Abstract
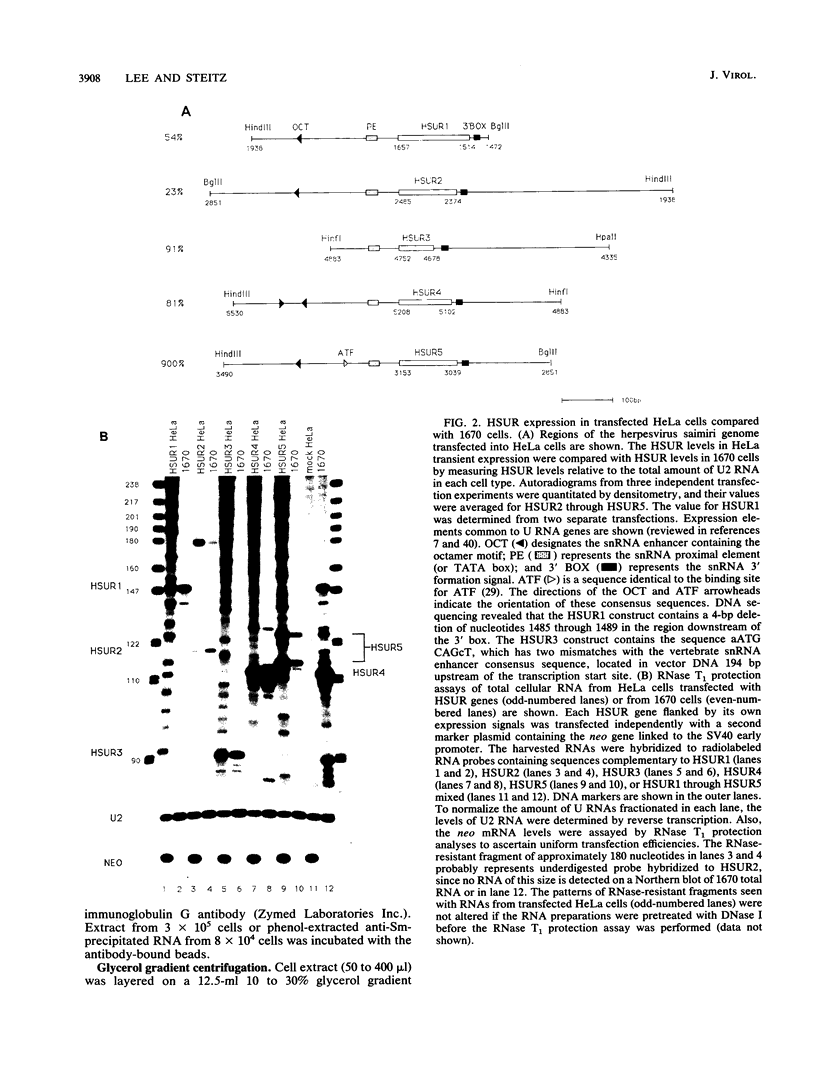
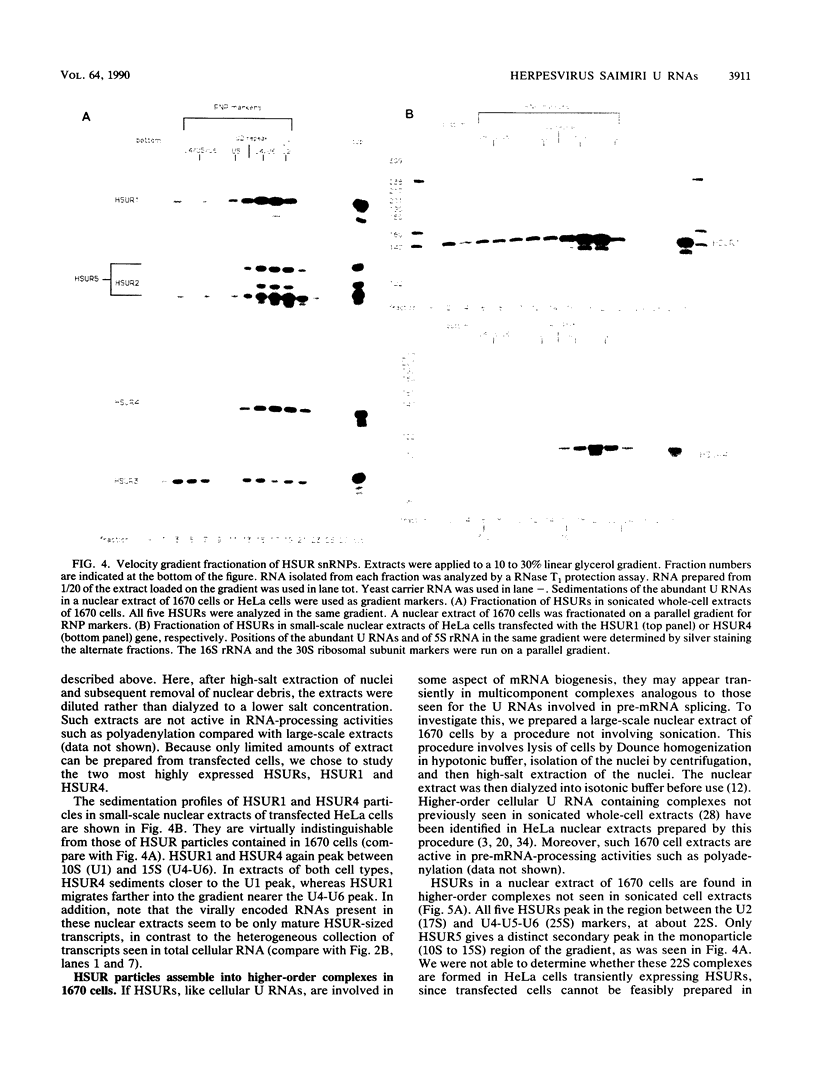
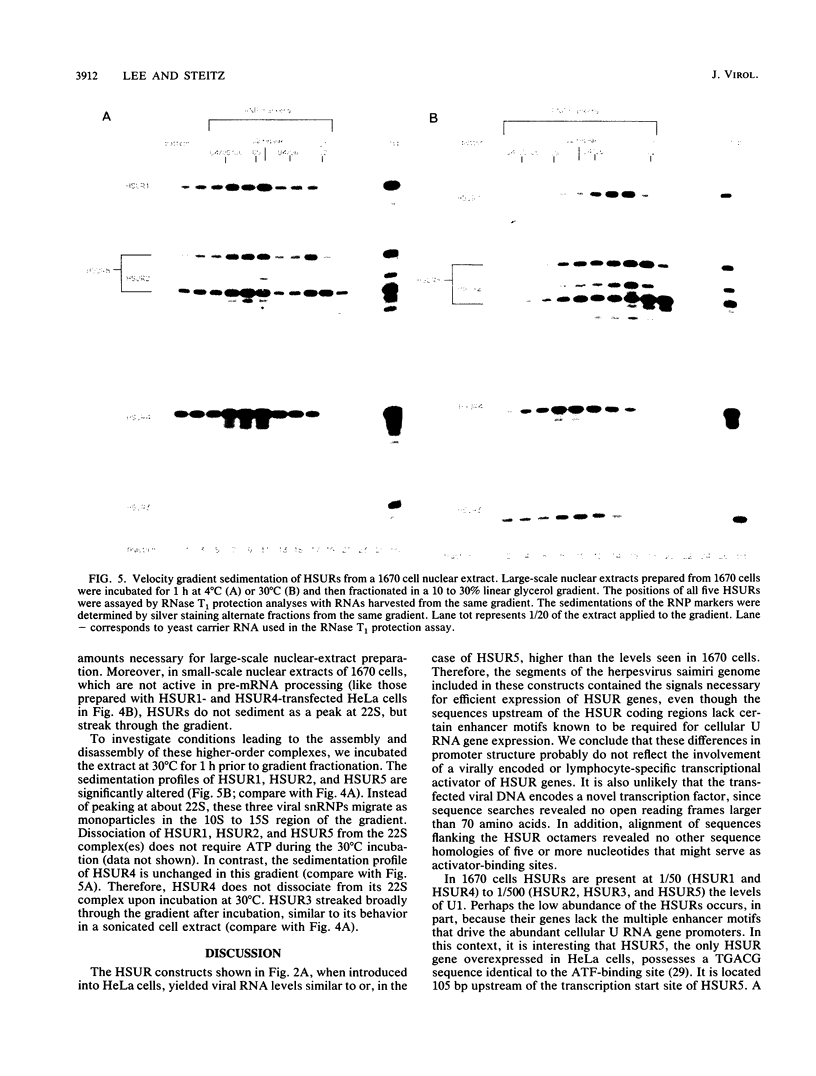
Marmoset T lymphocytes transformed by herpesvirus saimiri contain a set of five virally encoded U RNAs called HSUR1 through HSUR5. HSUR genes have been individually transfected into a nonlymphoid, nonsimian cell line (HeLa cells) in the absence of any other coding regions of the herpesvirus saimiri genome. The levels of HSUR1 through HSUR4 in HeLa transient-expression systems are comparable to those found in virally transformed T cells (23 to 91%). In contrast, HSUR5 is expressed at ninefold-higher levels in transfected HeLa cells. Immunoprecipitation experiments show that HSURs expressed in transfected cells bind proteins with Sm determinants and acquire a 5' trimethylguanosine cap structure, as they do in transformed T cells. HSUR1 or HSUR4 particles from transfected HeLa cells migrate between 10S and 15S in velocity gradients, identical to the sedimentation of "monoparticles" produced in virally transformed lymphocytes. We conclude from these transfection experiments that no other herpesvirus saimiri or host-cell-specific gene products appear to be required for efficient expression of the HSUR genes or for subsequent assembly of the viral U RNAs into small nuclear ribonucleoprotein particles. In lymphocytes transformed by herpesvirus saimiri, HSUR small nuclear ribonucleoprotein particles are involved in higher-order complexes that sediment between 20S and 25S. HSUR1, HSUR2, and HSUR5 dissociate from such complexes upon incubation at 30 degrees C, whereas the complex containing HSUR4 is stable to incubation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Chung J. S., Giglio L., Weiner A. M. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1987 Oct;1(8):808–817. doi: 10.1101/gad.1.8.808. [DOI] [PubMed] [Google Scholar]
- Black D. L., Pinto A. L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989 Aug;9(8):3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988 Nov;7(11):3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Burghoff R. L., Bakker A., Kamine J. Construction of replication-competent Herpesvirus saimiri deletion mutants. J Virol. 1984 Feb;49(2):343–348. doi: 10.1128/jvi.49.2.343-348.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R. C., Silva D. P., Waldron L. M., Letvin N. L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986 Feb;57(2):701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoeur A. M., Mathews M. B. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6772–6776. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann R., Fleckenstein B. Selectable recombinant herpesvirus saimiri is capable of persisting in a human T-cell line. J Virol. 1989 Apr;63(4):1818–1821. doi: 10.1128/jvi.63.4.1818-1821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. G., Steitz J. A. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9006–9010. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschka-Dierich C., Werner F. J., Bauer I., Fleckenstein B. Structure of nonintegrated, circular Herpesvirus saimiri and Herpesvirus ateles genomes in tumor cell lines and in vitro-transformed cells. J Virol. 1982 Oct;44(1):295–310. doi: 10.1128/jvi.44.1.295-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M. G., DeCorato D., Safer B., Galabru J., Hovanessian A. G. Adenovirus VAI RNA complexes with the 68 000 Mr protein kinase to regulate its autophosphorylation and activity. EMBO J. 1987 Mar;6(3):689–697. doi: 10.1002/j.1460-2075.1987.tb04809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmaier M., Tebb G., Mattaj I. W. Functional characterization of X. laevis U5 snRNA genes. EMBO J. 1987 Oct;6(10):3071–3078. doi: 10.1002/j.1460-2075.1987.tb02614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajewski J., Schneider R. J., Safer B., Munemitsu S. M., Samuel C. E., Thimmappaya B., Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986 Apr 25;45(2):195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Krainer A. R. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988 Oct 25;16(20):9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. Fractionation of HeLa cell nuclear extracts reveals minor small nuclear ribonucleoprotein particles. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8408–8412. doi: 10.1073/pnas.84.23.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
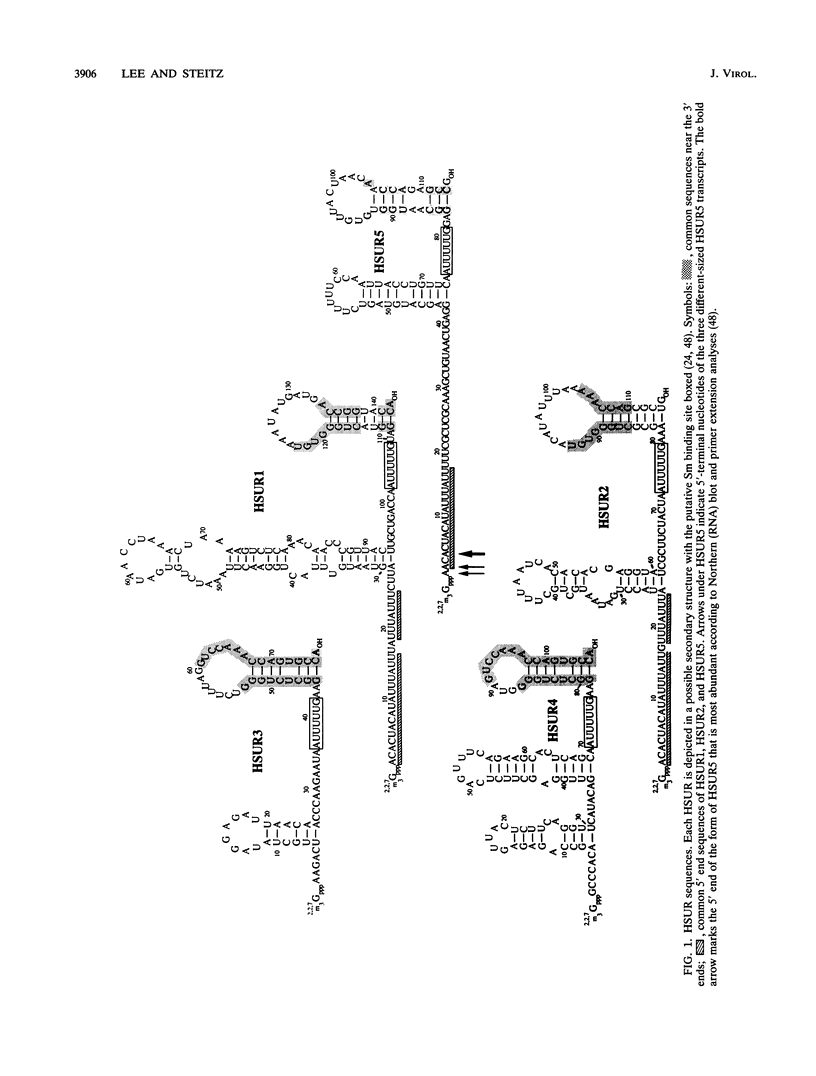
- Lee S. I., Murthy S. C., Trimble J. J., Desrosiers R. C., Steitz J. A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988 Aug 26;54(5):599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Jr, Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981 May;78(5):2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Andrews N. C., Miller G., Steitz J. A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981 Feb;78(2):805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Hardin J. A., Steitz J. A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981 Jan 23;211(4480):400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Interaction of a common cellular transcription factor, ATF, with regulatory elements in both E1a- and cyclic AMP-inducible promoters. Proc Natl Acad Sci U S A. 1988 May;85(10):3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin M., Ares M., Jr, Weiner A. M. U1 small nuclear RNA genes are subject to dosage compensation in mouse cells. Science. 1985 Jul 19;229(4710):272–275. doi: 10.1126/science.2409601. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Montzka K. A., Steitz J. A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. snRNP mediators of 3' end processing: functional fossils? Trends Biochem Sci. 1988 Nov;13(11):447–451. doi: 10.1016/0968-0004(88)90220-4. [DOI] [PubMed] [Google Scholar]
- Murthy S. C., Trimble J. J., Desrosiers R. C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989 Aug;63(8):3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S., Kamine J., Desrosiers R. C. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986 Jul;5(7):1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- O'Malley R. P., Mariano T. M., Siekierka J., Mathews M. B. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell. 1986 Feb 14;44(3):391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Bindereif A., Green M. R. In vitro reconstitution of snRNPs: a reconstituted U4/U6 snRNP participates in splicing complex formation. Genes Dev. 1989 Apr;3(4):479–487. doi: 10.1101/gad.3.4.479. [DOI] [PubMed] [Google Scholar]
- Roebuck K. A., Szeto D. P., Green K. P., Fan Q. N., Stumph W. E. Octamer and SPH motifs in the U1 enhancer cooperate to activate U1 RNA gene expression. Mol Cell Biol. 1990 Jan;10(1):341–352. doi: 10.1128/mcb.10.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin G. M., Bannwarth W., Birnstiel M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3' end of H3 messenger RNA. 1986 Oct 30-Nov 5Nature. 323(6091):777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Schneider R. J., Safer B., Munemitsu S. M., Samuel C. E., Shenk T. Adenovirus VAI RNA prevents phosphorylation of the eukaryotic initiation factor 2 alpha subunit subsequent to infection. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4321–4325. doi: 10.1073/pnas.82.13.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. RNA splicing and genes. JAMA. 1988 Nov 25;260(20):3035–3041. [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Lee S. I., Steitz J. A. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 1989 Feb 11;17(3):1258–1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Weller P., Bark C., Janson L., Pettersson U. Transcription analysis of a human U4C gene: involvement of transcription factors novel to snRNA gene expression. Genes Dev. 1988 Nov;2(11):1389–1399. doi: 10.1101/gad.2.11.1389. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]