Abstract
Collisions with DNA tracking systems are critical for the conversion of transient topoisomerase-DNA cleavage complexes to permanent strand breaks. Since DNA is overwound ahead of tracking systems, cleavage complexes most likely to produce permanent strand breaks should be formed between topoisomerases and positively supercoiled molecules. Therefore, the ability of human topoisomerase IIα and β, and topoisomerase I to cleave positively supercoiled DNA was assessed in the absence or presence of anticancer drugs. Topoisomerase IIα and β maintained ∼4-fold lower levels of cleavage complexes with positively rather than negatively supercoiled DNA. Topoisomerase IIα also displayed lower cleavage with overwound substrates in the presence of non-intercalative drugs. Decreased drug efficacy was due primarily to a drop in baseline (i.e., non-drug) cleavage, rather than an altered interaction with the enzyme-DNA complex. Similar results were seen for topoisomerase IIβ, but the effects of DNA geometry on drug-induced scission were somewhat less pronounced. With both topoisomerase IIα and β, intercalative drugs displayed higher relative cleavage enhancement with positively supercoiled DNA. This appeared to result from negative effects of high concentrations of intercalative agents on underwound DNA. In contrast to the type II enzymes, topoisomerase I maintained ∼3-fold higher levels of cleavage complexes with positively supercoiled substrates and displayed an even more dramatic increase in the presence of camptothecin. These findings suggest that the geometry of DNA supercoils has a profound influence on topoisomerase-mediated DNA scission, and that topoisomerase I may be an intrinsically more lethal target for anticancer drugs than either topoisomerase IIα or β.
Topoisomerases are ubiquitous enzymes that regulate DNA over- and underwinding, and remove knots and tangles from the genetic material (1-9). There are two classes of topoisomerases that are distinguished by their mechanism of action. Type I enzymes alter DNA topology by generating transient single-stranded breaks in the double helix (5, 8, 10), while type II enzymes act by creating transient double-stranded breaks (4, 5, 7-9, 11). Topoisomerases play critical roles in virtually every major DNA process, including replication, transcription, and recombination. In addition, they are essential for proper chromosome structure and segregation (1-7, 11). Beyond the critical physiological functions of topoisomerases, these enzymes are targets for some of the most important anticancer drugs currently used for treating human malignancies (4, 7-10, 12-15).
Humans encode three different type I topoisomerases, topoisomerase I, IIIα, and IIIβ (8, 16-19). Of these enzymes, only topoisomerase I currently is exploited for cancer chemotherapy (8, 10, 20). This enzyme is the target for an emerging class of drugs based on the parent compound, camptothecin (8, 10, 20, 21). One important aspect of topoisomerase I-targeted agents is that they are active against a spectrum of cancers that currently have few, if any, treatment options.
In addition to the type I enzymes, humans encode two isoforms of topoisomerase II, α and β (8, 9, 22, 23). Although these isoforms are closely related, they are encoded by separate genes and display different patterns of expression and cellular functions. Topoisomerase IIα is growth-dependent and is expressed primarily in rapidly proliferating tissues (2, 24-27). This isoform is essential for cell survival and is required for proper DNA replication and chromosome segregation (2, 6, 28). In contrast, topoisomerase IIβ is growth-independent and appears to be expressed in all cell types, regardless of proliferative status (2, 26, 29, 30). The cellular functions of this isoform have yet to be fully defined. Although topoisomerase IIβ is dispensable at the cellular level, it is required for proper neural development (31).
Type II topoisomerases are targets for a number of well-established chemotherapeutic agents (4, 7-10, 12, 13, 32). One of these drugs, etoposide, has been used in the clinic since the 1960’s (12, 13, 15). It is prescribed as treatment for a wide spectrum of leukemias, lymphomas, and solid tumors. The relative contributions of topoisomerase IIα and β to therapeutic outcomes are not clear at the present time. However, drug interactions with topoisomerase IIβ in differentiated tissues, such as heart, are believed to contribute to the dose-limiting toxicity of some agents (33-36).
Although topoisomerase-targeted anticancer drugs come from several different structural classes, they work through a common mechanism. All of these agents act by increasing levels of covalent topoisomerase-cleaved DNA complexes (i.e., cleavage complexes) that are requisite intermediates in the catalytic cycles of these enzymes (4, 7, 10, 32, 37). Cleavage complexes are transient in nature. However, they are converted to permanent DNA strand breaks when nucleic acid tracking systems, such as replication or transcription complexes, attempt to traverse these covalent topoisomerase-DNA roadblocks in the genetic material (4, 7, 10, 32, 37). The resulting strand breaks, as well as the inhibition of essential DNA processes, initiate multiple recombination/repair pathways (4, 7, 38-40). If the accumulation of strand breaks becomes overwhelming, they trigger cell death pathways (39).
Globally, DNA in all eukaryotic cells is underwound (i.e., negatively supercoiled) (3, 41-43). This puts energy into the genetic material and makes it easier to separate the two strands of the double helix. Because of the universal nature of underwound DNA, negatively supercoiled substrates have been used for many previous studies on the actions of anticancer drugs against topoisomerases. Other studies have utilized linear nucleic acid substrates, especially for mapping sites of drug-induced DNA scission.
In contrast to global underwinding, the movement of DNA tracking systems through the genetic material results in an acute overwinding (i.e., positive supercoiling) immediately ahead of replication or transcription enzymes (3, 6, 41, 43-47). Therefore, collisions with tracking systems, which are critical for the conversion of topoisomerase-DNA cleavage complexes to permanent strand breaks (4, 7, 10, 32, 37), most likely occur in overwound rather than underwound regions of the genome.
Despite the importance of DNA tracking enzymes to drug efficacy, relationships between the geometry of DNA supercoils and the activity of topoisomerase-targeted anticancer agents have not been characterized. Thus, the ability of drugs to induce DNA cleavage mediated by human topoisomerase IIα, topoisomerase IIβ, and topoisomerase I was assessed with positively supercoiled substrates. Results indicate that the geometry of DNA supercoils has a profound influence on topoisomerase-mediated nucleic acid cleavage and can significantly diminish or enhance the actions of anticancer drugs in an enzyme-dependent manner. Finally, results provide an explanation for the differential effects of intercalative and non-intercalative drugs on DNA scission mediated by type II topoisomerases.
EXPERIMENTAL PROCEDURES
Enzymes and Materials
Human topoisomerase IIα and topoisomerase IIβ were expressed in Saccharomyces cerevisiae (48) and purified as described previously (49). Human topoisomerase I was a gift from Dr. Mary Ann Bjornsti (St. Jude Children’s Research Hospital). Negatively supercoiled pBR322 DNA was prepared from Eschericia coli using a Plasmid Mega Kit (Qiagen) as described by the manufacturer.
Positively supercoiled pBR322 DNA was prepared by incubating negatively supercoiled plasmids with Archaeoglobus fulgidus reverse gyrase (50, 51). The average number of superhelical twists present in DNA substrates and the resulting σ values were determined by electrophoretic band counting relative to fully relaxed molecules (51). For negatively supercoiled substrates, time courses for the relaxation of pBR322 by topoisomerase I were resolved by electrophoresis in 1% agarose gels containing 1-2 μg/ml chloroquine (Sigma) in the running buffer. The initial plasmid contained ∼15 to 17 negative superhelical twists per molecule (σ ≈ -0.035 to -0.039). This superhelical density is typical of plasmids isolated from E. coli. For positively supercoiled substrates, time courses for the generation of positive superhelical twists by reverse gyrase were resolved by electrophoresis as above in running buffer containing 5-15 μg/ml netropsin B (Boehringer Mannheim). These plasmids contained ∼15 to 17 positive superhelical twists per molecule (σ ≈ +0.035 to +0.039). The handedness of positively supercoiled DNA was confirmed by two-dimensional gel electrophoresis (51). Thus, the supercoiled substrates employed for this study contained equivalent numbers of superhelical twists, but were of opposite handedness.
It should be noted that positively supercoiled plasmids bind less ethidium bromide than negatively supercoiled molecules. To ensure that equal amounts of plasmid were used in all experiments, DNA concentration was assessed by spectrophotmetric analysis and confirmed by ethidium bromide staining of linearized plasmid substrates (see insets, Figures 1, 6, and 8).
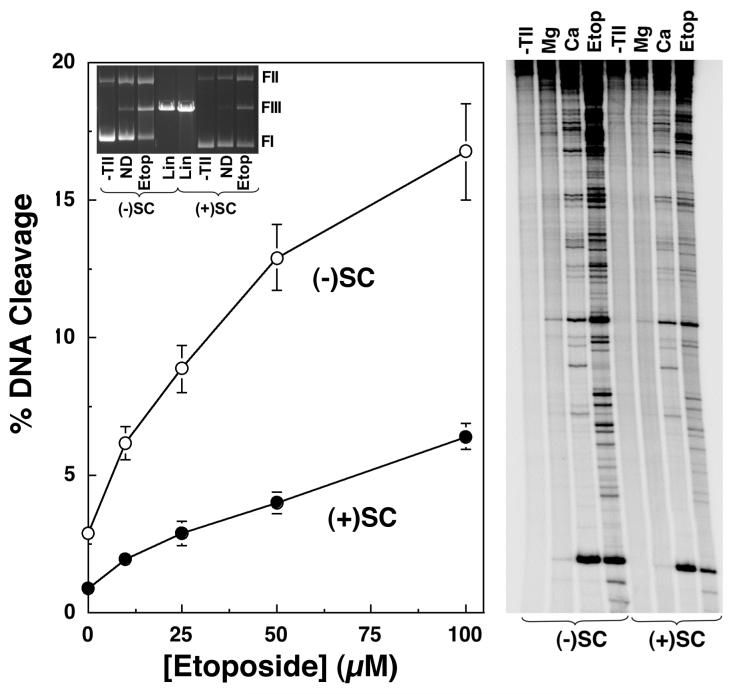
FIGURE 1.
Human topoisomerase IIα maintains lower levels of DNA cleavage complexes with positively supercoiled plasmids in the absence or presence of etoposide. The ability of topoisomerase IIα to cleave negatively [(-)SC, open circles] or positively [(+)SC, closed circles] supercoiled pBR322 plasmid DNA in the presence of 0-100 μM etoposide is shown (left panel). Assays employed Mg2+ as the divalent cation. Error bars represent the standard deviation of four independent assays. The inset shows a representative ethidium bromide-stained agarose gel of DNA cleavage assays with negatively and positively supercoiled plasmids in the absence of topoisomerase IIα (-TII) and in the absence (no drug, ND) or presence (Etop) of etoposide. Linear DNA standards (Lin) also are shown. The positions of supercoiled (form I, FI), nicked circular (form II, FII), and linear molecules (form III, FIII) are indicated. DNA sites cleaved by topoisomerase IIα were mapped in negatively or positively supercoiled plasmids (right panel). Products of DNA cleavage assays were linearized and singly-end labeled with [32P]phosphate. The autoradiogram is representative of three independent assays. Reactions were carried out in the presence of 0 or 100 μM etoposide, and utilized either Mg2+ or Ca2+ as the divalent cation. DNA from reactions that lacked topoisomerase IIα (-TII) are shown.
FIGURE 6.
Human topoisomerase IIβ maintains lower levels of DNA cleavage complexes with positively supercoiled plasmids in the absence or presence of etoposide. The ability of topoisomerase IIβ to cleave negatively [(-)SC, open circles] or positively [(+)SC, closed circles] supercoiled pBR322 plasmid DNA is shown (left panel). Error bars represent the standard deviation of four independent assays. DNA cleavage in the presence of 0-100 μM etoposide also is depicted (right panel). Error bars represent the standard deviation of three independent assays. The inset shows a representative ethidium bromide-stained agarose gel of DNA cleavage assays with negatively and positively supercoiled plasmids in the absence of topoisomerase IIβ (-TII) and in the absence (no drug, ND) or presence (Etop) of etoposide. Linear DNA standards (Lin) also are shown. The positions of supercoiled (FI), nicked circular (FII), and linear molecules (FIII) are indicated.
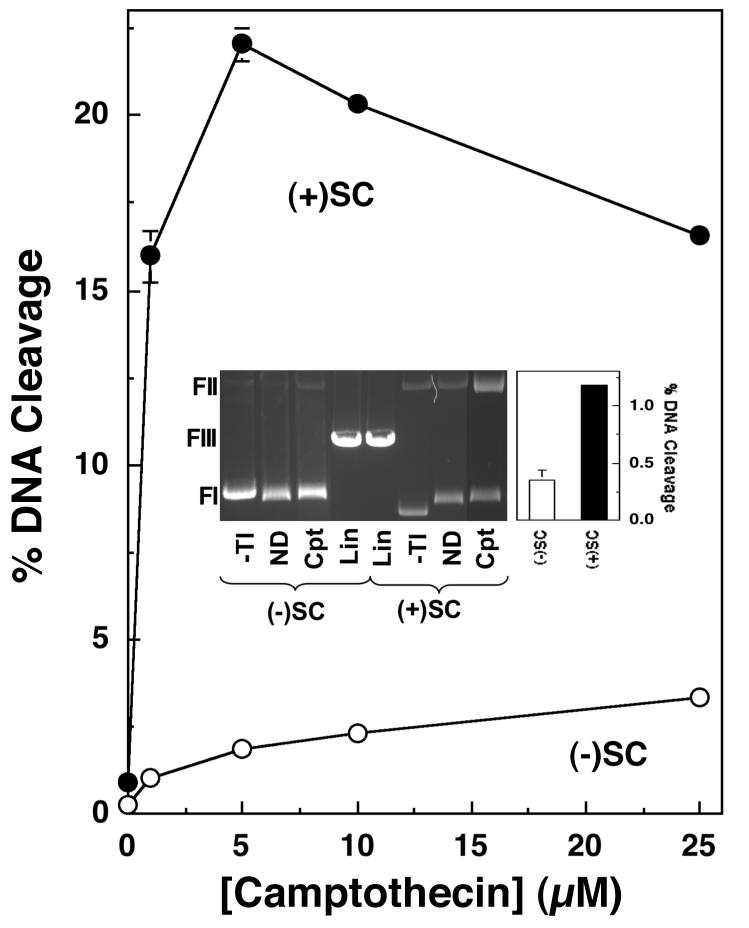
FIGURE 8.
Human topoisomerase I maintains higher levels of DNA cleavage complexes with positively supercoiled plasmids in the absence or presence of camptothecin. The ability of topoisomerase I to cleave negatively [(-)SC, open circles] or positively [(+)SC, closed circles] supercoiled pBR322 plasmid DNA in the presence of 0-25 μM campthothecin is shown. Error bars represent the standard deviation of three independent assays. The inset shows a representative ethidium bromide-stained agarose gel of DNA cleavage assays with negatively and positively supercoiled plasmids in the absence of topoisomerase I (-TI) and in the absence (no drug, ND) or presence (Cpt) of 5 μM camptothecin. Linear DNA standards (Lin) also are shown. The positions of supercoiled (FI), nicked circular (FII), and linear molecules (FIII) are indicated. The inset also shows a bar graph highlighting topoisomerase I-mediated DNA cleavage of negatively (open bar) and positively (closed bar) supercoiled DNA in the absence of drug.
[γ-32P]ATP (∼5000 Ci/mmol) and genistein were obtained from ICN, etoposide and camptothecin were from Sigma, TOP-53 and TAS-103 were gifts from Taiho Pharmaceuticals, amsacrine was a gift from Bristol-Myers Squibb, and CP-115,953 was a gift from Pfizer Global Research. Etoposide, camptothecin, genistein, TOP-53, amsacrine, and CP-115,953 were stored at 4 ° C as 10 or 20 mM stock solutions in 100% DMSO. TAS-103 was stored at 4 ° C as a 10 mM stock solution in water. All other chemicals were analytical reagent grade.
Plasmid DNA Cleavage
DNA cleavage reactions were carried out using the procedure of Fortune and Osheroff (52). Unless stated otherwise, topoisomerase II DNA cleavage assays contained 220 nM human topoisomerase IIα or β and 10 nM negatively or positively supercoiled pBR322 DNA in a total of 20 μL of topoisomerase II cleavage buffer [10 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, and 2.5% glycerol (v/v)]. Assays were carried out in the absence of compound or in the presence of 0-100 μM etoposide, 50 μM genistein or TOP-53, 5 μM CP-115,953, 0-500 μM amsacrine, 0-200 μM TAS-103, or 0-25 μM ethidium bromide. Topoisomerase I DNA cleavage assays contained 11 nM human topoisomerase I and 10 nM negatively or positively supercoiled pBR322 DNA in a total of 20 μL of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, 30 μg/ml BSA. Assays were carried out in the presence of 0-25 μM camptothecin. In all cases, reaction mixtures were incubated at 37 °C for 6 min and enzyme-DNA cleavage complexes were trapped by the addition of 2 μL of 5% SDS followed by 1 μL of 375 mM EDTA (pH 8.0). Proteinase K (2 μL of 0.8 mg/mL) was added and samples were incubated at 45 °C for 30 min to digest the enzyme. Samples were mixed with 2 μL of agarose gel loading buffer [60% sucrose in 10 mM Tris-HCl (pH 7.9)], heated at 45 °C for 5 min, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris-acetate (pH 8.3), 2 mM EDTA containing 0.5 μg/mL ethidium bromide. DNA bands were visualized with ultraviolet light and quantified using an Alpha Innotech digital imaging system. DNA cleavage was monitored by the conversion of supercoiled plasmid DNA to linear molecules.
Site-specific DNA Cleavage by Human Topoisomerase IIα
DNA sites cleaved by human topoisomerase IIα in negatively and positively supercoiled DNA were mapped as described previously (51). DNA cleavage mixtures contained 2.2 μM human topoisomerase IIα, 10 nM negatively or positively supercoiled pBR322 DNA in a total of 160 μL of topoisomerase II cleavage buffer. In some cases, the MgCl2 was replaced with CaCl2 or reaction mixtures included 100 μM etoposide. Samples were incubated at 37 °C for 6 min and enzyme-DNA cleavage complexes were trapped by the addition of 16 μL of 1% SDS followed by 8 μL of 375 mM EDTA (pH 8.0). Proteinase K (16 μL of 0.8 mg/mL) was added and mixtures were incubated at 45 °C for 30 min to digest the type II enzyme. Reaction products were purified by passage through Qiaquick Spin Columns (Qiagen) as described by the manufacturer. DNA cleavage complexes were linearized by treatment with HindIII. Terminal 5′-phosphates were removed by treatment with calf intestinal alkaline phosphatase and replaced with [32P]phosphate using T4 polynucleotide kinase and [γ-32P]ATP. Samples were treated with EcoRI and the singly-end labeled DNA products were purified by passage through a CHROMA SPIN+TE-100 column (Clontech). Loading buffer (40% formamide (w/v), 8.4 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanole FF) was added and samples were subjected to electrophoresis in 6% polyacrylamide sequencing gels. Gels were fixed in 10% methanol/10% acetic acid (v/v) and dried in vacuo. DNA cleavage products were visualized with a BioRad Molecular Imager FX.
The effects of ethidium bromide (0-10 μM) on the cleavage of linear DNA by human topoisomerase IIα was monitored using the above protocol with the exception that a singly-end labeled linear pBR322 fragment (53, 54) was used as the initial substrate. DNA cleavage was monitored by the loss of full-length linear molecules.
DNA Ligation by Human Topoisomerase IIα
DNA ligation assays were carried out by a modification of the procedure of Kingma et al. (55). DNA cleavage/ligation equilibria were established in topoisomerase II cleavage buffer as described above except that MgCl2 in the reaction buffer was replaced by 5 mM CaCl2. Assays were carried out in the absence of compound or in the presence of 100 μM etoposide. Topoisomerase II-DNA cleavage complexes were trapped by the addition of EDTA (pH 8.0) to a 6 mM final concentration. NaCl was added to a 500 mM final concentration in order to prevent re-cleavage of the DNA substrate. Ligation was initiated by the addition of MgCl2 at a 0.25 mM final concentration and terminated at times up to 60 s by the addition of 2 μL of 5% SDS. Samples were processed and analyzed as described for plasmid DNA cleavage. The percent DNA cleavage at time 0 was set to 100% and the rate of ligation was determined by quantifying the loss of cleaved DNA over time.
DNA Intercalation
Intercalation reaction mixtures contained 220 nM topoisomerase IIα and 5 nM pBR322 DNA in a total of 20 μL of 10 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 175 mM KCl, 5 mM MgCl2, 1 mM ATP, and 2.5% glycerol (v/v) that contained 0-25 μM ethidium bromide, 0-500 μM amsacrine, or 0-200 μM TAS-103. Mixtures were incubated at 37°C for 6 min, extracted with phenol:chloroform:isoamyl alcohol (25:24:1), and added to 3 μl of 0.77% SDS, 77 mM EDTA (pH 8.0). Samples were mixed with 2 μL of agarose gel loading buffer, heated at 45 °C for 5 min, and subjected to electrophoresis in a 1% agarose gel in 100 mM Tris-borate (pH 8.3), 2 mM EDTA. Gels were stained with 1 μg/mL ethidium bromide, and DNA bands were visualized as described for plasmid DNA cleavage.
The DNA intercalation assay is based on the fact that intercalative agents induce constrained negative supercoils and compensatory unconstrained positive superhelical twists in covalently closed circular DNA. Therefore, as the concentration of an intercalative compound increases, a plasmid that is negatively supercoiled or relaxed (i.e., contains no superhelical twists) appears to become positively supercoiled. Treatment of an intercalated plasmid with topoisomerase IIα removes the unconstrained positive DNA superhelical twists. Subsequent extraction of the compound allows the local drug-induced unwinding to redistribute in a global manner and manifest itself as a net negative supercoiling of the plasmid. Thus, in the presence of an intercalative agent, topoisomerase treatment converts relaxed plasmids to negatively supercoiled molecules (see insets, Figures 4 and 5).
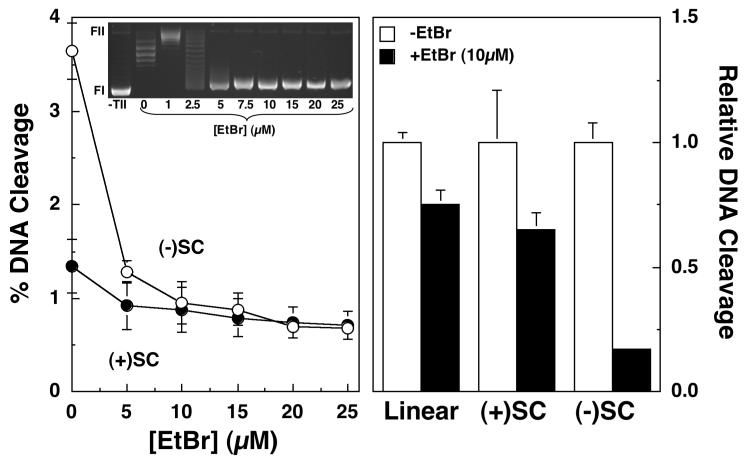
FIGURE 4.
Effects of ethidium bromide intercalation on DNA cleavage mediated by human topoisomerase IIα. The ability of topoisomerase IIα to cleave negatively [(-)SC, open circles] or positively [(+)SC, closed circles] supercoiled pBR322 plasmid DNA in the presence of 0-25 μM ethidium bromide is shown (left panel). Error bars represent the standard deviation of three independent assays. The inset shows a representative gel of DNA intercalation assays using negatively supercoiled plasmids in the absence of topoisomerase IIα (-TII), or in the absence (0 μM) or presence (1-25 μM) of ethidium bromide (see Experimental Procedures for the interpretation of intercalation assays). The positions of supercoiled (FI) and nicked circular (FII) molecules are indicated. Relative DNA cleavage of linear plasmid, or negatively or positively supercoiled molecules in the absence (-EtBr, open bars) or presence (+EtBr, closed bars) of 10 μM eithdium bromide also is depicted (right panel). Relative DNA cleavage was calculated by normalizing levels of scission in the absence of ethidium bromide to a value of 1.0. Error bars represent the standard deviation of three independent assays for supercoiled substrates, or the standard error of the mean for two independent assays for linear DNA.
FIGURE 5.
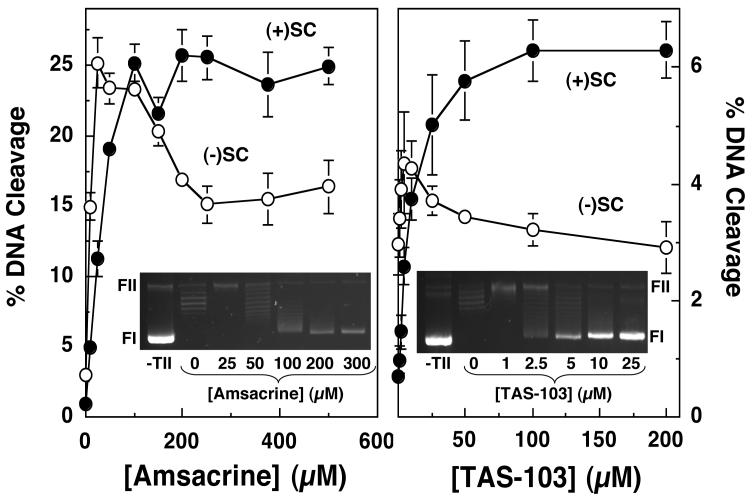
Effects of amsacrine and TAS-103 intercalation on DNA cleavage mediated by human topoisomerase IIα. The ability of topoisomerase IIα to cleave negatively [(-)SC, open circles] or positively [(+)SC, closed circles] supercoiled pBR322 plasmid DNA in the presence of 0-500 μM amsacrine (left panel) or 0-200 μM TAS-103 (right panel) is shown. Error bars represent the standard deviation of three independent assays. The insets show representative DNA intercalation assays in the absence of topoisomerase IIα (-TII), or in the absence (0 μM) or presence of the respective drug (see Experimental Procedures for the interpretation of intercalation assays). The positions of supercoiled (FI) and nicked circular (FII) molecules are indicated.
RESULTS
Effects of DNA Supercoil Geometry on Drug-induced DNA Cleavage Mediated by Human Topoisomerase IIα
Collisions with DNA tracking systems are critical for the conversion of transient topoisomerase-DNA cleavage complexes to permanent strand breaks (4, 7, 10, 32, 37). Since the double helix is acutely overwound immediately ahead of tracking systems (3, 6, 41, 43-47), cleavage complexes most likely to produce permanent strand breaks should be formed between topoisomerases and positively supercoiled DNA. Unfortunately, relatively little is known about the interaction between these enzymes and positively supercoiled substrates.
A recent study demonstrated that human topoisomerase IIα, which is involved in replicative processes in vivo, relaxes positive superhelical twists >10-fold faster than it does negative supercoils (51). Furthermore, the enzyme maintains a lower concentration of DNA cleavage complexes with these overwound substrates (51). Levels of DNA cleavage mediated by topoisomerase IIα in the presence of either Mg2+ or Ca2+ are ∼3- to 4-fold lower with positively supercoiled DNA (Figure 1). The decreased baseline levels of DNA cleavage observed with overwound DNA may reflect, at least in part, somewhat lower binding affinity (∼2-fold) that topoisomerase IIα displays for positively over negatively supercoiled molecules (51). The lower level of cleavage notwithstanding, the site specificity of DNA scission is not affected by the handedness of the substrate (Figure 1, right panel) (51).
Many anticancer drugs kill cells by increasing levels of double-stranded DNA breaks generated by topoisomerase II (4, 7, 10, 32, 37). As a first step towards characterizing the effects of these drugs on enzyme-DNA cleavage complexes formed with positively supercoiled substrates, the ability of etoposide to induce DNA cleavage by human topoisomerase IIα was assessed. Etoposide was chosen for initial experiments because its mechanism of action is well defined (4, 7, 15, 56-59). In addition, the drug is not intercalative in nature and displays little, if any, affinity for DNA (4, 7, 15, 56, 60). Over a range of drug concentrations, DNA cleavage induced by etoposide was ∼3- to 4-fold lower with positively as compared to negatively supercoiled molecules (Figure 1). Moreover, as observed in the absence of drug, the geometry of DNA supercoils did not affect the site specificity of etoposide-induced scission (Figure 1, right panel).
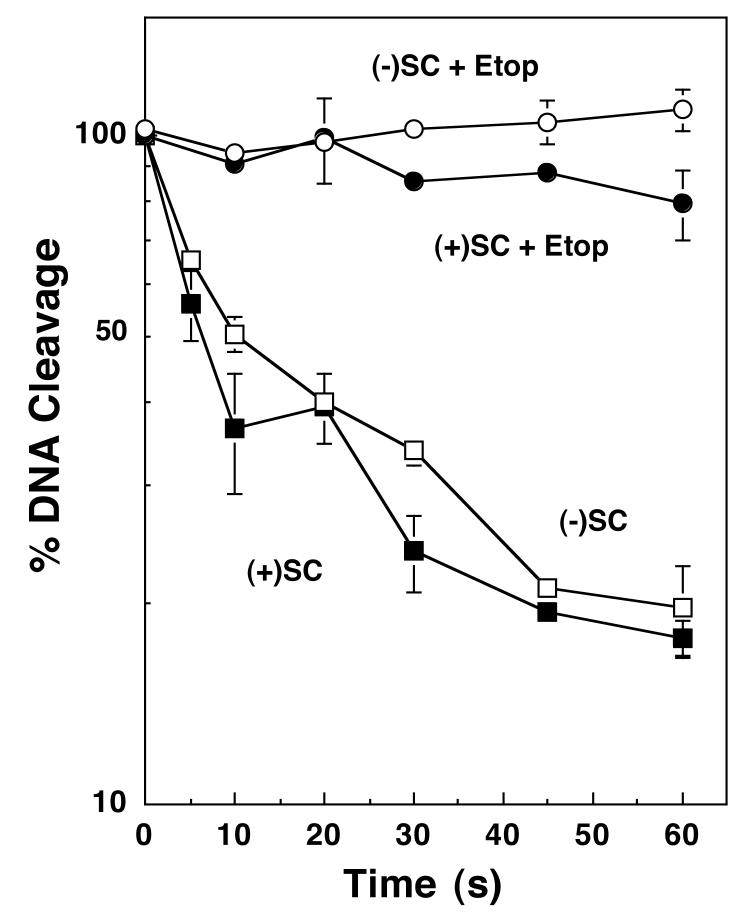
Etoposide increases levels of topoisomerase II-DNA cleavage complexes primarily by inhibiting the ligation of cleaved strands (4, 7, 15, 57, 58). Therefore, the effects of etoposide on the ability of topoisomerase IIα to ligate positively or negatively supercoiled molecules were determined. As seen in Figure 2, the handedness of superhelical twists had little impact on rates of DNA ligation in either the presence or absence of etoposide. Thus, the decreased levels of DNA strand breaks induced by etoposide in positively supercoiled substrates do not result from a decreased inhibition of ligation.
FIGURE 2.
Etoposide inhibits the ligation of negatively or positively supercoiled DNA by human topoisomerase IIα. A time course of ligation in the presence of negatively [(-)SC, open symbols] or positively [(+)SC, closed symbols] supercoiled pBR322 plasmid is shown. DNA ligation was monitored in the absence (boxes) or presence (circles) of 100 μM etoposide (+Etop). The initial level of DNA cleavage was set to 100% and the rate of ligation was determined by quantifying the loss of the cleaved DNA over time. Error bars represent the standard deviation of three independent assays.
Despite the lower absolute percent DNA cleaved with positively supercoiled substrates, the relative enhancement of scission by etoposide with overwound and underwound substrates was similar. For example, at 50 μM drug, this enhancement was ∼4.5-fold and ∼3.6-fold for positively and negatively supercoiled DNA, respectively (Figures 1 and 3, Table 1). These data yield a “superhelical specificity” for etoposide (i.e., relative cleavage enhancement observed with positively supercoiled DNA divided by the relative cleavage enhancement with negatively supercoiled DNA) of ∼1.3 (Table 1). It should be noted that the superhelical specificity for etoposide varied little (1.0-1.3) over the entire range of drug concentration examined. These correlations, together with the ligation and site-specificity data, imply that interactions between etoposide and topoisomerase IIα are not altered significantly by the handedness of DNA. Rather, the decrease in drug-induced cleavage of positively supercoiled DNA reflects the reduced baseline (i.e., non-drug) level of enzyme-mediated scission with this substrate.
FIGURE 3.
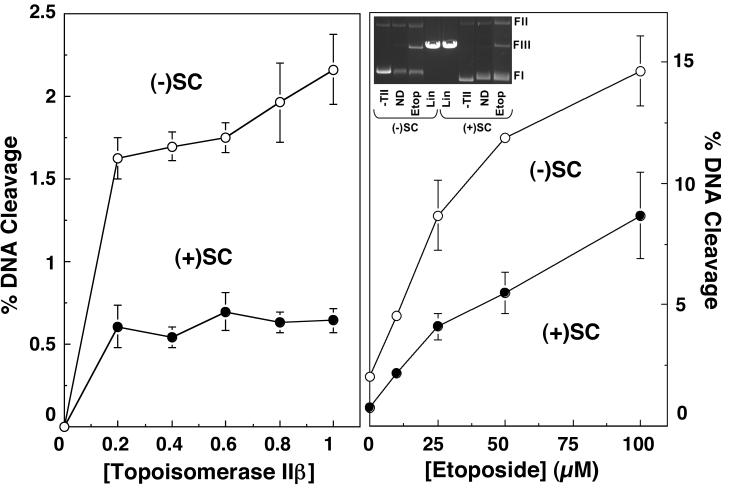
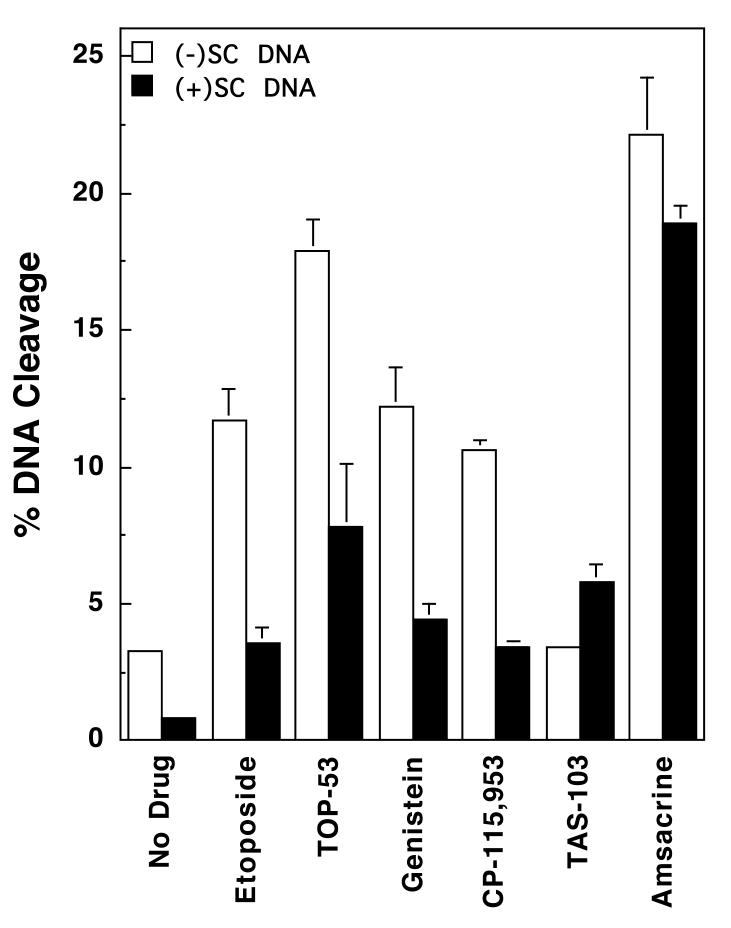
Effects of DNA superhelical geometry on drug-induced DNA cleavage mediated by human topoisomerase IIα. The ability of topoisomerase IIα to cleave negatively [(-)SC DNA, open bars] or positively [(+)SC DNA, closed bars] supercoiled pBR322 plasmid DNA in the presence of various topoisomerase II-targeted drugs is shown. All drugs were used at a concentration of 50 μM, except for CP-115,953, which was used at 5 μM (multiple DNA cleavage events per plasmid were observed at higher concentrations of CP-115,953). Error bars represent the standard deviation of 3 or 4 independent assays.
Table 1.
Relative DNA cleavage enhancement of human topoisomerase IIα by anticancer agents
| Druga | Relative Cleavage Enhancement (-)SC DNA | Relative Cleavage Enhancement (+)SC DNA | Superhelical Specificity [(+)SC/(-)SC] |
|---|---|---|---|
| Etoposide | 3.6 ± 0.4 | 4.5 ± 0.4 | 1.3 |
| TOP-53 | 5.4 ± 0.5 | 9.8 ± 1.1 | 1.8 |
| Genistein | 3.7 ± 0.4 | 5.7 ± 0.5 | 1.5 |
| CP-115,953 | 3.2 ± 0.2 | 4.4 ± 0.6 | 1.4 |
| TAS-103 | 1.1 ± 0.1 | 7.8 ± 0.9 | 7.1 |
| Amsacrine | 7.5 ± 0.3 | 22.6 ± 0.7 | 3.0 |
All drugs were at 50 μM, with the exception of CP-115-953, which was at 5 μM (multiple DNA cleavage events per plasmid were observed at higher concentrations of CP-115,953).
In light of this finding, the influence of DNA geometry on the actions of other topoisomerase II-targeted drugs was characterized. The agents employed represent a structurally diverse group of natural and synthetic compounds. TOP-53 is a non-intercalative drug that is a more potent derivative of etoposide (61, 62). Genistein and CP-115,953 are non-intercalative compounds that share a similar core ring structure. Genistein is a naturally occurring bioflavanoid that is believed to possess chemopreventative properties (63-65). CP-115,953 belongs to the quinolone family (4, 66-69), a drug class that includes a number of widely used antibacterials that target prokaryotic type II topoisomerases. TAS-103 displays strong interactions with DNA and is both an outside binder and an intercalating agent (70-72). Finally, amsacrine is an intercalative compound that is in clinical use (4, 73, 74).
All of the above compounds increased DNA cleavage mediated by human topoisomerase IIα. Data for 50 μM drugs are shown in Figure 3 and Table 1. The only exception is CP-115,953, for which data are shown for 5 μM drug (higher drug concentrations were not used because they induced multiple DNA cleavage events per plasmid). As found for etoposide, most induced higher absolute levels of scission with negatively supercoiled substrates, but displayed greater relative cleavage enhancement with positively supercoiled DNA. The non-intercalative drugs displayed superhelical specificities between 1.3 and 1.8. These values remained constant from the lowest drug concentrations examined (5 or 10 μM) up to concentrations that induced multiple DNA cleavage events per plasmid (50 or 100 μM). It was not possible to determine superhelical specificities for CP-115,953 above 5 μM due to the reason discussed above. Taken together, these results suggest that the superhelical specificity for DNA cleavage with non-intercalative topoisomerase II poisons is independent of drug concentration.
The superhelical specificities of the two intercalative drugs (TAS-103 and amsacrine) at 50 μM, 7.1 and 3.0, respectively, were greater than those seen for the non-intercalative compounds. These higher values suggest that DNA geometry may influence the actions of intercalative compounds towards topoisomerase IIα beyond their effects on baseline DNA cleavage mediated by the enzyme.
Drugs that target topoisomerases are believed to work at the enzyme-nucleic acid interface (4, 7, 59, 75-79). However, intercalative agents have two additional effects on DNA that could impact levels of topoisomerase-mediated scission in a geometry-specific manner. First, since these compounds locally underwind DNA, they induce compensatory unconstrained positive superhelical twists in distal regions of covalently closed circular molecules (80, 81). Thus, as the concentration of an intercalating agent increases, a plasmid that is topologically negatively supercoiled would appear to contain positive superhelical twists. As discussed above, baseline levels of DNA cleavage mediated by topoisomerase IIα are lower with positively supercoiled substrates. Consequently, the apparent change in DNA topology induced by intercalation could diminish the ability of a compound to enhance cleavage with underwound substrates. In contrast, the apparent geometry of a positively supercoiled plasmid (which already is overwound) would not change substantially upon addition of an intercalative drug.
Second, the accumulation of drugs in the double helix has the potential to inhibit enzyme binding or activity. Because the generation of positive superhelical twists by DNA intercalation induces torsional stress in the double helix, the ability of covalently closed molecules to absorb these compounds is limited. Since overwound plasmids are under positive torsional stress even in the absence of drugs, they cannot bind as many intercalative molecules as underwound DNA. Therefore, enzyme activity on positively supercoiled substrates is less likely to be inhibited by the accumulation of bound drug.
Two independent experiments were carried out to determine whether the above effects contribute to the higher superhelical specificity of intercalative agents. The first utilized ethidium bromide, a classical intercalating agent that does not enhance topoisomerase II-mediated DNA scission. When ethidium bromide was included in reaction mixtures, there was a precipitous drop in the ability of human topoisomerase IIα to cleave negatively supercoiled pBR322 (Figure 4). Levels of cleavage decreased ∼6-fold at 10 μM ethidium bromide, which corresponds to the concentration at which “full” intercalation was observed (see inset). Beyond 10 μM, little additional inhibition was observed. Thus, the decrease in DNA cleavage induced by ethidium bromide correlates with the change in the apparent supercoiled state of the plasmid substrate. Consistent with this conclusion, ethidium bromide had a much smaller effect on DNA cleavage when the initial substrate was positively supercoiled (Figure 4). Furthermore, once the concentration of ethidium bromide exceeded 10 μM, DNA cleavage levels for positively and negatively substrates were virtually identical.
To determine whether ethidium bromide accumulation on the double helix affects the DNA cleavage activity of topoisomerase IIα independent of changes in DNA geometry, a linear substrate was employed (Figure 4, right panel). Since linear molecules are not topologically constrained, DNA intercalation does not induce torsional stress or positive supercoiling in these substrates. The addition of 10 μM ethidium bromide only had a minor effect on levels of DNA cleavage with linear pBR322. These results suggest that ethidium bromide inhibits DNA cleavage mediated by human topoisomerase IIα primarily by altering the apparent topology of the DNA substrate and to a lesser extent by accumulation within the double helix.
The second experiment examined the effects of a broad concentration range of amsacrine or TAS-103 on DNA scission mediated by topoisomerase IIα. In contrast to ethidium bromide, these two intercalating drugs are potent topoisomerase II poisons that enhance DNA cleavage (4, 70-74). The initial study examined amsacrine. When negatively supercoiled plasmids were employed as the substrate, peak levels of cleavage were observed between 25 and 50 μM drug (Figures 3 and 5, left panel). This is the amsacrine concentration range in which changes in DNA topology begin to appear (Figure 5, left panel inset). Cleavage levels dropped ∼50% between 25 and 250 μM drug and plateaued thereafter (Figure 5, left panel). It is notable that “full” intercalation is observed at an amsacrine concentration of ∼200 μM (see inset). As discussed above, these findings are consistent with the suggestion that the rise in drug-induced DNA cleavage by amsacrine is attenuated by the concomitant fall in baseline scission caused, at least in part, by the apparent change in the geometry of the plasmid substrate.
A different pattern was seen with positively supercoiled substrates. Levels of DNA scission peaked between 100 and 200 μM amsacrine and remained constant at higher drug concentrations (Figure 5, left panel). Because of the changes in cleavage enhancement seen with negatively and positively supercoiled substrates, the superhelical specificity of amsacrine-induced DNA cleavage was concentration-dependent. Values were in the range of 1.0-1.5 at drug concentrations that did not substantially alter the apparent topology of negatively supercoiled substrates (<25 μM), increased to 3.0-3.5 at concentrations that began to affect apparent topology (25-150 μM), and plateaued at ∼5.1 at concentrations that induced full intercalation.
It is notable that the plateau level of cleavage observed with positively supercoiled DNA was higher than that observed with negatively supercoiled molecules. It is unlikely that amsacrine intercalation could make negatively supercoiled plasmids appear to be more overwound than positively supercoiled molecules at a comparable drug concentration (see Figure 4). Therefore, it is concluded that the preferential accumulation of amsacrine in underwound substrates also contributes to the decrease in DNA cleavage observed with negatively supercoiled molecules at high drug concentrations.
To determine whether the concentration dependence of superhelical specificity is a general feature of intercalative drugs, the ability of TAS-103 to induce DNA cleavage by topoisomerase IIα was examined (Figure 5, right panel). Results were similar to those seen with amsacrine. Scission of negatively supercoiled plasmid rose initially, peaked at drug concentrations that began to alter the apparent topology of the substrate, and fell to approximately baseline thereafter. Conversely, scission of positively supercoiled plasmids increased and remained high over the entire range of TAS-103 examined. As a result, the superhelical specificity of TAS-103, like amsacrine, was concentration dependent. Values were 1.2-1.6 at drug concentrations that did not substantially alter the apparent topology of negatively supercoiled substrates (<5 μM), increased to 2.6-3.7 at concentrations that begin to affect apparent topology (5-10 μM), and plateaued at 7.1-9.3 at concentrations that induced full intercalation. This concentration dependence for superhelical specificity is in sharp contrast to those calculated for non-intercalative compounds and may be a defining distinction between intercalative and non-intercalative topoisomerase II poisons.
Effects of DNA Supercoil Geometry on Drug-induced DNA Cleavage Mediated by Human Topoisomerase IIβ
In contrast to human topoisomerase IIα, topoisomerase IIβ is not required for DNA replication and does not preferentially remove positive superhelical twists (2, 26, 28-30, 51). However, the effects of nucleic acid geometry on DNA cleavage mediated by the β isoform have never been examined. Since topoisomerase IIβ is an important target for anticancer drugs (35) and also appears to mediate some of the toxic effects of these agents in differentiated tissues (33-35), the ability of the enzyme to cleave positively vs. negatively supercoiled substrates was assessed.
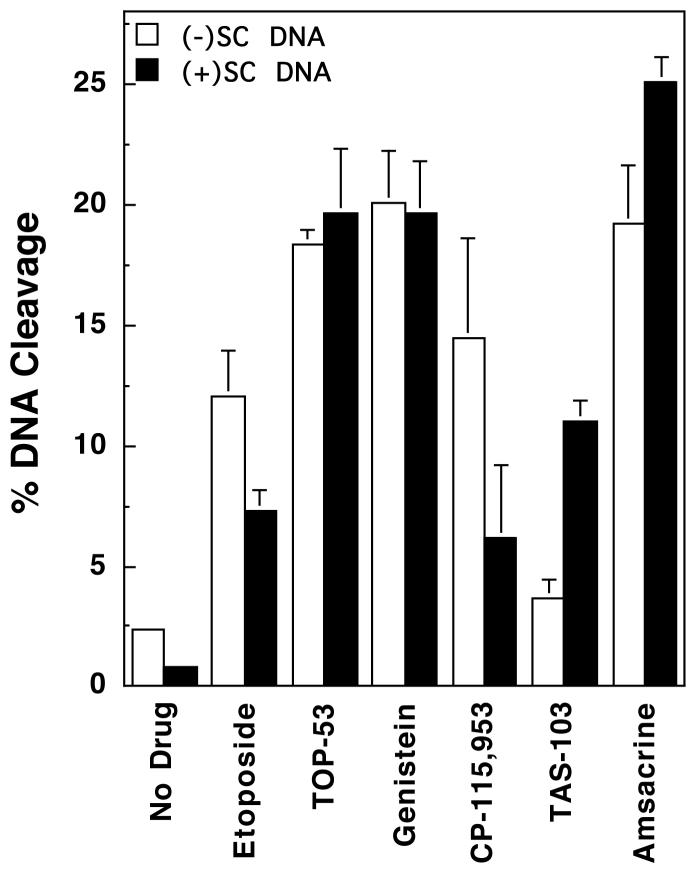
Similar to topoisomerase IIα, topoisomerase IIβ maintained lower (∼3- to 4-fold) levels of cleavage complexes with positively supercoiled DNA in the absence of drug (Figure 6, left panel). The enzyme also displayed lower (∼2-fold) cleavage activity with positively supercoiled substrates over a range of etoposide concentrations (Figure 6, right panel). However, the effects of DNA geometry on drug-induced scission were not as pronounced with topoisomerase IIβ as they were with the α isoform. As seen in Figure 7, the percentage of positively supercoiled molecules cleaved in the presence of several drugs approached or actually exceeded that observed with negatively supercoiled substrates. Consequently, the calculated superhelical specificities for most drugs were somewhat higher for topoisomerase IIβ than they were for topoisomerase IIα (Table 2). Once again, the superhelical specificity of the non-intercalative compounds was concentration-independent and remained constant at all drug concentrations examined. Furthermore, the highest superhelical specificities were observed for the two intercalative drugs, TAS-103 and amsacrine.
FIGURE 7.
Effects of DNA superhelical geometry on drug-induced DNA cleavage mediated by human topoisomerase IIβ. The ability of topoisomerase IIβ to cleave negatively [(-)SC DNA, open bars] or positively [(+)SC DNA, closed bars] supercoiled pBR322 plasmid DNA in the presence of various topoisomerase II-targeted drugs is shown. All drugs were used at a concentration of 50 μM, except for CP-115,953, which was used at 5 μM (multiple DNA cleavage events per plasmid were observed at higher concentrations of CP-115,953). Error bars represent the standard deviation of 3 or 4 independent assays.
Table 2.
Relative DNA cleavage enhancement of human topoisomerase IIβ by anticancer agents
| Druga | Relative Cleavage Enhancement (-)SC DNA | Relative Cleavage Enhancement (+)SC DNA | Superhelical Specificity [(+)SC/(-)SC] |
|---|---|---|---|
| Etoposide | 4.9 ± 0.3 | 8.2 ± 0.5 | 1.6 |
| TOP-53 | 7.9 ± 0.6 | 20.9 ± 0.9 | 2.7 |
| Genistein | 8.5 ± 0.2 | 22.0 ± 2.1 | 2.6 |
| CP-115,953 | 5.6 ± 1.8 | 10.4 ± 4.8 | 1.9 |
| TAS-103 | 1.7 ± 0.3 | 12.6 ± 1.3 | 7.3 |
| Amsacrine | 7.8 ± 1.1 | 31.7 ± 4.7 | 4.1 |
All drugs were at 50 μM, with the exception of CP-115-953, which was at 5 μM (multiple DNA cleavage events per plasmid were observed at higher concentrations of CP-115,953).
To further analyze this finding, the effects of amsacrine and TAS-103 on DNA scission mediated by topoisomerase IIβ was examined over a broad concentration range (data not shown). As found with the α isoform, levels of negatively supercoiled DNA cleaved by topoisomerase IIβ peaked at drug concentrations at which changes in apparent topology were obvious, dropped, and plateaued at concentrations that induced full intercalation. In contrast, levels of cleavage with positively supercoiled substrates increased and remained high over the entire drug ranges examined. Thus, as discussed above for topoisomerase IIα, the superhelical specificities for amsacrine and TAS-103 were concentration-dependent with topoisomerase IIβ, and rose from initial values of 1.2 and 1.7, respectively, at low concentrations, to 8.3 and 9.1, respectively, at high concentrations. These results further suggest that drug-induced cleavage of underwound substrates by topoisomerase IIβ is attenuated by the ability of intercalative agents to change the apparent geometry of DNA and by increased drug accumulation on negatively supercoiled molecules.
Effects of DNA Supercoil Geometry on Drug-induced DNA Cleavage Mediated by Human Topoisomerase I
Topoisomerase I is an important target for several new anticancer drugs that are based on camptothecin, a naturally occurring non-intercalative compound found in the Chinese tree Camptotheca acuminata (8, 10, 20, 21, 82). These drugs kill cells by increasing levels of single-stranded DNA breaks (i.e., nicks) that are generated by the type I enzyme (8, 10, 37). Since one of the major functions of topoisomerase I is to alleviate torsional stress that builds up ahead of replication forks and transcription complexes (2, 5, 10), the effects of nucleic acid geometry on enzyme-mediated DNA cleavage was examined in the absence or presence of camptothecin (Figure 8).
In sharp contrast to the type II enzymes, human topoisomerase I maintained a higher level of cleavage intermediates with overwound DNA. Three times more nicked molecules were generated with positively supercoiled plasmids than were observed with negatively supercoiled substrates (see bar graph inset). A far more striking effect of DNA geometry on topoisomerase I was seen in the presence of camptothecin (Figure 8). Over a concentration range of 1-25 μM drug, the enzyme generated dramatically higher levels of nicked DNA with positively supercoiled substrates. For example, at 1 μM camptothecin, ∼16 times more DNA cleavage was observed with positively as compared to negatively supercoiled pBR322, yielding a superhelical specificity of 4.7. Together with the findings for the type II enzymes, these results demonstrate that the handedness of DNA supercoils has a profound influence on DNA cleavage reactions mediated by human topoisomerases.
DISCUSSION
Beyond their critical physiological functions, topoisomerases are targets for a number of important anticancer drugs (4, 7-10, 12-15). While these agents all increase the concentration of topoisomerase-generated breaks in the genetic material (4, 7, 10, 32, 37), their ability to kill cells requires the actions of DNA tracking systems, such as replication or transcription complexes (4, 7, 10, 32, 37). Collisions between tracking systems and cleavage complexes convert these transient enzyme intermediates to permanent DNA strand breaks either directly, or by the induction of recombination (4, 7, 38-40). It is the accumulation of these permanent strand breaks that ultimately triggers cell death pathways (39).
Previous studies on the interaction of anticancer drugs and topoisomerases have used negatively supercoiled or linear DNA as cleavage substrates. However, the movement of enzymes through the double helix leads to the formation of overwound DNA ahead of tracking systems (3, 6, 41, 43-47). As result, the cleavage complexes most likely to produce permanent DNA strand breaks should form between topoisomerases and positively supercoiled DNA. Therefore, the ability of human topoisomerase IIα and β, and topoisomerase I to cleave positively supercoiled molecules was assessed in the absence or presence of anticancer drugs.
Topoisomerase IIα (51) and β both maintain ∼3- to 4-fold lower levels of cleavage complexes with positively supercoiled DNA than with negatively supercoiled molecules. This decrease in nucleic acid scission benefits the cell, because it makes it less likely that the actions of a DNA tracking system will generate permanent topoisomerase II-associated strand breaks during normal cellular processes.
Topoisomerase IIα also maintains lower concentrations of drug-induced cleavage intermediates with positively supercoiled substrates. With the non-intercalative agents examined, the relative enhancement of DNA scission seen with overwound DNA is similar to that observed with underwound molecules. Furthermore, the superhelical specificity for DNA cleavage appears to be independent of drug concentration. Consequently, it is proposed that decreased drug efficacy is due primarily to a drop in baseline levels of cleavage mediated by topoisomerase IIα, rather than an altered interaction with the enzyme-DNA complex.
Results were somewhat different for topoisomerase IIβ. Whereas some non-intercalative drugs such as etoposide and CP-115,953, follow trends similar to those seen for the α isoform, others, such as genistein and TOP-53, induced equivalent levels of cleavage with positively and negatively supercoiled plasmids. The underlying reason for the increased drug effect with positively supercoiled DNA is not known. However, this finding suggests that in some cases, drug-induced stimulation of DNA cleavage by topoisomerase IIβ is more likely to generate permanent strand breaks than would topoisomerase IIα under comparable circumstances. This difference notwithstanding, the superhelical specificity for DNA cleavage with non-intercalative drugs was once again concentration-independent.
Consistently, with both topoisomerase IIα and β, intercalative drugs displayed higher relative cleavage enhancement in the presence of positively rather than negatively supercoiled DNA. This superhelical specificity did not correlate with a greater intrinsic drug activity with overwound substrates. Rather, it appears to result from the apparent positive supercoiling of underwound molecules and the preferential accumulation of intercalative drugs in negatively supercoiled substrates. At higher drug concentrations, these effects attenuate the stimulation of topoisomerase II-mediated DNA cleavage of negatively supercoiled molecules and result in an increased superhelical specificity. As a result, they probably are responsible for the characteristic “bell-shaped curve” observed for the enhancement of scission by intercalative anticancer drugs. In contrast, DNA cleavage by topoisomerase II rises and plateaus with positively supercoiled substrates. This finding implies that intercalative agents are able to maintain their effectiveness ahead of DNA tracking systems, even at high drug concentrations.
Results with topoisomerase I were unexpected. Despite the fact that this enzyme characteristically functions to alleviate overwinding ahead of DNA tracking systems (5, 8, 10), it maintains ∼3 times higher levels of cleavage complexes with positively supercoiled substrates. Thus, under normal physiological circumstances, topoisomerase I is inherently more likely to trigger the formation of permanent DNA strand breaks than either type II isoform. The cellular ramifications of this enzyme feature are unclear at the present time.
The influence of DNA topology on topoisomerase I-mediated scission is even more dramatic in the presence of camptothecin. This finding suggests that topoisomerase I is an intrinsically more lethal target for anticancer drugs than is topoisomerase IIα or β.
In summary, numerous factors influence the effectiveness of topoisomerase-targeted anticancer drugs, including the concentration, localization, and roles played by each enzyme in specific nucleic acid processes. Results of the present study indicate that the topological state of the genetic material also has a profound influence on topoisomerase-mediated DNA cleavage and the response of topoisomerase IIα, topoisomerase IIβ, and topoisomerase I to anticancer drugs. Although topoisomerase IIα is the enzyme that is most frequently targeted by chemotherapeutic regimens, it may actually be the topoisomerase that is least likely to generate cleavage complexes that are converted to permanent DNA strand breaks in treated cells. Alternatively, all other things being equal, topoisomerase I appears to be the enzyme most likely to fragment the genome. This may be one of the reasons why camptothecin-based drugs display a spectrum of activity that exceeds those of other established anticancer agents.
ACKNOWLEDGEMENTS
We are grateful to Jo Ann Byl for preparing the human topoisomerase IIβ used for experiments in this study, to Dr. Mary-Ann Bjornsti (St. Jude Children’s Research Hospital) for her gift of human topoisomerase I, and to Renier Vélez-Cruz and Ryan P. Bender for critical reading of the manuscript.
Footnotes
This work was supported by National Institutes of Health research grant GM33944. A.K.M. was a trainee under grant 5 T32 HD07043 from the National Institutes of Health.
REFERENCES
- 1.Osheroff N. DNA topoisomerases. Biochim. Biophys. Acta. 1998;1400:1–2. doi: 10.1016/s0167-4781(98)00123-7. [DOI] [PubMed] [Google Scholar]
- 2.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 5.Champoux JJ. DNA topisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:1349–1364. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 8.Sabourin M, Osheroff N. Wiley Encyclopedia of Molecular Medicine. John Wiley & Sons, Inc.; 2002. Topoisomerases; pp. 3192–3197. [Google Scholar]
- 9.Velez-Cruz R, Osheroff N. Encyclopedia of Biological Chemistry. Elsevier Inc.; 2004. DNA topoisomerases: type II; pp. 806–811. [Google Scholar]
- 10.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 11.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 12.Hande KR. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 13.Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 14.Takimoto CH, Wright J, Arbuck SG. Clinical applications of the camptothecins. Biochim. Biophys. Acta. 1998;1400:107–119. doi: 10.1016/s0167-4781(98)00130-4. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anti-Cancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 16.Liu LF, Miller KG. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc. Natl. Acad. Sci. USA. 1981;78:3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halligan BD, Davis JL, Edwards KA, Liu LF. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J. Biol. Chem. 1982;257:3995–4000. [PubMed] [Google Scholar]
- 18.Hanai R, Caron PR, Wang JC. Human TOP3: A single-copy gene encoding DNA topoisomerase III. Proc. Natl. Acad. Sci. USA. 1996;93:3653–3657. doi: 10.1073/pnas.93.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulaouic H, Roulon T, Flamand O, Grondard L, Lavelle F, Riou J-F. Purification and characterization of human DNA topoisomerase IIIalpha. Nucleic Acids Res. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zunino F, Pratesi G. Camptothecins in clinical development. Expert Opin. Investig. Drugs. 2004;13:269–284. doi: 10.1517/13543784.13.3.269. [DOI] [PubMed] [Google Scholar]
- 21.Sriram D, Yogeeswari P, Thirumurugan R, Bal TR. Camptothecin and its analogues: a review on their chemotherapeutic potential. Nat. Prod. Res. 2005;19:393–412. doi: 10.1080/14786410412331299005. [DOI] [PubMed] [Google Scholar]
- 22.Drake FH, Zimmerman JP, McCabe FL, Bartus HF, Per SR, Sullivan DM, Ross WE, Mattern MR, Johnson RK, Crooke ST, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 23.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 24.Heck MM, Earnshaw WC. Topoisomerase II: a specific marker for cell proliferation. J. Cell. Biol. 1986;103:2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiang YH, Wu HY, Liu LF. Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 1988;48:3230–3235. [PubMed] [Google Scholar]
- 26.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 27.Bauman ME, Holden JA, Brown KA, Harker WG, Perkins SL. Differential immunohistochemical staining for DNA topoisomerase II alpha and beta in human tissues and for DNA topoisomerase II beta in non-Hodgkin’s lymphomas. Mod. Pathol. 1997;10:168–175. [PubMed] [Google Scholar]
- 28.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, Boege F. Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells. J. Biol. Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 29.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIβ. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.Isaacs RJ, Davies SL, Sandri MI, Redwood C, Wells NJ, Hickson ID. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIβ and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 32.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 33.Baguley BC, Leteurtre F, Riou JF, Finlay GJ, Pommier Y. A carbamate analogue of amsacrine with activity against non-cycling cells stimulates topoisomerase II cleavage at DNA sites distinct from those of amsacrine. Eur. J. Cancer. 1997;33:272–279. doi: 10.1016/s0959-8049(96)00410-8. [DOI] [PubMed] [Google Scholar]
- 34.Moreland N, Finlay GJ, Dragunow M, Holdaway KM, Baguley BC. Cellular responses to methyl-N-[4-9-acridinylamino)-2-methoxyphenyl] carbamate hydrochloride, an analogue of amsacrine active against non-proliferating cells. Eur. J. Cancer. 1997;33:1668–1676. doi: 10.1016/s0959-8049(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 35.Gatto B, Leo E. Drugs acting on the beta isoform of human topoisomerase II (p180) Curr. Med. Chem. Anti-Canc. Agents. 2003;3:173–185. doi: 10.2174/1568011033482486. [DOI] [PubMed] [Google Scholar]
- 36.Sehested M, Holm B, Jensen PB. Dexrazoxane for protection against cardiotoxic effects of anthracyclines. J. Clin. Oncol. 1996;14:2884. [PubMed] [Google Scholar]
- 37.Liu LF, D’Arpa P. Topoisomerase-targeting antitumor drugs: mechanisms of cytotoxicity and resistance. Important Adv. Oncol. 1992:79–89. [PubMed] [Google Scholar]
- 38.Baguley BC, Ferguson LR. Mutagenic properties of topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:213–222. doi: 10.1016/s0167-4781(98)00137-7. [DOI] [PubMed] [Google Scholar]
- 39.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 40.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 41.Cozzarelli NR, Wang JC. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1990. [Google Scholar]
- 42.Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol. 1992;2:369–379. [Google Scholar]
- 43.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 46.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 47.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 49.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIα and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez AC. Studies of a positive supercoiling machine. Nucleotide hydrolysis and a multifunctional “latch” in the mechanism of reverse gyrase. J. Biol. Chem. 2002;277:29865–29873. doi: 10.1074/jbc.M202853200. [DOI] [PubMed] [Google Scholar]
- 51.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005 doi: 10.1074/jbc.M503320200. in press, Epub September 27, 2005. [DOI] [PubMed] [Google Scholar]
- 52.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIalpha by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 53.O’Reilly EK, Kreuzer KN. A unique type II topoisomerase mutant that is hypersensitive to a broad range of cleavage-inducing antitumor agents. Biochemistry. 2002;41:7989–7997. doi: 10.1021/bi025897m. [DOI] [PubMed] [Google Scholar]
- 54.Baldwin EL, Byl JA, Osheroff N. Cobalt enhances DNA cleavage mediated by human topoisomerase IIα in vitro and in cultured cells. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- 55.Kingma PS, Osheroff N. Apurinic sites are position-specific topoisomerase II-poisons. J. Biol. Chem. 1997;272:1148–1155. doi: 10.1074/jbc.272.2.1148. [DOI] [PubMed] [Google Scholar]
- 56.Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984;44:5857–5860. [PubMed] [Google Scholar]
- 57.Osheroff N. Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry. 1989;28:6157–6160. doi: 10.1021/bi00441a005. [DOI] [PubMed] [Google Scholar]
- 58.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 59.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti M-A, Patchan MW, Thompson RB, Osheroff N. Topoisomerase II-etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J. Biol. Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 60.Chow KC, Macdonald TL, Ross WE. DNA binding by epipodophyllotoxins and N-acyl anthracyclines: implications for mechanism of topoisomerase II inhibition. Mol. Pharmacol. 1988;34:467–473. [PubMed] [Google Scholar]
- 61.Utsugi T, Shibata J, Sugimoto Y, Aoyagi K, Wierzba K, Kobunai T, Terada T, Ohhara T, Tsuruo T, Yamada Y. Antitumor activity of a novel podophyllotoxin derivative (TOP-53) against lung cancer and lung metastatic cancer. Cancer Res. 1996;56:2809–2814. [PubMed] [Google Scholar]
- 62.Byl JA, Cline SD, Utsugi T, Kobunai T, Yamada Y, Osheroff N. DNA topoisomerase II as the target for the anticancer drug TOP-53: mechanistic basis for drug action. Biochemistry. 2001;40:712–718. doi: 10.1021/bi0021838. [DOI] [PubMed] [Google Scholar]
- 63.Barnes S, Peterson TG, Coward L. Rationale for the use of genistein-containing soy matrices in chemoprevention trials for breast and prostate cancer. J. Cell. Biochem. Suppl. 1995;22:181–187. doi: 10.1002/jcb.240590823. [DOI] [PubMed] [Google Scholar]
- 64.Stoll BA. Eating to beat breast cancer: potential role for soy supplements. Ann. Oncol. 1997;8:223–225. doi: 10.1023/a:1008237505645. [DOI] [PubMed] [Google Scholar]
- 65.Lamartiniere CA. Protection against breast cancer with genistein: a component of soy. Am. J. Clin. Nutr. 2000;71:1705S–1707S. doi: 10.1093/ajcn/71.6.1705S. discussion 1708S-1709S. [DOI] [PubMed] [Google Scholar]
- 66.Robinson MJ, Martin BA, Gootz TD, McGuirk PR, Moynihan M, Sutcliffe JA, Osheroff N. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J. Biol. Chem. 1991;266:14585–14592. [PubMed] [Google Scholar]
- 67.Spitzner JR, Chung IK, Gootz TD, McGuirk PR, Muller MT. Analysis of eukaryotic topoisomerase II cleavage sites in the presence of the quinolone CP-115,953 reveals drug-dependent and -independent recognition elements. Mol. Pharmacol. 1995;48:238–249. [PubMed] [Google Scholar]
- 68.Bromberg KD, Burgin AB, Osheroff N. Quinolone action against human topoisomerase IIα: stimulation of enzyme-mediated double-stranded DNA cleavage. Biochemistry. 2003;42:3393–3398. doi: 10.1021/bi027383t. [DOI] [PubMed] [Google Scholar]
- 69.Elsea SH, McGuirk PR, Gootz TD, Moynihan M, Osheroff N. Drug features that contribute to the activity of quinolones against mammalian topoisomerase II and cultured cells: correlation between enhancement of enzyme-mediated DNA cleavage in vitro and cytotoxic potential. Antimicrob. Agents Chemother. 1993;37:2179–2186. doi: 10.1128/aac.37.10.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Utsugi T, Aoyagi K, Asao T, Okazaki S, Aoyagi Y, Sano M, Wierzba K, Yamada Y. Antitumor activity of a novel quinoline derivative, TAS-103, with inhibitory effects on topoisomerases I and II. Jpn. J. Cancer Res. 1997;88:992–1002. doi: 10.1111/j.1349-7006.1997.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byl JAW, Fortune JM, Burden DA, Nitiss JL, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: primary cellular target and DNA cleavage enhancement. Biochemistry. 1999;38:15573–15579. doi: 10.1021/bi991791o. [DOI] [PubMed] [Google Scholar]
- 72.Fortune JM, Velea L, Graves DE, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: DNA interactions and topoisomerase catalytic inhibition. Biochemistry. 1999;38:15580–15586. doi: 10.1021/bi991792g. [DOI] [PubMed] [Google Scholar]
- 73.Nelson EM, Tewey KM, Liu LF. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4′-(9-acridinylamino)-methanesulfon-m-anisidide. Proc. Natl. Acad. Sci. USA. 1984;81:1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pommier Y, Minford JK, Schwartz RE, Zwelling LA, Kohn KW. Effects of the DNA intercalators 4′-(9-acridinylamino)methanesulfon-m-anisidide and 2-methyl-9-hydroxyellipticinium on topoisomerase II mediated DNA strand cleavage and strand passage. Biochemistry. 1985;24:6410–6416. doi: 10.1021/bi00344a015. [DOI] [PubMed] [Google Scholar]
- 75.Beck WT, Danks MK, Wolverton JS, Kim R, Chen M. Drug resistance associated with altered DNA topoisomerase II. Adv. Enzyme Reg. 1993;33:113–127. doi: 10.1016/0065-2571(93)90012-3. [DOI] [PubMed] [Google Scholar]
- 76.Vassetzky YS, Alghisi GC, Gasser SM. DNA topoisomerase II mutations and resistance to anti-tumor drugs. Bioessays. 1995;17:767–774. doi: 10.1002/bies.950170906. [DOI] [PubMed] [Google Scholar]
- 77.Kingma PS, Burden DA, Osheroff N. Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance. Biochemistry. 1999;38:3457–3461. doi: 10.1021/bi982855i. [DOI] [PubMed] [Google Scholar]
- 78.Leroy D, Kajava AV, Frei C, Gasser SM. Analysis of etoposide binding to subdomains of human DNA topoisomerase II alpha in the absence of DNA. Biochemistry. 2001;40:1624–1634. doi: 10.1021/bi0019141. [DOI] [PubMed] [Google Scholar]
- 79.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. Anti-Cancer Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 80.Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J. Mol. Biol. 1970;54:247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- 81.Waring MJ. Drugs and DNA: uncoiling of the DNA double helix as evidence of intercalation. Humangenetik. 1970;9:234–236. doi: 10.1007/BF00279229. [DOI] [PubMed] [Google Scholar]
- 82.Wall ME, Wani MC. Camptothecin and taxol: discovery to clinic--thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1995;55:753–760. [PubMed] [Google Scholar]