (TIPs; polymorphisms consisting of the presence/absence of a TE at a particular chromosomal location) in closely related species, subspecies, and haplotypes and served as ongoing sources of genomic and genetic variation (Bennett et al., 2004).
Different from DNA transposons (class II TEs), which can be deleted precisely at a relatively low frequency, the vast majority of retrotransposon insertions (class I TEs) are irreversible, rarely undergoing precise excision. Hence, the absence of retrotransposon is regarded to be the ancestral state. Moreover, the probability that different retrotransposons would independently insert into the exact same location is negligible. Consequently, retrotransposon-based insertion polymorphisms (RBIPs), as an important subset of TIPs, are very useful in the study of deeper phylogeny in wide germplasm pools. RBIPs were developed using the PCR-based method for retrotransposon isolation (Pearce et al., 1999) as well as comparative genomics approaches. RBIPs can detect individual insertions by PCR with flanking host sequence primers and a retrotransposon-specific primer (Flavell et al., 1998). They have been applied in the study of population genetics and phylogenetic analyses of both plants and animals (Stoneking et al., 1997; Batzer and Deininger, 2002; Vitte et al., 2004; Jing et al., 2005).
Although TIPs are abundant and also informative (Du et al., 2006), a genome-wide survey of TIPs has remained scarce in plants. In rice (Oryza sativa), indica and japonica represent two major types of rice cultivars with highly diverged genomic backgrounds (Sang and Ge, 2007; Kovach et al., 2007). The sequencing of indica and japonica rice genomes provides a powerful resource for comparative and functional genomic analyses. The International Rice Genome Sequencing Project has generated highly accurate genome sequences of japonica Nipponbare using a map-based strategy (International Rice Genome Sequencing Project, 2005), and the Beijing Genomics Institute (BGI) used a shotgun approach to sequence the indica 93-11 genome with coverage of 6.28× (Yu et al., 2005). Moreover, we sequenced an approximately 23-Mb region on chromosome 4 from another indica cultivar, Guangluai 4, using the bacterial artificial chromosome (BAC)-based approach, which allowed for an in-depth comparative analysis of cultivated rice genome variations and a high-quality assessment of polymorphisms between indica and japonica cultivars. With the available genome sequences, candidate DNA polymorphisms across the rice genome were discerned to develop molecular markers (Feltus et al., 2004; Shen et al., 2004). A handful of polymorphisms were also shown to be important sources of evolutionary changes, such as functional variations in key domestication-related genes cloned in rice (Kovach et al., 2007). Therefore, it is likely that the examination of genome-wide sequence differences between the two subspecies of cultivated rice will help us understand the nature of mutations and their evolutionary potentials (Ma and Bennetzen, 2004; Tang et al., 2006).
Recent studies have found that more than 10% of the structural genes contained TEs in rice (Sakai et al., 2007), implying that TIPs would also represent significant sources of genetic variation. Previous work has revealed substantial differences in genome sizes (Han and Xue, 2003), gene content (Ding et al., 2007), and transcript levels (Liu et al., 2007) between the two subspecies. Our question is whether mobile elements played an important role in the genetic differentiation. To address this question, we performed a systematic study of recent transposon insertion events (both class I and class II TEs). In this study, a comparative approach was adopted to detect TIPs between the genomes of indica and japonica, which to our knowledge represents the first genome-wide survey of TIPs in plants. We also used RBIPs in the 23-Mb collinear regions of chromosome 4 to analyze the divergence of three sequenced rice varieties: japonica Nipponbare, indica 93-11, and indica Guangluai 4. We show that transposon insertions affected a large number of genes, potentially serving as an important driving force for intraspecific variation of cultivated rice.
RESULTS
The Abundance of TIPs between Cultivated Rice Genomes
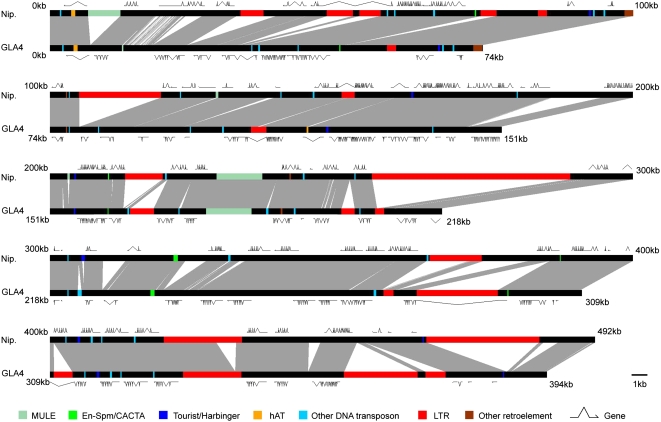
To investigate the difference between japonica Nipponbare and indica Guangluai 4 genome sequences, we selected an indica-japonica collinear region on chromosome 4, where both cultivars have BAC-based sequences and differ substantially in size (Fig. 1). The total length of this region is 492 kb in Nipponbare and 394 kb in Guangluai 4. We analyzed TEs and non-TE related genes and compared the differences between the two genome sequences in this region. Consistent with the conclusions from studies of other organisms (Britten et al., 2003), the divergence is mainly due to large insertions or deletions (indels). We counted all large indels of greater than 100 bp between the two genomes and found a total length of 147.7 kb (30.0% of the DNA sequence in the region of Nipponbare) for inserts in Nipponbare and 47.5 kb (12.1% of the DNA sequence in the region of Guangluai 4) for inserts in Guangluai 4. Surprisingly, over 67% of these indels resulted from TIPs, which generated a total length of 100.5 kb for inserts in Nipponbare and 42.3 kb for inserts in Guangluai 4. We also examined small indels of less than 100 bp and found no indels resulting from intact transposon insertions.
Figure 1.
Sequence comparison of an orthologous region between japonica Nipponbare and indica Guangluai 4. The region is approximately 492 kb in Nipponbare, from 31,616,657 to 32,108,476 bp on chromosome 4 (TIGR pseudomolecule 5.0), and approximately 394 kb in Guangluai 4. Light gray shading indicates the homologous regions, and the white areas show the indels of more than 100 bp. TEs are represented by bars of designated colors. All non-TE genes are indicated by dark lines with arrows. Exons are depicted as horizontal lines, and introns are depicted as the lines connecting exons.
To identify the differences induced by the TIPs, we performed a systematic analysis of approximately 23-Mb sequences of chromosome 4. Our approach for detecting TIPs in rice involved identifying all indels of more than 100 bp between the two genomes and then screening these insert regions to identify de novo transposon insertions. We reasoned that this approach should be effective, because many indels were related to TIPs, and, also, the lengths of most transposon insertions were longer than 100 bp, as indicated in the orthologous region mentioned above.
We aligned all of the orthologous regions between Nipponbare and Guangluai 4, and mined all indels of more than 100 bp. The results were the same when individual BACs and constructed contigs of Guangluai 4 were used for alignment. We found that there were 821 insertions (>100 bp) in Nipponbare relative to Guangluai 4 and 751 insertions (>100 bp) in Guangluai 4 relative to Nipponbare, with a total length of 3.2 Mb and 2.4 Mb, respectively (Table I). Overall, the 1,572 insertions were distributed throughout these regions, ranging from 100 to 118,675 bp in length. Large indels of greater than 2 kb were primarily responsible for the different sizes of orthologous regions between Nipponbare and Guangluai 4 (Fig. 2). The homology-based approach was used to identify indels that were caused by de novo transposon insertions. We regarded an indel as a TE insertion by employing the following criteria: first, it should have similarity to a known TE family and possess the structure of a transposon; second, it should be bound by target site duplication (TSD). With these criteria, 691 insertions of transposons were identified in the approximately 23-Mb orthologous regions of Nipponbare and Guangluai 4 (Supplemental Table S1). Among them, the most abundant polymorphisms identified were Ty3/gypsy insertion polymorphisms. A total of 110 insertions of Ty3/gypsy retrotransposons were detected in Nipponbare, while 127 insertions were detected in Guangluai 4, equivalent to 0.95 and 0.89 Mb of the sequences investigated, respectively (Table II). Other abundant transposon insertions included Ty1/copia, En-Spm/CACTA, and MULE, which were consistent with their content in the Nipponbare genome.
Table I.
Number of indels and TIPs between the two rice varieties
| Variety | Coverage of Insert Regionsa (Copy No.) | Coverage of TE Insertionsb (Copy No.) | Proportionc |
|---|---|---|---|
| Nipponbare | 3,209.096 kb (821) | 1,731.500 kb (342) | 53.96% |
| Guangluai 4 | 2,359.033 kb (751) | 1,475.964 kb (349) | 62.57% |
| Total | 5,568.426 kb (1,572) | 3,207.464 kb (691) | 57.60% |
Insert regions of >100 bp.
TE insertions of >100 bp.
The proportion of coverage of insert regions to TE insertions.
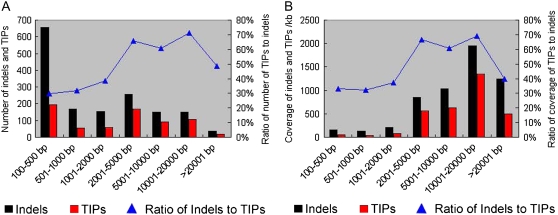
Figure 2.
Contribution of TIPs to large indels. TIPs and indels in the approximately 23-Mb orthologous regions of chromosome 4 are classified into seven groups according to their sizes, as shown at the bottom of the histograms. A, Bars show the number of TIPs and indels, in black and red, respectively. The blue line indicates the proportion of TIPs to indels. B, Bars show the coverage of TIPs and indels. The blue line denotes the ratio of TIPs to indels.
Table II.
Comparison of polymorphic transposon insertions between the two rice varieties
| Class | TE Insertion in Nipponbare
|
TE Insertion in Guangluai 4
|
||
|---|---|---|---|---|
| Copy No. | Coverage | Copy No. | Coverage | |
| bp | bp | |||
| Class I | ||||
| Ty1/copia | 46 | 278,712 | 35 | 226,678 |
| Ty3/gypsy | 110 | 953,718 | 127 | 889,838 |
| Unclassified LTR | 23 | 105,781 | 23 | 85,415 |
| Other class I | 10 | 26,293 | 8 | 39,133 |
| Total class I | 189 | 1,364,504 | 193 | 1,241,064 |
| Class II | ||||
| hAT | 13 | 23,541 | 13 | 30,542 |
| En-Spm/CACTA | 23 | 179,208 | 10 | 74,619 |
| MULE | 40 | 133,731 | 27 | 84,644 |
| Tourist/Harbinger | 21 | 6,627 | 41 | 11,170 |
| Mariner/Stowaway | 20 | 4,394 | 23 | 5,251 |
| Other MITE | 13 | 3,941 | 24 | 7,562 |
| Other class II | 11 | 7,879 | 5 | 2,544 |
| Total class II | 141 | 359,321 | 143 | 216,332 |
| Other TEs | 12 | 7,675 | 13 | 18,568 |
| Total TEs | 342 | 1,731,500 | 349 | 1,475,964 |
Although the total number of TE insertions is nearly equal in Nipponbare and Guangluai 4, the size of long terminal repeat (LTR)-retrotransposon insertions varies substantially between Nipponbare and Guangluai 4. The average length of LTR-retrotransposons is 7.5 kb in Nipponbare and 6.5 kb in Guangluai 4, which may suggest that internal deletions of LTR-retrotransposons occur more frequently in Guangluai 4. Moreover, some DNA transposon families seemed to have insertion bias in the two subspecies. The insertions of En-Spm/CACTA and MULE were more abundant in Nipponbare than in Guangluai 4, while Tourist/Harbinger insertions were more abundant in Guangluai 4.
In the approximately 23-Mb orthologous regions, there are at least 179 “young LTR-retrotransposons” in the Nipponbare genome (covering about 1.34-Mb sequences), which accumulated after the divergence of japonica and indica from a common ancestor. As the total length of the rice nuclear genome was calculated to be 389 Mb and chromosome 4 had a relatively modest retrotransposon content (International Rice Genome Sequencing Project, 2005), we estimate that there are more than 3,000 young LTR-retrotransposons with a total length of 22.6 Mb in the rice nuclear genome (equivalent to approximately 6% of the rice genome). Compared with all LTR-retrotransposons in the rice genome, young LTR-retrotransposons occupy less than 10% in number but more than 40% in size, mainly because fewer deletions occurred in the newly inserted LTR-retrotransposons.
RBIPs as Reagents to Reveal an Evolutionary History
To determine the evolutionary history of three cultivated rice varieties, Nipponbare, Guangluai 4, and 93-11, whose genomic sequences are available, we tested for the presence/absence of RBIPs between Nipponbare/Guangluai 4 in the BGI 93-11 genome by searching against the BGI 93-11 contigs. An insertion of a TE was considered to be present in 93-11 rice when the corresponding region of 93-11 had the TE insertion. Alternatively, an insertion was judged to be absent in the 93-11 genome if the TE sequences did not exist in the orthologous region of 93-11 (see “Materials and Methods” for details).
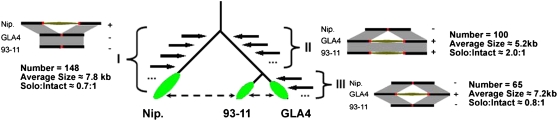
In total, 163 retrotransposon insertions present in the Nipponbare genome and 165 retrotransposon insertions present in the Guangluai 4 genome were investigated in the 93-11 genome (Table III). Of the 163 retrotransposon insertions present in Nipponbare, 148 insertions are absent in the 93-11 genome (consistent with Guangluai 4), while only 15 insertions are present in 93-11 (consistent with Nipponbare; Fig. 3, type I). This result indicates that the radiation between gene pools of Guangluai 4 and 93-11 probably occurred after the divergence between indica and japonica. The 15 exceptions reflect introgression between the two genome pools that may have occurred hundreds of years ago, as reported previously (Feltus et al., 2004). Based on these data, we estimated that the introgression rate would be about 9.2% (15 of 163 = 9.2%). As for the 165 retrotransposon insertions in the Guangluai 4 genome, 100 insertions are present in the 93-11 genome (Fig. 3, type II), while 65 insertions are absent in 93-11 (Fig. 3, type III). Furthermore, the two distinct states (presence or absence) are correlated with the ages of the insertions. This is largely based on the following evidence. First, the average length of the former 100 retrotransposon insertions is 5,212 bp, while that of the latter 65 retrotransposon insertions is 7,163 bp. Second, among the former retrotransposons, the ratio of solo LTRs to intact LTR elements is about 2:1, while the ratio of solo LTRs to intact LTR elements among the latter is about 0.8:1. These results suggest that most of the 100 retrotransposon insertions present in the 93-11 genome were inserted into the Guangluai 4 gene pool before the divergence of the Guangluai 4 and 93-11 gene pools and after the divergence of indica and japonica, whereas most of the 65 retrotransposon insertions absent in 93-11 were inserted into the Guangluai 4 gene pool after its divergence from a common ancestor with the 93-11 gene pool, although a few exceptions existed, possibly due to introgression between the two gene pools. It remains unclear whether the other 54 insertions (26 insertions in Nipponbare and 28 insertions in Guangluai 4) are present in 93-11, because there were no corresponding sequences found in 93-11 contigs or the flanking sequences of the insertions were repetitive in the genome.
Table III.
Summary of in silico analysis of RBIP patterns in three varieties
ND, Not determined.
| Variety | Present in Nipponbare Only | Absent in Guangluai 4 Only | Not Determined | Total No. | |||
|---|---|---|---|---|---|---|---|
| Nipponbare | + | 148a | + | 15 | + | 26 | 189 |
| Guangluai 4 | − | − | − | ||||
| 93-11 | − | + | ND | ||||
| Variety
|
Absent in Nipponbare Only
|
Present in Guangluai 4 Only
|
Not Determined
|
Total No.
|
|||
| Nipponbare | − | 100b | − | 65c | − | 28 | 193 |
| Guangluai 4 | + | + | + | ||||
| 93-11 | + | − | ND | ||||
Figure 3.
The phylogenetic relationship of three varieties, japonica Nipponbare, indica 93-11, and indica Guangluai 4, characterized by in silico analysis of RBIPs. The first node represents the divergence between two subspecies, while the second node denotes the radiation of ancestral indica into two gene pools, the ancestors of the 93-11 and Guangluai 4 gene pools, which are represented by the green ellipses. The dashed line represents the introgression between the two. I, II, and III are the expected patterns of RBIPs in the three varieties. Type I indicates the insertions that occurred in the Nipponbare genome after the divergence between the two subspecies. Type II occurred in the common ancestor of the Guangluai 4 and 93-11 gene pools, after the divergence. Type III happened in the Guangluai 4 gene pool, after the radiation of indica into at least two gene pools. Copy number, average length, and ratio of solo LTR to intact LTR are also listed.
In addition, we examined DNA transposon polymorphisms in 93-11. Because of the possibility of the excision of DNA transposons and the lack of ancestor information, we could not determine whether an individual TIP was an insertion or a precise excision event. However, it was found that, of 119 DNA transposon insertions present in Nipponbare, only 23 insertions were also present in 93-11 (Supplemental Table S5). The 23 insertions present in 93-11 can result from the introgression events or the excision events. Deducting from the introgression portion (9.2%), there were likely only 10.1% resulting from the excision events (23/119 − 9.2% = 10.1%). According to these results, we propose that the precise excision of DNA transposons is not frequent in rice.
Genome-Wide Detection of TIPs between Nipponbare and 93-11
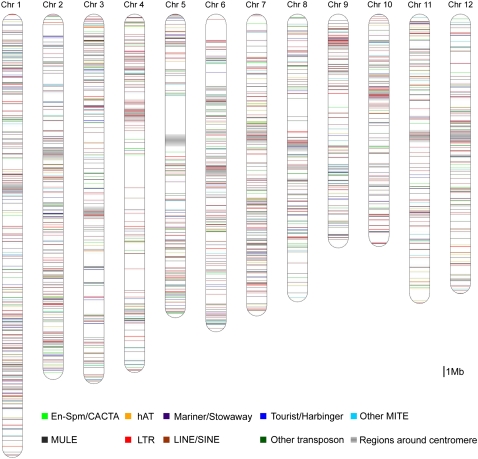
With the availability of two rice whole genome sequences and whole genome alignment, we started our mining from the alignment result of BGI 93-11 contigs with The Institute for Genomic Research (TIGR) Nipponbare pseudomolecule 5.0 (Ouyang et al., 2007). Because of the assembly problem of the 93-11 repetitive regions caused by the whole genome shotgun strategy, we only mined all insert regions in Nipponbare, and those insert regions in the 93-11genome had to be neglected. For each candidate insert region in Nipponbare, we also checked whether any 93-11 contig covers both the partial insert region and its flanking sequence, and, if found, those inserts were excluded from further analysis (see “Materials and Methods” for details). Following the algorithm, overall, 4,348 insert regions of more than 100 bp were found in the Nipponbare genome. The average length of the insert regions in Nipponbare is 2,681 bp, with the longest insert of 58,750 bp, which is filled with LTR-retrotransposons around the centromeric region of chromosome 7. After applying the approach to detect transposon insertions described above, we identified 2,041 TE insertions in the Nipponbare genome (Fig. 4; Supplemental Table S2).
Figure 4.
Distribution of 2,041 TIPs in the rice genome. Individual transposon insertions are represented by horizontal lines, and different kinds of transposons are shown in different colors. The light gray bars on the chromosomes indicate the position of centromeres. Detailed information for each TIP is listed in Supplemental Table S2.
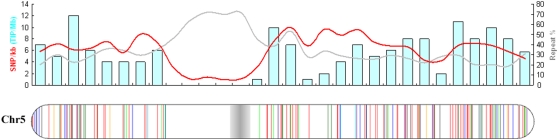
TIPs are not randomly distributed on five of the 12 rice chromosomes (chromosomes 1, 3, 4, 5, and 8; P < 0.01; Supplemental Table S6). The uneven distribution is to some extent caused by the position bias of TE insertions. It was also found that some regions lacking TIPs are also the regions of low polymorphism between Nipponbare and 93-11. For instance, the longest region lacking TE insertions is also the longest single-nucleotide polymorphism (SNP)-poor region, which is located on chromosome 5, as shown in Figure 5 (9–13 Mb in the pseudomolecules [Feltus et al., 2004]). This may reflect the introgression of chromosomal segments between Nipponbare and 93-11.
Figure 5.
Densities of SNPs, TIPs, and repeats on rice chromosome 5 (approximately 30 Mb). At top, the azure bars indicate the numbers of TIPs per megabase. The red line shows SNP rate (per kilobase) after subtraction of repetitive regions, and the gray line shows the percentage of repetitive DNA. The distribution of TIPs on chromosome 5 is shown at bottom.
Types of TE-Induced Genetic Variations
TIPs have considerable effect on genome structure and size, as described above. Moreover, they also contribute to the variation of individual genes. Various ways have been discovered in which TIPs can affect the intraspecific variation of individual genes (Fig. 6). To explore the evolutionary significance of TIPs in genetic variation, we examined all of the TIPs in the expressed gene regions and determined whether any variation caused by TIPs existed between indica and japonica (Supplemental Tables S3 and S4). Since EST and cDNA sequences can provide direct evidence for gene expression and because they are currently the most important resources for transcriptome exploration in rice, we considered a TIGR gene locus as an expressed gene region if it had at least one corresponding EST (or cDNA) in the database. The variations in these regions were classified into three types: (1) the alteration of cDNA sequence; (2) the change of intron size; and (3) the rearrangement of the promoter region. We counted the number of TIPs that were associated with the three types of genetic variation and observed that at least 10% of TIPs occurred in the expressed gene regions leading to changes ranging from subtle to dramatic (Tables IV and V).
Figure 6.
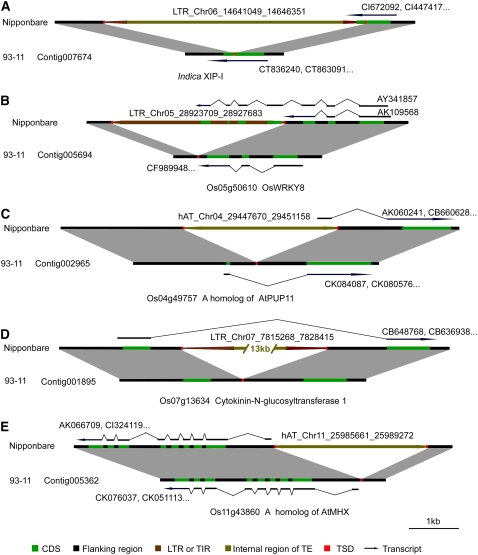
Examples of genetic variation types associated with TIPs. A, Two gene fragments were separated by a Dasheng insertion into the coding region of XIP-I. B, The insertion of copia in the 3′ UTR of OsWRKY8 created an alternative isoform in Nipponbare, which was a chimeric transcript possessing three additional exons from the TE. C, The first intron of a homolog of AtPUP11 was inserted by a hAT transposon, resulting in the loss of its original first exon and the gain of an additional exon deriving from the TE. D, Transposition of a large gypsy into a rice glucosyltransferase gene generated an intron of 15 kb. E, A 5′ upstream region of a gene was inserted by a hAT transposon. Homologous regions are indicated by light gray shading. Horizontal lines and arrows over/below the genomic region represent the corresponding Fl-cDNA or EST. LTR and internal sequences of transposons, TSDs, and coding regions are indicated by designated colors. The transcripts of indica in E are not found in the rice EST or Fl-cDNA database.
Table IV.
TIPs found in the expressed gene region
| Type | TIPs Identified between Nipponbare and 93-11
|
TIPs Identified between Nipponbare and Guangluai 4
|
TIPs Estimated between Rice Subspecies
|
|||
|---|---|---|---|---|---|---|
| No. | Proportion | No. | Proportion | No.a | Proportionb | |
| % | % | % | ||||
| Alteration of cDNA sequence | 87 | 4.3 | 27 | 3.9 | 449–495 | 3.9–4.3 |
| Rearrangement of promoter region | 77 | 3.8 | 16 | 2.3 | 265–438 | 2.3–3.8 |
| Change of intron size | 109 | 5.3 | 28 | 4.1 | 472–610 | 4.1–5.3 |
| Total in the expressed gene region | 273 | 13.4 | 71 | 10.3 | 1,186–1,543 | 10.3–13.4 |
| Other TIPs | 1,768 | 86.6 | 620 | 89.7 | 9,974–10,331 | 86.6–89.7 |
| Total TIPs | 2,041 | 100.0 | 691 | 100.0 | Approximately 11,517c | 100.0 |
The number was obtained by multiplying the proportion by total TIPs estimated.
The proportion was derived from two subsets of TIPs identified.
This estimation relies on 23-Mb collinear regions of chromosome 4, which have continuous BAC-based sequences.
Table V.
A partial list of identified genetic variations induced by TIPs
| Transposon
|
Insertion Site
|
Gene
|
Transcripts
|
Outcome | ||||
|---|---|---|---|---|---|---|---|---|
| Class | Length | BGI Contig Identifier | Position | TIGR_LOC | Annotation | japonica | indica | |
| bp | ||||||||
| CACTA | 1,388 | Contig006839 | 9,800 | Os01g45020 | F-box protein | CI766880 | CK040274, CK048023 | Gene inactivation; changing of expression |
| hAT | 959 | Contig000154 | 13,948 | Os01g57350 | d-Erythro-sphingosine kinase/diacylglycerol kinase | AK107062, CI279975 | – | Isoforms; creation or loss of exons |
| hAT | 3,489 | Contig002965 | 17,271 | Os04g49757a | A homolog of AtPUP11 | AK060241, CB660628 | CK084087, CK080576 | |
| MULE | 243 | Contig006815 | 3,983 | Os06g15170 | 3-Ketoacyl-CoA synthase | CI446376, CI443637 | – | |
| MULE | 1,453 | Contig007131 | 14,019 | Os07g38890 | Indole-3-acetic acid-amido synthetase GH3.9 | AK106839, CI095296 | – | |
| Tourist | 273 | Contig004245 | 15,911 | Os08g34150 | OsTIL-2, temperature-induced lipocalin-2 | AK105927, AK105927 | CT852268 | |
| Tourist | 288 | Contig000182 | 56,896 | Os10g10990 | Transcription initiation factor IIF, α-subunit | AK100664, AK243052 | CB628339, BI797383 | |
| Ty1/copia | 5,675 | Contig000123 | 69,888 | Os02g55550 | Ubiquitin protein ligase | AK068929, CI338600 | – | |
| Ty1/copia | 6,249 | Contig002530 | 4,510 | Os05g05620 | Glutathione S-transferase, GSTF1 | AK120912, CI344777 | CT830741 | |
| Ty1/copia | 6,432 | Contig005393 | 13,567 | Os05g47750 | Ser/Thr protein kinase | AK243156, CI392921 | CB619204, CB619241 | |
| Ty1/copia | 3,975 | Contig005694 | 14,785 | Os05g50610a | OsWRKY8 | CI162452 | – | |
| Ty1/copia | 3,054 | Contig009065 | 7,833 | Os06g01934 | BEL1-related homeotic protein 14 | AK111490, AK111490 | – | |
| Ty1/copia | 6,419 | Contig005468 | 9,437 | Os07g44380 | Disease resistance response protein 206 | – | CR292117, CR292358 | |
| Ty3/gypsy | 443 | Contig012046 | 2,913 | Os03g23980 | 3-β-Hydroxysteroid dehydrogenase/isomerase family protein | CI749065 | CX112394, CB624403 | |
| Ty3/gypsy | 5,303 | Contig007674 | 8,175 | Os06g24990a | Xylanase inhibitor protein 1 precursor | AK241892, CI447417 | CT863091, CT836240 | |
| Ty3/gypsy | 3,364 | Contig004822 | 6,929 | Os07g09814 | F-box protein | CX106642, CA764781 | CI140279, CI078113 | |
| Ty3/gypsy | 5,211 | Contig005981 | 7,187 | Os11g34970 | NB-ARC protein | – | CB628201 | |
| Ty1/copia | 5,704 | Contig004208 | 1,838 | Os01g09384 | Ankyrin-like protein | AK071981, CI574303 | – | Creation of long introns; potential |
| Ty1/copia | 6,433 | Contig014492 | 5,088 | Os01g59819 | Noncyanogenic β-glucosidase precursor | AK241229, CI428297 | BI306425 | Influence on gene expression level |
| Ty1/copia | 5,013 | Contig002407 | 33,107 | Os02g29510 | Nonimprinted in Prader-Willi/Angelman syndrome region protein 1 | AK070515, CI592449 | CK081087, CK039442 | |
| Ty1/copia | 5,755 | Contig011823 | 3,476 | Os06g23504 | DTW domain protein | – | CR279627, CX110997 | |
| Ty1/copia | 5,751 | Contig005631 | 7,546 | Os09g32450 | DNA topoisomerase 3-β-1 | AK066999, CI617601 | CT856568 | |
| Ty3/gypsy | 12,895 | Contig000177 | 86,127 | Os01g48874 | Expressed protein | CB685172, AK099862 | CT860454 | |
| Ty3/gypsy | 5,204 | Contig003484 | 18,429 | Os07g08300 | Major facilitator superfamily antiporter | AK065334 | CB634135, CB634363 | |
| Ty3/gypsy | 13,148 | Contig001895 | 27,445 | Os07g13634a | Cytokinin-N-glucosyltransferase 1 | CB648768, CB636938 | – | |
| hAT | 3,621 | Contig001630 | 9,196 | Os02g39030 | Myelin transcription factor 1 | AK065609, CI565963 | CK041254, CT845751 | Insertion in 5′ upstream region of gene; potential alteration of gene regulation |
| hAT | 3,612 | Contig005362 | 4,432 | Os11g43860a | A homolog of ATMHX | AK066709 | CK051113 CK076037 | |
| MULE | 3,594 | Contig000401 | 10,796 | Os01g56350 | Expressed protein | AK065563 | CT851672 | |
| MULE | 7,863 | Contig000016 | 85,384 | Os04g58140 | HBS1-like protein | AK110947, CI616732 | – | |
| MULE | 1,411 | Contig000053 | 89,119 | Os05g48240 | Dynamin-related protein 1A | CI567809, CI767272 | CF958238, CK054223 | |
| MULE | 7,808 | Contig014467 | 1,250 | Os07g18050 | Pre-mRNA splicing factor SLT11 | CI755183, CI581483 | CK008072, CK066987 | |
| MULE | 9,178 | Contig007231 | 9,414 | Os07g32590 | Met aminopeptidase 1B, chloroplast precursor | AK066776 | CK064717, CK007243 | |
| Ty3/gypsy | 3,169 | Contig001596 | 7,633 | Os05g47700 | Nonspecific lipid-transfer protein precursor | CI761209, CI296121 | CF989490, CT84609 | |
The cases in Figure 6.
Alteration of cDNA Sequence
After the divergence, TIPs within gene regions are likely to result in a variety of outcomes, including the alteration of gene structure and expression. To investigate these TE-induced changes in transcription level, we searched transcripts (including Fl-cDNA and ESTs) around the insertion sites. If there was a cDNA or EST match, gene annotation was inspected in Nipponbare and 93-11 on the basis of rice transcript alignments and TIGR annotation release 5. Then, individual examinations were conducted to identify the difference caused by TE insertion. TE insertions into TE-related genes were excluded manually.
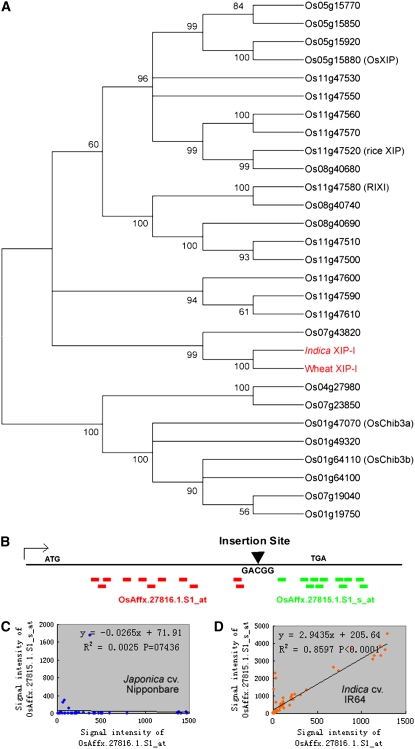
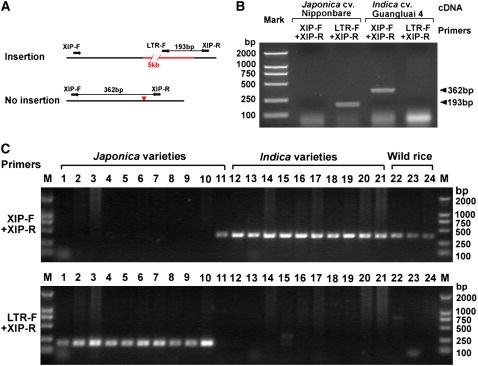
Overall, 4.3% of TIPs between Nipponbare and 93-11 and 3.9% of TIPs between Nipponbare and Guangluai 4 resulted in abnormal termination or alterative splicing, respectively. TEs that insert within coding regions are most likely to result in null mutations. For example, in hexaploid wheat (Triticum aestivum), the xylanase inhibitor protein I gene (XIP-I), whose crystal structure, expression pattern, and function have been studied in detail, was shown to function in plant defense against secreted fungal pathogen xylanases by its competitive inhibiting activity against fungal endo-1,4-β-d-xylanases (Elliott et al., 2002; Flatman et al., 2002; Payan et al., 2004; Igawa et al., 2005). Although several XIP-type xylanase inhibitors, riceXIP (Goesaert et al., 2005), OsXIP (Tokunaga and Esaka, 2007), and RIXI (Durand et al., 2005), were recently isolated from rice, there is no orthologous gene of wheat XIP-I reported to date in rice. We found its ortholog on chromosome 6 of indica 93-11, named indica XIP-I here (Fig. 7A). It does not have an ortholog in the Nipponbare genome. The mutation is caused by the insertion of a Dasheng (a type of LTR-retrotransposon) into the coding region of the XIP-I gene locus in japonica Nipponbare (Fig. 6A). Further analysis of Nipponbare's transcripts revealed that the transcription stops at the LTR of the TE, creating a truncated open reading frame (ORF), with the loss of the second half of the host XIP-I gene. As expected, no transcriptional activity can be observed in the second half of the gene in Nipponbare, according to both transcript evidence and Affymetrix microarray data of different rice cultivars (Fig. 7, B–D). Expression analysis of the gene was also carried out by reverse transcription (RT)-PCR, and the result is shown in Figure 8. We further detected whether the XIP-I gene is present in the genomes of other indica, japonica, and wild rice varieties using PCR. The LTR insertion in the XIP-I gene was detected in nearly all japonica varieties (except japonica Xuehehanzao; Fig. 8), and no insertion was found in any indica varieties or three wild Oryza species, indicating that the truncated XIP-I gene is unique to japonica varieties.
Figure 7.
Phylogenetic and GeneChip expression analyses of the XIP-I gene. A, Phylogenetic relationship of wheat XIP-I and its homologous proteins in rice. Both wheat XIP-I and indica XIP-I are highlighted with red. OsXIP, riceXIP, RIXI, OsChib3a, and OsChib3b (Park et al., 2002) are proteins that have been identified and studied in rice. B, The small colored boxes represent the positions of the probes in the two probe sets of the Affymetrix GeneChip. The probes of OsAffx.27816.1.S1_at and OsAffx.27815.1.S1_s_at are shown with red and green, respectively. The insertion position in the gene is indicated with a black triangle. The transcription initiation site is also indicated. C and D, Plot and correlation of hybrid intensity between OsAffx.27816.1.S1_at and OsAffx.27815.1.S1_s_at in different samples from Nipponbare (C) or IR64 (D). The horizontal axis shows the intensity of the probe set OsAffx.27816.1.S1_at calculated based on the hybrid intensity of its 11 probes, while the vertical axis shows the intensity of OsAffx.27815.1.S1_s_at. Pearson's correlation coefficient was used in linear correlation analysis. The significance of the slope of the regression lines is determined from the t statistic.
Figure 8.
Detection of the LTR insertion in the XIP-I gene using PCR. A, Small arrows indicate the locations of the primers used in PCR amplification. The expected sizes of PCR products in different patterns (insertion or no insertion) are also shown. B, RT-PCR analysis of XIP-I gene expression in japonica Nipponbare and indica Guangluai 4. C, Detection of the insertion in the genomic DNA of 10 indica, 11 japonica, and three wild rice varieties.
It was found that 3′ untranslated regions (UTRs) in exons are preferentially inserted, which can be easily understood because insertions in 3′ UTRs seem to be less destructive than insertions in other locations of the coding region. On the other hand, they also provide the raw material for new protein-coding regions. For example, we found that TE insertions in 3′ UTRs created an alternative spliceosome. OsWRKY8, a member of the WRKY gene family encoding transcription factors that are involved in the regulation of various biological processes (Xie et al., 2005), was inserted by a copia in the 3′ UTR (Fig. 6B). Two alternative transcript isoforms coexist in Nipponbare: one is identical to the gene isoform of indica 93-11, while the other acquired four additional exons in the transposon region, thus giving rise to a chimeric gene containing both a principal part of the host OsWRKY8 gene and a fraction of the LTR.
Insertions in introns could also have an influence on gene splicing sites. For instance, we found that a putative rice purine permease, which is a homologous gene of AtPUP11, shifted its transcription start site to the transposon hAT, thus generating a truncated ORF lacking its original first exon (Fig. 6C).
Change of Intron Size
We aligned all of the KOME Fl-cDNA and National Center for Biotechnology Information EST sequences with the genome sequences of Nipponbare using BLASTN and found that 5.3% of TIPs between Nipponbare and 93-11 and 4.1% of TIPs between Nipponbare and Guangluai 4 occurred in intron regions, respectively. TEs that insert in intron regions are less harmful relatively and have a greater chance to survive as a consequence. Generally, this is the cause of intron length polymorphisms (Wang et al., 2005). Despite the fact that small indels are usually found in introns, some transposon insertions could change intron length greatly, engendering an intron longer than 15 kb (Fig. 6D).
Modification of Expression Level and Rearrangement of Promoter Region
Considering that many promoters have fragments of TEs in plants (White et al., 1994), we investigated the TIPs in the promoter regions. The insertion of TEs could potentially modify the expression of adjacent genes, through the disruption of native promoter regulation or the donation of new regulatory signals (Kang et al., 2001; Pooma et al., 2002; Kashkush et al., 2003). In comparison with exons and introns, which can be identified precisely, it is less certain at defining regulatory regions. Here, we chose the genomic sequences that were 250 bp upstream from the predicted transcription start site of an expressed gene as the potential promoter region. A total of 3.8% of TIPs between Nipponbare and 93-11 and 2.3% of TIPs between Nipponbare and Guangluai 4 were found in such upstream regions (Fig. 6E).
We then experimentally compared relative expression levels of 15 genes that possessed TIPs in the defined upstream regions between japonica Nipponbare and indica 93-11. The results of real-time RT-PCR analyses of 14-d-old seedlings are shown in Supplemental Figure S1. Of 15 genes examined, five genes showed greater than 2-fold differences in relative expression levels between Nipponbare and 93-11. In particular, two of them, Os01g49110 and Os12g23754, showed 23-fold down-regulation and 18-fold up-regulation with the TE insertion, respectively.
Interestingly, the majority of TIPs in the upstream region of expressed genes are DNA transposons (80.5%, i.e. 62 of 77 TIPs between Nipponbare and 93-11 in the promoter region [Supplemental Table S3]), significantly higher than the average proportion (49.9%, i.e. 1,018 of 2,041 TIPs between Nipponbare and 93-11). Of these, MULEs also account for a relatively higher portion (36.4% in the promoter region versus 13.8% on average). Given the report that the vast majority of Pack-MULE transcripts is initiated from promoters in element sequences (Jiang et al., 2004), we proposed that promoters in the terminal inverted repeat (TIR) region of DNA transposons would play a complementary role. We did not find any new non-TE-related genes created by newly inserted transposons themselves, although it has been suggested that some transposons, like MULEs, can pack host gene fragments and form novel protein-coding genes in a new locus of the genome.
DISCUSSION
Utility of the TIPs
We have identified 691 TIPs between Nipponbare and Guangluai 4 in the 23-Mb collinear regions of chromosome 4 and 2,041 TIPs between the Nipponbare and 93-11 genomes. These TIPs can be used to develop molecular markers. Of the transposon insertions, about half of them were less than 1.5 kb. For these small TE insertions, a single PCR would be feasible, using primers derived from its flanking regions, resembling simple sequence repeat polymorphisms. For larger TE insertions, two rounds of PCR need to be performed. In the first reaction, amplification is a test using primers flanking the insertion. In the second reaction, one primer is designed from the flanking sequence and the other recognizes the LTR/TIR sequence of the corresponding TE. Then, as a codominant marker system, the different allelic states (presence and absence of the transposon insertion) at a locus will be revealed (Flavell et al., 1998).
Although the TIPs identified here are based on differences between only one japonica and two indica varieties, a large portion of those TIPs could be applicable to combinations of japonica and its related wild species (e.g. Oryza rufipogon) or other combinations of japonica and indica cultivars, because RBIPs and numerous DNA TIPs identified here can be regarded as events occurring in the recent past (after the divergence between indica and japonica). For example, among 2,041 TIPs between Nipponbare and 93-11, 94 are located in regions that have corresponding Guangluai 4 BAC sequences. After a comparison with Guangluai 4, we found that 85.1% (80 of 94 polymorphisms) were also polymorphic between Nipponbare and Guangluai 4.
The marker system based on TE insertions offers an ideal tool to evaluate the transposition history, frequency, and timing of mobile elements in rice. Since the patterns of the RBIPs can reveal the relationship among observed cultivars in a phylogenetically meaningful way, phylogenetic and biodiversity studies can be carried out using RBIPs. Vitte et al. (2004) tested 13 RBIPs in 66 rice varieties of both indica and japonica types and suggested that there were at least two independent domestication events of rice in Asia. More RBIPs would be needed to study genetic diversity in Oryza species and to determine the extent to which the introgression has occurred within/between cultivated and wild species in rice.
History of Rice Evolution: Early Radiation followed by Introgression
TE insertion polymorphisms distribute quite unevenly. It may reflect the local variation in TE insertions caused by differences in chromosome physiology (e.g. chromatin features, euchromatin region, or heterochromatin region). But we also observed that regions of low TE insertion polymorphism appeared to be correlated with regions of low SNPs. Occasional crosses between ancestors of 93-11 and Nipponbare may have happened, leading to the introgression of chromosomal segments. This may explain why there are 15 retrotransposon insertions absent in Guangluai 4 but shared by Nipponbare and 93-11.
In this study, two indica varieties, 93-11 and Guangluai 4, were investigated; they were the paternal cultivar of a superhybrid and a cultivar widely grown in China several decades ago, respectively. To our surprise, the TIPs between them are not rare. We found that a number of de novo transposon insertions occurred only in Guangluai 4, most of which date back to more than 0.1 million years ago. Although there may exist limited introgression, it still cannot account for the deep divergence between 93-11 and Guangluai 4 genomes. Therefore, the radiation of the indica genomes occurred unambiguously earlier than the domestication of rice, supporting multiple domestications of O. sativa.
Estimating the Level of Genomic Variation Caused by TIPs in Rice
After the completion of rice genome sequencing, the content of all types of transposons in the rice genome is estimated to be 35%. Now, our mining provided an opportunity to measure the level of variation caused by TIPs in rice varieties. The 23-Mb collinear regions of Nipponbare and Guangluai 4 are both derived from high-quality BAC-based sequences; therefore, the number of TIPs identified between Nipponbare and Guangluai 4 can be used as a gold standard to estimate the number of TIPs in the rice genome. Because the 23-Mb regions of chromosome 4 represent about 6% of the rice genome, there would be more than 11,517 TIPs in the rice genome on average (691/6% = 11,517), accounting for 53.5 Mb of DNA sequence (3.21/6% = 53.5 Mb). Hence, more than 14% of the genomic DNA sequences, which are different between indica and japonica, are due to the movements of TE. We propose that the average density of TIPs is relatively comparable between genomes of different varieties, although the 2,041 polymorphic transposon insertions identified between Nipponbare and 93-11 account for about one-sixth of the expected number. This is mainly due to the shotgun assemblies of 93-11. Despite the 6.28× coverage, the International Rice Genome Sequencing Project estimated that the nonredundant coverage of the indica 93-11 assembly was 69%. Moreover, it consists of thousands of small pieces of contigs, and misassembly of large pieces is also likely to happen. In our study, we found that the same contigs of 93-11 can be aligned to different regions in the Nipponbare genome, and several polymorphic contigs of 93-11 can be aligned to one region of the Nipponbare genome. So we had to apply relatively strict selection criteria in order to improve the accuracy of our investigation; consequently, we missed some TIPs. These observations indicate that the draft sequences of 93-11, although providing a genome-wide survey of TIPs, fell short of ascertaining all variation between subspecies.
As described above, the approach we used to recognize transposons primarily relied on sequence similarity with known repeats, and the increased improvement of the rice TE database allowed the identification of most TEs. Although this homology-based method with TSD detection performed well here, the genome comparison followed by inner structure analysis provided an innovative and complementary method for TE discovery, especially in detecting new TE families and instances (Caspi and Pachter, 2006). This is because TEs are highly enriched in these insert regions. In fact, we found that at least 56.7% of large insertion regions (>100 bp) are associated with transposon insertions. From 777 large insertion regions with direct repeats whose terminal sequences did not have any similarity to known repeat databases, we found that at least 19 of them showed clear structural features of TE elements (including six LTR-retrotransposons, four MITEs, and one MULE). These small amounts of elements are transposon insertions missed in our survey. Therefore, an integrated approach including both comparative genomic methods and structure-based methods would be desirable, given the existence of transposons with low copy numbers and the anticipated availability of multiple genome sequences of closely related species, subspecies, and varieties (Bergman and Quesneville, 2007).
Transposon Insertions as Important Sources of Genetic Variation in Rice
In this study, we showed that more than 10% of TIPs occurred in expressed gene regions. We provided a number of cases to exemplify a wide spectrum of changes induced by transposon insertions, involving deleterious effects, alternative splicing, shift of the transcription initiation site, loss or gain of exons, and so on. We estimated that the alterations at the level of the cDNA sequences between rice subspecies could add up to more than 400 (approximately 1% of all rice genes; Table IV). This is still a conservative estimation, because the variations identified in our study were mainly based on rice Fl-cDNA or EST sequences, and those lacking transcript evidence in the database were not examined.
Moreover, we used quantitative RT-PCR to examine the relative transcription levels of 15 genes that possessed TIPs in the upstream regions between Nipponbare and 93-11. At least two genes showed dramatic changes in expression levels between the two cultivars. Therefore, the TIP-influenced expression difference could potentially serve as an important source of genetic variation. An explicit experimental evaluation of the impact of TIPs on global gene expression, however, awaits full-scale transcriptional profiling in future work.
Among thousands of polymorphic TE insertions identified, we did not find any elements carrying a gene fragment and creating a new gene, if the TE-related genes, like transposases taken along by them, were neglected. To our surprise, two transcription factors regulating light signaling in Arabidopsis (Arabidopsis thaliana) were reported to be co-opted from a transposase (Lin et al., 2007). So we cannot exclude the possibility that some transposases brought by TE insertions have important functions and may explain intraspecific variation.
MATERIALS AND METHODS
Genomic Sequence Alignments and Identification of Indels
Physical mapping of the rice (Oryza sativa) indica Guangluai 4 chromosome 4 was conducted by an integrated approach (Zhao et al., 2002), and the sequenced BACs were assembled, forming 87 contigs (http://www.ncgr.ac.cn/english/edatabasei_ctg.htm). The overlap regions of Guangluai 4 BACs were noted to avoid double-counting in the following analysis. The BAC sequences and 87 contigs of indica Guangluai 4 that were more than 100 kb in length were aligned with rice pseudomolecules (TIGR release 5) to determine their corresponding regions in japonica Nipponbare by BLASTN search with a threshold e-value of 10−100. The identified collinear regions in the japonica chromosome 4 were extracted for further comparison. Candidate indels were identified using the diffseq program (using the default parameter) in the EMBOSS package (Rice et al., 2000), and indels of more than 100 bp were further confirmed by BLAST2 (Altschul et al., 1997). Two types of comparisons with the corresponding japonica sequences, by BACs directly or by 87 assembled contigs, were performed.
The alignment results of BGI 93-11 contigs and Nipponbare pseudomolecules, which were generated by the software nucmer, were downloaded using the GFF Dumper on the TIGR Genome Browser. We found that a small quantity of anchor results were self-contradictory; that is, two 93-11 contigs that localized on the same location yielded opposite patterns (insertion or no insertion in japonica). Hence, a perl script was written to wipe off all of these abnormal anchor results. We used only maximal exact matches that were unique in both the query and reference sequences as the alignment anchors to avoid potential errors caused by misassembly or inaccurate anchoring. Then, another script was developed to mine all of the indels of more than 100 bp based on the renewed anchor results. The indels of more than 100 bp were further confirmed by BLAST2.
The indica Guangluai 4 BACs were obtained from http://www.ncgr.ac.cn/chinese/databasei.htm. The genomic sequences of japonica Nipponbare were downloaded from http://www.tigr.org/tdb/e2k1/osa1, and the indica 93-11 sequences were downloaded from ftp://ftp.genomics.org.cn.
Mining of TIPs in the Rice Genome
For each insertion region identified above, the query sequence, composed of the insertion region and its flanking DNA (both 100 bp upstream and 100 bp downstream), was extracted and used to screen against all known TE sequences using RepeatMasker (open version 3.0.5). The known TE sequences included all transposons and transposon-like elements collected by Repbase (volume 12, issue 9; http://www.girinst.orgthe RTEdb (Juretic et al., 2004), the TIGR Rice Repeat Database (ftp://ftp.tigr.org/pub/data/TIGR_Plant_Repeats/TIGR_Oryza_Repeats.v3.3), and the MULE TIR library (Juretic et al., 2005). We used a Smith-Waterman cutoff score of 225 calculated by the cross_match program (other settings: -nolow, -no_is, -nocut). After that, the insertion regions were set aside unless they were recognized as intact transposon elements or both of their terminal sequences belonged to the same transposon family, which were distinct from their flanking sequences. Meanwhile, all indels were examined by a perl script to determine whether potential TSDs (2–18 bp) were present. All candidate transposon insertions satisfying both criteria (i.e. with homology to known TE sequences and the detection of TSDs) were further inspected. The classification of the identified transposon insertions was based on the descriptions in the repeat databases. Those transposon insertions, which had different definitions in different repeat databases, were then removed.
Characterization of RBIPs between Nipponbare and Guangluai 4 in 93-11 Targeted Regions
To determine the states (presence or absence) of transposon insertions identified between Nipponbare and Guangluai 4 in the corresponding regions of indica 93-11, we conducted sequence comparisons targeting the transposon insertion sites. For each transposon insertion identified between Nipponbare and Guangluai 4, three unique 200-bp sequences were extracted and used to search against the assembled indica 93-11 contigs, using BLASTN with a threshold e-value of 10−20. For the first two unique 200-bp sequences, each was composed of 100 bp of one transposon terminal sequence and 100 bp of its flanking DNA, from one genome with the TE insertion. The third one was a 200-bp sequence free of transposon insertion, from the other genome without the TE insertion. An insertion of a TE was considered to be shared in the indica 93-11 genome when either of the first two unique sequences was found in assembled indica 93-11 contigs (a threshold identity percentage of 95%). Alternatively, an insertion was judged to be absent in indica 93-11 when the third unique sequence was found in 93-11 contigs, with the same threshold value. We regarded the insertion as not having its explicit target region if the BLAST search did not yield any expected result or yielded two equally perfect hits, indicating both the presence and absence of the insertion in the 93-11 genome. Then, the extracted region and its clear ortholog were aligned using BLAST2 to check for the presence or absence of the insertion. Meanwhile, we also used the anchor result mentioned above to seek the corresponding locations of 93-11 contigs to confirm the states of the TE insertions in 93-11 targeted regions and eliminate all potential artifacts.
Classification of LTR-Retrotransposons
The LTR-retrotransposon insertions, which were identified between Nipponbare and Guangluai 4, were taken out for further analysis. Sequence comparisons and structural analysis were used to classify solo LTRs, intact LTR elements, and other truncated elements. Intact LTR retrotransposons were identified by the LTR_Finder program (Xu and Wang, 2007; http://tlife.fudan.edu.cn/ltr_finder/, using the default parameter) and the alignment results of their terminal sequences using the BLAST2 program. The paired length between two terminal sequences of a retrotransposon must be longer than 100 bp, and the identity must be greater than 85%. Solo LTR retroelements were identified by sequence homology search against all known TE repeat databases using RepeatMasker, as described above. Those elements, which were composed of a single LTR, were recognized as solo LTR retroelements.
EST Analysis and Gene Prediction
All publicly available rice ESTs were obtained from the National Center for Biotechnology Information EST database (http://www.ncbi.nlm.nih.gov/projects/dbEST/). Full-length cDNAs of both KOME (http://red.dna.affrc.go.jp/cDNA/; japonica Nipponbare; Rice Full-Length cDNA Consortium, 2003) and National Center for Gene Research (http://www.ncgr.ac.cn/cDNA/; indica Guangluai 4; Liu et al., 2007) were also included. Transposon insertions and their flanking regions were used to search against the EST/Fl-cDNA database using BLASTN with a threshold e-value of 10−20. The candidate transcripts were then aligned with genomic sequences using GMAP (Wu and Watanabe, 2005) with a cutoff of minimum 95% identity over 70% of the length of the transcript. Gene predictions in Nipponbare were mainly based on the annotation provided by TIGR. The exon-intron structure and various transcript isoforms of the genes were reexamined individually via alignment with their corresponding cDNA/EST. If a cDNA or EST transcribes through an insertion site in one genome without the TE insertion, or possesses a truncated gene and a fragment of its flanking transposon sequences in the other genome with the TE insertion, the gene was considered to have different transcript structures between the two genomes. In addition, if various transcript isoforms around the insertion site were found in the genome with the TE insertion at the site (i.e. if two transcripts from the same genome showed different exon-intron structures in the insertion site; Fig. 6B), it would be determined as alternative splicing. For each expressed gene, the transcription start site was determined by comparing UTR sequences (TIGR release 5) with the corresponding genomic sequence. TIPs in the region of less than 250 bp upstream from the transcription start site were defined to be in the potential regulatory region.
Phylogenetic Analysis
BLASTp search against all of the annotated proteins in the whole rice genome at TIGR (release 5) was conducted using the wheat (Triticum aestivum) XIP-I protein (GenBank accession no. CAD19479) as the query. The search resulted in the identification of 30 proteins with an e-value cutoff of 1E-5. Among them, Os06g25010 and Os06g24990, as two gene fragments after TE insertion, were replaced by indica XIP-I. The protein is coded by the longest ORF within an indica rice full-length cDNA (GenBank accession no. CT836240), and there are no nucleotide differences between the indica cDNA and 93-11 genome sequences. Os12g18750 was removed because it shows an incomplete domain and low homology when checked individually. Those protein sequences were aligned using ClustalW (Thompson et al., 1994). Unrooted phylogenetic trees were generated in MEGA4 (Tamura et al., 2007) by the neighbor-joining method (Saitou and Nei, 1987) using the Poisson correction method (Zuckerkandl and Pauling, 1965). The 50% majority rule condensed tree is shown in Figure 7. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are indicated next to the branches (Felsenstein, 1985). For convenience, we have removed the LOC prefix from all TIGR locus identifiers.
Microarray Data Extraction and Statistical Analysis
From Rice Multi-platform Microarray Search, we got the two Affymetrix probe set identifiers, OsAffx-27816-1-S1_at and OsAffx.27815.1.S1_s_at, which represent the two gene fragments of indica XIP-I separated by a TE insertion. The probes in the two probe sets were remapped to the rice genomes, Nipponbare pseudomolecules and 93-11 contigs, by BLASTN. We downloaded the microarray data files of each experiment from the GEO Web site (http://www.ncbi.nlm.nih.gov/geo/). Overall, there are 57 chips of indica IR64 (45 from GSE6893 and 12 from GSE6901) and 45 chips of japonica Nipponbare (13 from GSE7951, 4 from GSE6908, 24 from GSE6719, and 4 from GSE6720). The signal intensity data were extracted using a perl script. Pearson's correlation coefficient was applied in linear correlation analysis. The significance of the slope of the regression line was determined by the R language package.
Confirmation of the Insertion in the XIP-I Gene by PCR Assays
RNAs of japonica Nipponbare and indica Guangluai 4 were extracted from 14-d-old rice seedlings. First-strand cDNA was synthesized by SuperScript II reverse transcriptase (Invitrogen) at 42°C for 1 h. Genomic DNA of the 10 indica (Guangluai 4, 93-11, Teqing, Kasalath, TN1, Liantangzao, Zhefu 802, Nanjing 6, IR36, and Zhaiyeqing 8) and 11 japonica (Nipponbare, Yangguang, Qiuguang, Shiokari, Lansheng, Juangguang, Jinnanfeng, Suyunuo, Nongken 58, Yueguang, and Xueheaizao) varieties was prepared from 2-week-old rice seedling shoots as described (Liu et al., 2007). The genomic DNA of Oryza rufipogon, Oryza barthii, and Oryza longistaminata was provided by Wang Zixuan (Plant Genome Center). Gene-specific primers were designed around the insertion site of XIP-I genes: 5′-AGGTGGAGGATAGGCGATTGA-3′ and 5′-ACGACAACTGCGAGGCCTACT-3′. The primer in the LTR region was 5′-ACCAGGACTTTAGGCCGTTCC-3′. PCR was carried out by Taq DNA polymerase (TaKaRa Biotechnology) with the following program: an initial 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 15 s.
Real-Time PCR Analysis
RNAs of 14-d-old seedlings of indica 93-11 and japonica Nipponbare were extracted as described above. Quantitative PCR was performed on the Applied Biosystems 7500 real-time PCR System using SYBR Premix Ex Taq (TaKaRa). The PCR thermal cycle conditions were as follows: denaturing at 95°C for 10 s and 40 cycles at 95°C for 5 s and 60°C for 34 s. The two rice genes used as internal reference genes to calculate relative transcript levels were UBQ5 (AK061988) and eEF-1α (AK061464; Jain et al., 2006). The primer efficiency used to calculate the relative quantification was 2.0. The primer sequences are listed in Supplemental Table S7. Three technical replicates were used for real-time PCR analysis. We performed Student's t test (two tailed) to identify relative differences between Nipponbare and 93-11.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Real-time PCR analysis of 15 genes in the 14-d-old seedlings of both japonica Nipponbare and indica 93-11.
Supplemental Table S1. The 691 TIPs between Nipponbare and Guangluai 4 in the 23-Mb regions of chromosome 4.
Supplemental Table S2. The 2,041 TIPs between Nipponbare and 93-11 in the whole genome.
Supplemental Table S3. All identified genetic variations induced by TIPs between Nipponbare and 93-11.
Supplemental Table S4. All identified genetic variations induced by TIPs between Nipponbare and Guangluai 4.
Supplemental Table S5. Summary of in silico analysis of DNA TIP patterns in three rice varieties.
Supplemental Table S6. The nonrandom distribution of TIPs in the rice chromosomes.
Supplemental Table S7. Primer sequences used for real-time PCR analysis.
Supplementary Material
Acknowledgments
We thank Yiwen Yan for taking care of rice plants and Yajun Hu for help in the experiments. We also thank Dr. Thomas Bureau and his students from McGill University for their helpful comments on the detection of transposon MULEs.
This work was supported by the Ministry of Science and Technology of China (grant nos. 2006AA10A102 and 2005CB120805) and the Chinese Academy of Sciences (grant no. KSCW2–YW–N–024).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Bin Han (bhan@ncgr.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3 370–379 [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE (2004) Natural genetic variation caused by transposable elements in humans. Genetics 168 933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42 251–269 [PubMed] [Google Scholar]
- Bergman CM, Quesneville H (2007) Discovering and detecting transposable elements in genome sequences. Brief Bioinform 8 382–392 [DOI] [PubMed] [Google Scholar]
- Britten RJ, Rowen L, Williams J, Cameron RA (2003) Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci USA 100 4661–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Pachter L (2006) Identification of transposable elements using multiple alignments of related genomes. Genome Res 16 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Araki H, Wang Q, Zhang P, Yang S, Chen JQ, Tian D (2007) Highly asymmetric rice genomes. BMC Genomics 8 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du CG, Swigonova Z, Messing J (2006) Retrotranspositions in orthologous regions of closely related grass species. BMC Evol Biol 6 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, Juge N (2005) Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J 272 1745–1755 [DOI] [PubMed] [Google Scholar]
- Elliott GO, Hughes RK, Juge N, Kroon PA, Williamson G (2002) Functional identification of the cDNA coding for a wheat endo-1,4-beta-D-xylanase inhibitor. FEBS Lett 519 66–70 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence intervals on phylogenies: an approach using the bootstrap. Evolution Int J Org Evolution 39 783–791 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies Indica and Japonica genome alignments. Genome Res 14 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatman R, McLauchlan WR, Juge N, Furniss C, Berrin JG, Hughes RK, Manzanares P, Ladbury JE, O'Brien R, Williamson G (2002) Interactions defining the specificity between fungal xylanases and the xylanase-inhibiting protein XIP-I from wheat. Biochem J 365 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Knox MR, Pearce SR, Ellis THN (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16 643–650 [DOI] [PubMed] [Google Scholar]
- Goesaert H, Gebruers K, Courtin CM, Delcour JA (2005) Purification and characterization of a XIP-type endoxylanase inhibitor from rice (Oryza sativa). J Enzyme Inhib Med Chem 20 95–101 [DOI] [PubMed] [Google Scholar]
- Han B, Xue YB (2003) Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol 6 134–138 [DOI] [PubMed] [Google Scholar]
- Igawa T, Tokai T, Kudo T, Yamaguchi I, Kimura M (2005) A wheat xylanase inhibitor gene, Xip-I, but not Taxi-I, is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci Biotechnol Biochem 69 1058–1063 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345 646–651 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao ZR, Zhang XY, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573 [DOI] [PubMed] [Google Scholar]
- Jing RC, Knox MR, Lee JM, Vershinin AV, Ambrose M, Ellis THN, Flavell AJ (2005) Insertional polymorphism and antiquity of PDR1 retrotransposon insertions in Pisum species. Genetics 171 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic N, Bureau TE, Bruskiewich RM (2004) Transposable element annotation of the rice genome. Bioinformatics 20 155–160 [DOI] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res 15 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Lebrun MH, Farrall L, Valent B (2001) Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol Plant Microbe Interact 14 671–674 [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33 102–106 [DOI] [PubMed] [Google Scholar]
- Kazazian HH (2004) Mobile elements: drivers of genome evolution. Science 303 1626–1632 [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA 94 7704–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23 578–587 [DOI] [PubMed] [Google Scholar]
- Lin RC, Ding L, Casola C, Ripoll DR, Feschotte C, Wang HY (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Lu TT, Yu SL, Li Y, Huang YC, Huang T, Zhang L, Zhu JJ, Zhao Q, Fan DL, et al (2007) A collection of 10,096 indica rice full-length cDNAs reveals highly expressed sequence divergence between Oryza sativa indica and japonica subspecies. Plant Mol Biol 65 403–415 [DOI] [PubMed] [Google Scholar]
- Ma JX, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1948) Mutable loci in maize. Carnegie Inst Washington Year Book 47 155–169 [PubMed] [Google Scholar]
- Meyers BC, Tingley SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35 D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Kim DH, Truong NH, Itoh Y (2002) Heterologous expression and characterization of class III chitinases from rice (Oryza sativa L.). Enzyme Microb Technol 30 697–702 [Google Scholar]
- Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, et al (2004) The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J Biol Chem 279 36029–36037 [DOI] [PubMed] [Google Scholar]
- Pearce SR, Stuart-Rogers C, Knox MR, Kumar A, Ellis THN, Flavell AJ (1999) Rapid isolation of plant Ty1-copia group retrotransposon LTR sequences for molecular marker studies. Plant J 19 711–717 [DOI] [PubMed] [Google Scholar]
- Pooma W, Gersos C, Grotewold E (2002) Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16 276–277 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei N (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Sakai H, Tanaka T, Itoh T (2007) Birth and death of genes promoted by transposable elements in Oryza sativa. Gene 392 59–63 [DOI] [PubMed] [Google Scholar]
- Sang T, Ge S (2007) Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev 17 533–538 [DOI] [PubMed] [Google Scholar]
- Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY, Wang G, Wang C, Qian LL, Li X, et al (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M, Fontius JJ, Clifford SL, Soodyall H, Arcot SS, Saha N, Jenkins T, Tahir MA, Deininger PL, Batzer MA (1997) Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res 7 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tang T, Lu J, Huang J, He J, McCouch SR, Shen Y, Kai Z, Purugganan MD, Shi S, Wu CI (2006) Genomic variation in rice: genesis of highly polymorphic linkage blocks during domestication. PLoS Genet 2 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga T, Esaka M (2007) Induction of a novel XIP-type xylanase inhibitor by external ascorbic acid treatment and differential expression of XIP-family genes in rice. Plant Cell Physiol 48 700–714 [DOI] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Namthamat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Ishii T, Lamy F, Brar D, Panaud O (2004) Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol Genet Genomics 272 504–511 [DOI] [PubMed] [Google Scholar]
- Wang XS, Zhao XQ, Zhu J, Wu WR (2005) Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res 12 417–427 [DOI] [PubMed] [Google Scholar]
- White SE, Habera LF, Wessler SR (1994) Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci USA 91 11792–11796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou XL, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35 W265–W268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li SG, Li H, Zhou J, Ni PX, Dong W, Hu SN, Zeng CQ, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 266–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Cheng ZK, Chen MS, Wang SY, Feng Q, Huang YC, Li Y, Tang YS, Zhou B, et al (2002) A fine physical map of the rice chromosome 4. Genome Res 12 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In V Bryson, HJ Vogel, eds, Evolving Genes and Proteins. Academic Press, New York, pp 97–166
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.