Abstract
The positive transcription elongation factor b (P-TEFb) (CDK9/cyclin T (CycT)) promotes mRNA transcriptional elongation through phosphorylation of elongation repressors and RNA polymerase II. To understand the regulation of a transcriptional CDK by its cognate cyclin, we have determined the structures of the CDK9/CycT1 and free cyclin T2. There are distinct differences between CDK9/CycT1 and the cell cycle CDK CDK2/CycA manifested by a relative rotation of 26° of CycT1 with respect to the CDK, showing for the first time plasticity in CDK cyclin interactions. The CDK9/CycT1 interface is relatively sparse but retains some core CDK–cyclin interactions. The CycT1 C-terminal helix shows flexibility that may be important for the interaction of this region with HIV TAT and HEXIM. Flavopiridol, an anticancer drug in phase II clinical trials, binds to the ATP site of CDK9 inducing unanticipated structural changes that bury the inhibitor. CDK9 activity and recognition of regulatory proteins are governed by autophosphorylation. We show that CDK9/CycT1 autophosphorylates on Thr186 in the activation segment and three C-terminal phosphorylation sites. Autophosphorylation on all sites occurs in cis.
Keywords: CDK9, cyclin T, flavopiridol, protein kinase, P-TEFb
Introduction
The positive transcription elongation factor b (P-TEFb) coordinates the elongation phase of transcription. After transcription initiation, elongation by RNA polymerase (Pol) II is halted by the DRB (5,6-dichlorobenzimidazole 1-β-D-ribofuranoside)-sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF). These multimeric repressors bind to the polymerase and in their presence RNA Pol II produces short aborted transcripts. P-TEFb phosphorylates DSIF and NELF (Fujinaga et al, 2004; Yamada et al, 2006), and the RNA Pol II carboxy-terminal domain (CTD) on position Ser2 in the heptad repeat sequence Y1S2P3T4S5P6S7 (using the single letter amino-acid code) (Marshall et al, 1996). These phosphorylations release the repressor actions and allow productive transcription elongation. The hyperphosphorylated CTD recruits members of the elongation complex and the splicing and 3′-polyadenylation machinery (reviewed in Sims et al, 2004; Peterlin and Price, 2006).
P-TEFb is composed of cyclin-dependent kinase 9/cyclin T (CDK9/CycT). CDK9 activation requires binding of a T family cyclin (CycT1, CycT2a or CycT2b) or CycK. CycT1 was first identified as an 87-kDa (726 residues) protein that interacts specifically with the transactivation domain of the HIV-1 TAT protein (Peng et al, 1998; Wei et al, 1998). The N-terminal ∼260 residue cyclin domain of CycT activates CDK9 and is both necessary and sufficient to support HIV transcription (Bieniasz et al, 1999). In vivo, more than half of P-TEFb is associated with the small 7SK nuclear RNA (snRNA), an abundant and evolutionarily conserved RNA (Nguyen et al, 2001; Yang et al, 2001). The association reduces P-TEFb CTD kinase activity by at least 15-fold. Inactive P-TEFb also contains an additional protein, HEXIM1 (hexamethylene bisacetamide (HMBA)-induced protein; Michels et al, 2003; Yik et al, 2003). HMBA is a potent inducer of cell differentiation and suppressor of cell proliferation. The observed regulation of P-TEFb by HEXIM1/7SK RNA suggests a role for P-TEFb in directing cells towards either proliferation or differentiation. Binding of the bromodomain protein Brd4 to CycT counteracts the inhibitory effects of HEXIM/7SK RNA and recruits P-TEFb to sites of transcription (reviewed in Zhou and Yik, 2006). During cell division, this serves to mark epigenetically genes whose active transcription status needs to be preserved (Yang et al, 2008).
As in other CDKs, phosphorylation has a major function in CDK9 regulation. Phosphorylation of Thr186 in the activation segment is important for CDK9 kinase activity (Li et al, 2005). Unlike other CDKs, Thr186 is not phosphorylated by the CDK-activating kinase CAK (CDK7/CycH/Mat1) (Kim and Sharp, 2001), instead it has been suggested that it is an autophosphorylation site (Li et al, 2005). Phosphorylation on Thr186 is also important for binding of 7SK RNA to CDK9/CycT (Chen et al, 2004). In addition, autophosphorylation on the C-terminal region of CDK9 enhances the binding of TAR RNA to the CDK9/CycT1/TAT complex (Garber et al, 2000) and ensures the nuclear localization of CDK9/CycT1 (Napolitano et al, 2003).
Flavopiridol is a broad specificity CDK inhibitor with a distinct preference for CDK9. Ki values for CDK9/CycT (3 nM) are approximately 10-fold lower than those for other CDKs (40–70 nM) (Chao et al, 2000). Flavopiridol administration has profound effects on transcription and results in decreased levels of labile mRNAs, including those for early transcription factors, cytokines, cell cycle regulators and kinases, as well as those proteins that mediate the antiapoptotic response such as Mcl-1 and XIAP. Flavopiridol is currently in clinical trials as a CDK9 inhibitor for a number of cancers (Shapiro, 2006), with promising results for the treatment of chronic lymphocytic leukaemia (CLL) (Byrd et al, 2007).
The structures of the cell cycle CDK, CDK2, in complex with cyclins A, B and E have given insight into the mechanism of cyclin CDK activation and substrate recognition (Brown et al, 1995, 1999, 2007; Jeffrey et al, 1995; Russo et al, 1996a, 1996b; Honda et al, 2005). For the transcriptional CDKs, there are separate structures of CDK7 (Lolli et al, 2004) and the individual cyclins CycH, CycC, CycK and CycT1. To date there is no structure of a mammalian transcriptional CDK/cyclin complex. Here, we describe structures of human CDK9/CycT1, CDK9/CycT1 in complex with AMPPNP or flavopiridol and the structure of free CycT2. There are distinct differences between the cell cycle CDKs and the transcriptional CDKs that are defined by a different orientation of the cyclin. The CDK9/CycT1/flavopiridol complex reveals unanticipated structural rearrangements that bury this potent inhibitor at the ATP-binding site. We characterize CDK9 autophosphorylation: autophosphorylation occurs on Thr186 and three C-terminal sites (Ser347, Thr262 and Thr363). Autophosphorylation on both the activation segment and the C-terminal sites occurs in cis. Although T186 phosphorylation is activatory, phosphorylation on C-terminal sites does not appear to directly modulate CDK9 activity.
Results
The CDK9/CycT1 complex
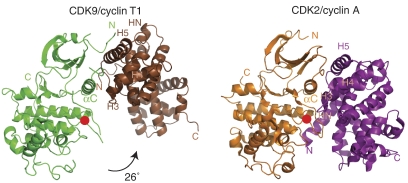
The structures of CDK9 and CycT1 in the CDK9/CycT1 complex are similar to previously determined structures for other CDKs and cyclins. However, there is a dramatic difference in the association of the CDK and cyclin between CDK9/CycT1 and the cell cycle CDK/cyclin complexes (e.g. CDK2/CycA). The orientation of the cyclin with respect to the CDK is rotated by about 26° (Figure 1). This results in a comparatively sparse number of contacts between CDK9/CycT1. The buried molecular surface area (1763 Å2) is only 60% of the molecular surface area buried on complex formation of CDK2/CycA (2892 Å2) (Supplementary Table S1). Nevertheless, the CDK9/CycT1 surface area buried is not untypical for tightly interacting protein–protein complexes (Lo Conte et al, 1999). In agreement with a smaller interaction interface surface, plasmon resonance measurements indicate a KD of approximately 300 nM for CDK9/CycT1 (S Baumli and LN Johnson, unpublished data) as compared with 48–55 nM for CDK2/CycA measured in solution (Heitz et al, 1997).
Figure 1.
The structure of CDK9/CycT1 and comparison with CDK2/CycA. Schematic overall representations of CDK9/CycT1 (left; CDK9 is in green, CycT1 in brown) and CDK2/CycA (right; CDK2 is in orange, CycA in magenta). The phospho-threonine residues in the activation segments are marked by a red circle. The CDK structures have been aligned and the 26° rotation of CycT1 with respect to the position of CycA is indicated. This and other structure figures were prepared with PyMOL (WL DeLano, The PyMOL Molecular Graphics System; DeLano Scientific, San Carlos, CA, USA, 2002).
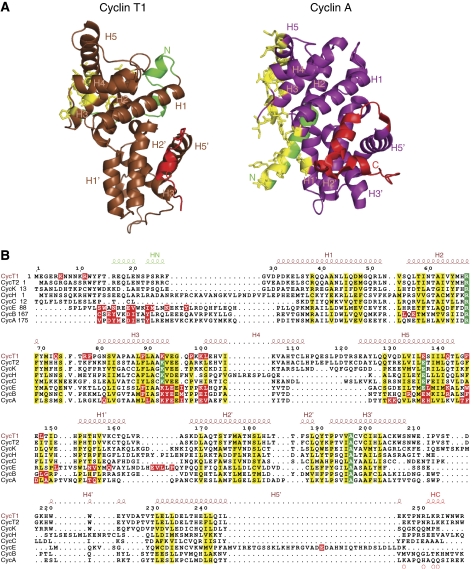
Cyclin T structure. In common with other cyclins, both CycT1 and CycT2 contain two canonical cyclin box folds each consisting of five helices and short N-terminal (HN) and C-terminal (HC) helices. The two cyclin box folds are arranged around the central H3 and H3′ helices (superscript prime refers to helices in the second cyclin box) (Figure 2A). There are no significant conformational changes in CycT1 between the free and CDK9-bound states, consistent with previous observations that the cyclin fold is rigid (Brown et al, 1995), with the exception of a few connecting loops and the C-terminal region (discussed below).
Figure 2.
Comparison of CycT1 and CycA. (A) Schematic diagrams of CycT1 (left) and CycA (right) with secondary structural elements labelled. The colours are the same as Figure 1 but with the HN and HC helices in green and red, respectively, and residues in contact with the CDK in yellow. The orientation of CycA is as in Figure 1. CycT1 has been aligned with CycA for comparison. (B) Structure-based sequence alignment of transcriptional and cell cycle cyclins. Residues conserved in all eight cyclins are in green and those that are similar in yellow. Cyclin A, B, E and T1 residues in contact with their cognate CDK (distance <3.5 Å) are shown highlighted in red. Secondary structure and numbering for CycT1 are shown above the sequences.
Despite their similarity in sequence and structure (Figure 2B and Supplementary Table S2), cyclins involved in cell cycle control (CycA, CycB and CycE) and those involved in transcription (CycT1, CycT2, CycC, CycH and CycK) differ significantly outside the classical cyclin fold in the length and orientation of the HN and HC helices (Figure 2A and Supplementary Figure S1). In the cell cycle kinases, the HN helix makes critical interactions with several regions of the CDK, which are important for the activatory function of the cyclin. In CDK9/CycT1, HN makes only a few interactions with CDK9 (discussed later). In the cell cycle kinases, the HC helix appears to have no function, whereas in CDK9/CycT1 the HC helix contributes to recognition of regulatory and recruitment proteins (HEXIM and TAT).
A further difference between the two families is in the ‘hydrophobic pocket' present in CycA and CycE (characterized by the sequence motif 210MRAILVDW217 in helix H1 in CycA) that serves as a recruitment site for CDK2 substrates (Cheng et al, 2006) and the p27Kip1 inhibitor (Russo et al, 1996a). In the transcriptional cyclins, CycT1 and CycT2, sequence differences at the H1 motif (37YRQQAANL44) and an extra turn of the H4 helix result in a non-functional recruitment site (Figure 2B).
The C-terminal helix of CycT1 is pliable. In the recognition of HIV TAT, residue CycT1 Cys261 contributes to the zinc-mediated interaction of CycT1 and TAT and is responsible for the species-specific nature of this interaction (Garber et al, 1998; Fujinaga et al, 1999). The free and bound CycT1 structures show differences in their HC helices. In free CycT1 and T2, the HC helix extends from Pro249 to Ala262. In CDK9-bound CycT1, the chain takes a different route in the H5′/HC linker region. It adopts a helical conformation for residues Asn250 to Trp256 and then an extended conformation to residue Arg259, the end of the construct (Supplementary Figure S2). CycT1 HC is not in contact with CDK9 but it is in contact with a symmetry related molecule in the CDK9/CycT1 crystal structure. The shift in conformation indicates that the C-terminal region of CycT1 is pliable. Flexibility in this region may contribute to the CycT1/TAT recognition process.
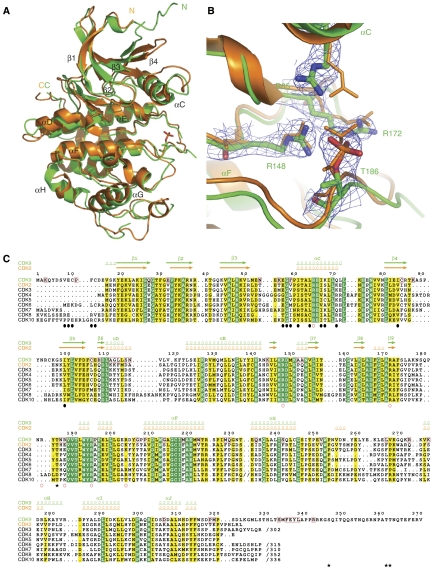
CDK9 structure. The CDK9 structure exhibits a typical kinase fold comprising the N-terminal lobe (residues 16–108), which is mainly β-sheet with one α-helix (αC), and a C-terminal lobe (residues 109–330) comprised mainly of α-helices. CDK9 is similar in structure to CDK2 (Figure 3A) and has 40% sequence identity. The major differences occur in loop regions and include the loop before the αC helix, corresponding to a CycA contact region in the CDK2/CycA complex, the loop between β4 and β5 where CDK9 has an insertion compared with CDK2 (not defined by electron density (residues 87–98)), the loop between the αD and αE helices (residues 114–121), part of the activation segment (where additional residues 179–182 are not defined) and the loop between αG and αH residues 259–267.
Figure 3.
CDK9 structure. (A) The structures of CDK9 (green) and CDK2 (orange) superimposed. The cyclins have been omitted for clarity. (B) A close-up of pThr186 in CDK9 with the final 2Fo–Fc electron density and side chain contacts. CDK2 pThr160 and contacts (orange) are superimposed. (C) Sequence comparison of CDK9, CDK2 and other CDKs. Secondary structural elements are indicated for CDK9 and CDK2 in green and orange, respectively. Residues conserved in all CDKs are shown in green and those that are similar in yellow. Residues invariant in CDK9s from human, mouse, chicken, worm, fly and fish but not conserved in the other CDKs are boxed with a red frame in the CDK9 sequence (Supplementary Figure S7). Black circles below the sequences indicate CDK9 residues in contact with CycT1 (<3.5 Å). Red open circles indicate basic residues that contribute to the positively charged surface of CDK9 (Supplementary Figure S4). Black asterisks correspond to autophosphorylation sites.
CDK9 is in the active kinase conformation, characterized by the orientation of the αC helix, the conformation of the residues important for phosphoryl transfer (Asp149, Lys151 and Thr165), the active conformation of the Asp167, Phe168 and Gly169 (the ‘DFG' motif) at the start of the activation segment and the conformation of the activation segment itself that leaves free access for the substrate to the catalytic site. Phospho-Thr186 in the activation segment is clearly visible in the electron density map and interacts with two arginines Arg148 and Arg172 (Figure 3B). A third arginine, Arg65, is just too distant (3.9 Å) to form a hydrogen bond (These residues correspond to Arg126, Arg150 and Arg50 that contact phospho-Thr160 in CDK2.) (Figure 3C). Of interest is the replacement in the activation segment of residue Glu162 in CDK2 by Arg188 in CDK9. In the non-phospho-CDK2/CycA complex (Jeffrey et al, 1995), Glu162 is turned inwards so that it partly contacts the arginine cluster. Phosphorylation of Thr160 results in exposure of Glu162 and its internal site is taken by the phospho-threonine leading to the correct alignment of the activation segment for substrate recognition. Although there is no structure of non-phospho-CDK9, we may anticipate that the activation segment would not adopt a conformation with Arg188 turned in, as this would place Arg188 close to the other three arginines.
Interactions between CycT1 and CDK9
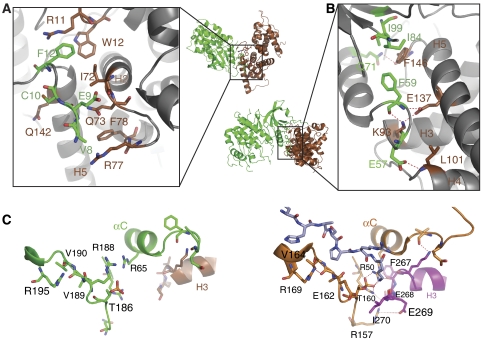
The most obvious difference between the CDK9/CycT1 and CDK2/CycA structures is in the role of the cyclin N-terminal helix HN. The HN helix in CycT1 adopts an orientation in which it is directed towards the solvent (Figure 2A). There is no change in conformation from its position in free CycT1. However, there are interactions from the N terminus of CycT1 to CDK9 and the N-terminal extension of CDK9 to CycT1 that are specific to the CDK9/CycT1 complex (Figure 4A and Supplementary Table S3). In the CDK2/CycA complex, the CycA HN helix contacts residues from several different elements of the kinase seemingly strengthening the complex (discussed in Brown et al, 2007) (Figure 1, right). These contacts contribute a buried molecular surface area corresponding to nearly 30% of the total buried surface in CDK2/CycA and are essential for the CycA activation of CDK2 (Kobayashi et al, 1992). The structures of the neuronal CDK5 in complex with its activator the single domain cyclin, p25 (Tarricone et al, 2001), and the yeast transcriptional Pho85/Pho80 complex (Huang et al, 2007) also show lack of involvement of the cyclin HN helix in the CDK/cyclin interactions, yet both complexes have a similar cyclin orientation to the CDK as in CDK2/CycA, although with differences in the interactions for the cyclin H3/H4 loop. Evidently it is not the lack of the HN helix alone that leads to the alternative orientation of the cyclin in CDK9/CycT1.
Figure 4.
Details of CDK9/CycT1 interactions. (A) The contacts between CycT1 and the N-terminal residues of CDK9. The view, as indicated in the small overall figure, is 120° rotated about the horizontal axis with respect to Figure 1. Secondary structural elements, shown for guidance but not in contact, are in grey. The colour code is as in Figure 1. (B) The major contacts between CDK9 and CycT1 in the vicinity of the CDK9 αC helix. For further details see text. (C) Selected residues showing the substrate-binding site and phospho-threonine environment for CDK9/CycT1 (left) and CDK2/CycA (right). The substrate heptapeptide bound to CDK2/CycA (Brown et al, 1999) is shown in cyan.
Despite the difference in cyclin orientation, there are core contacts that are common to both cell cycle and transcriptional CDK complexes (Figure 4B and Supplementary Table S3). These include contacts from the cyclin H3, H4 and H5 helices and the CDK αC helix and β4 strand. The cyclin H5 helix runs parallel to the CDK αC helix and assists in its alignment in the active conformation, a feature also used in the co-activation of the EGF receptor tyrosine kinase in the asymmetric dimer (Zhang et al, 2006). The hydrogen bonds from CycT1 residues in H3 (Lys93), H4 (Leu101) and H5 (Glu137) and the H5/H1′ loop (Phe146) to CDK9 are similar to those observed between CycA and CDK2 (Supplementary Table S3). Phe146 is the focus of a hydrophobic cluster comprising residues from CycT1 and CDK9 and similar interactions are also made in CDK2/CycA. Taken together, these interactions appear sufficient to stabilize the active conformation of the CDK9.
In an attempt to understand why the orientation of CycT1 with respect to the CDK is different in CDK9/CycT1, we modelled CycT1 in the position of CycA as in the CDK2/CycA structure. The results showed major clashes between the end of CycT1 H3 helix and the CDK9 loop at the start of αC helix. Although the CDK pre-αC helix loop in CDK2 has been observed to shift in response to binding other cyclins (Honda et al, 2005) and could exhibit some mobility in CDK9, a clash between the CDK9 αC helix Leu61 and conserved CycT1 Lys93 could not easily be removed by conformational changes. There is also a clash between the N-terminal CycT1 residues (Arg5 and Lys6) and the CDK9 αC/β4 loop. However, the N-terminal region of CycT1 is not well ordered in the crystal and could shift. This analysis suggests that whereas in CDK2/CycA the HN helix contributes to the defined binding mode, in CDK9/CycT1 that lacks this helix interaction and with a sequence change in the CDK αC helix that results in a clash, a different binding mode is possible and favoured by the unique conformation of the N-terminal extension of CDK9 (Figure 4A).
Mapping of identical and conserved residues between the CDK9 cyclins T1, T2 and K onto the surface of CycT1 reveals that only a small part of the transcriptional cyclin surface is conserved. The conserved region includes the interaction site with CDK9 (Supplementary Figure S3). This would suggest that all three cyclins share the same interaction mode with CDK9. The conserved surface patch is slightly bigger than the actual interaction surface and extends to the second cyclin box containing residues that appear to be important for structural stability. The absence of any other strongly conserved surface patches between CycT and CycK might be indicative of their association with different regulatory molecules and therefore evolving different functional roles within the cell.
CDK9 substrate recognition
In cell cycle CDK/cyclin complexes, the phospho-threonine in the activation segment has an organizing role bringing different elements of the structure together to create a special conformation of the activation segment for recognition of substrate (Russo et al, 1996b; Brown et al, 1999). Although we have not yet achieved substrate binding for CDK9/CycT1, it is possible to make certain deductions by comparison with CDK2/CycA structures. The conformations of the activation segments that determine substrate specificity for the Ser-Pro motif are very similar in CDK2 and CDK9 (Figure 4C). The left-handed conformation of CDK9 Val190 (CDK2 Val164) that is promoted by interactions with CDK9 Arg195 (CDK2 Arg169) creates a nonpolar pocket that can accommodate proline but no other residue (Brown et al, 1999).
In the CDK9/CycT1 complex, there are no interactions that connect the phospho-threonine site to the cyclin (Figure 4C, left). In CDK2/CycA, the main chain oxygens of residues Phe267 and Glu268 at the end of CycA H3 helix contact CDK2 side chains Arg50 and Arg150, respectively, which in turn contact the phospho-threonine pThr160 (Figure 4C, right; and Supplementary Table S3). The nonpolar CycA residue Ile270 (end H3) shields the phospho-threonine from solvent. These interactions are not made in the CDK9/CycT1 complex because of the change in orientation of the cyclin and there is no equivalent residue to Ile270 in CycT1 because of a sequence deletion (Figure 2B). As a result, the phospho-threonine in CDK9 is more exposed.
We conclude that CDK9 pThr186 has a limited function as an organizational centre, but nevertheless is able to promote the active conformation of the activation segment for substrate recognition of the S/T-P motif. Arg188 in the CDK9 apo structure (Figure 4C, left) partially blocks the assumed peptide substrate site but this residue is not well ordered and on binding AMPPNP or flavopiridol it shifts to an external position. Arg188 is part of one of the basic clusters (Supplementary Figure S4) that may contribute to recognition of CDK7 (as part of TFIIH)-catalysed phosphorylated sites on the serine position 5 of the CTD. Hierarchical phosphorylation has been established for glycogen synthase kinase, CK1 and CK2, whereby a priming substrate phosphorylation is a prerequisite for the generation of a consensus sequence but this has not been definitively established for CDK9.
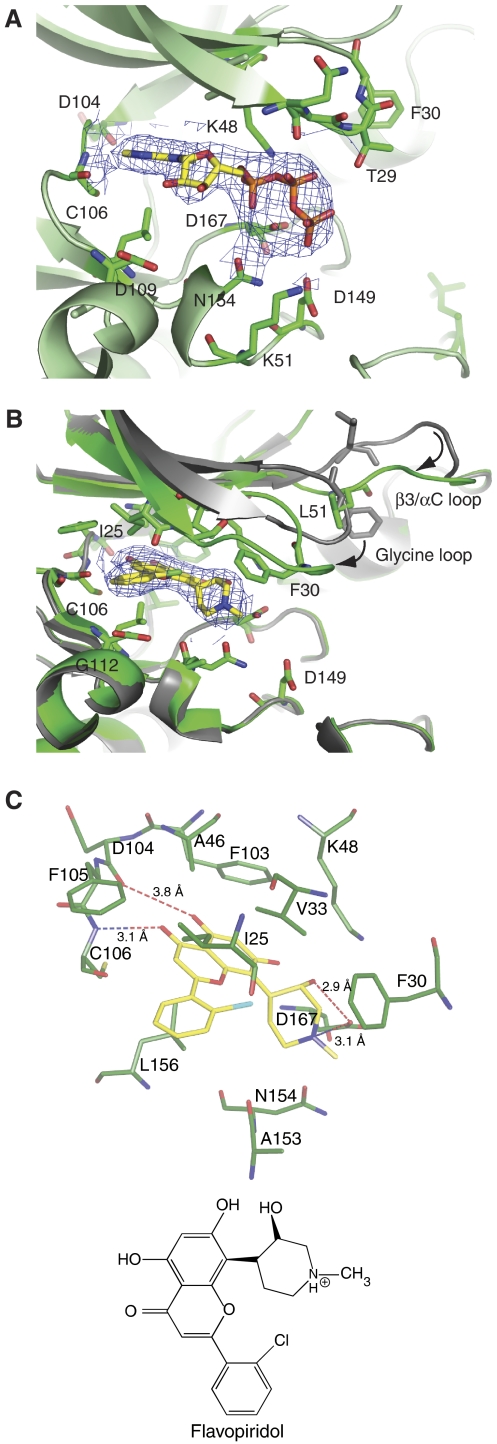
Flavopiridol binds to the ATP-binding site in CDK9
A study of the ATP analogue, AMPPNP, co-crystallized with CDK9/CycT1 at 2.9-Å resolution showed the nucleotide bound in a similar mode to that observed with CDK2/CycA (Figure 5A). The adenine N6 and N1 nitrogens participate in hydrogen bonds as donor and acceptor, respectively, to the hinge region main chain oxygen and main chain nitrogen of residues Asp104 and Cys106 (corresponding to CDK2 Glu81 and Leu83). There are similar constellations of aliphatic and aromatic groups that contact the adenine and the conformation of the triphosphate moiety resembles that in CDK2/CycA (PDB 1QMZ). This is not unexpected as the ATP-binding site is strongly conserved between CDKs.
Figure 5.
AMPPNP and flavopiridol binding. (A) Details of the interaction of AMPPNP with CDK9/CycT. The CDK9 main chain is represented in green together with the side chains of residues within 3.5 Å of AMPPNP. (B) Details of the interaction of flavopiridol with CDK9/CycT1 and conformational changes. CDK9 in the flavopiridol complex is in green and in the AMPPNP complex in grey. (C) Details of the flavopiridol CDK9 interactions and a schematic representation of flavopiridol. The electron density maps are the final 2Fo–Fc maps contoured at 1σ around the ligand (A) AMPPNP and (B) flavopiridol.
Flavopiridol was co-crystallized with CDK9/CycT1 and the structure solved at 2.8-Å resolution. Flavopiridol binds to CDK9 at the ATP-binding pocket in a similar manner to the binding of deschloro-flavopiridol to inactive monomeric CDK2 (De Azevedo et al, 1996) (Figure 5B). Flavopiridol is almost entirely buried in CDK9 with only 8% of its molecular surface area exposed. There are hydrogen bonds from the flavopiridol O4 oxygen and O5 hydroxyl to hinge residues Cys106 NH and Asp104 O (long) and contacts between the piperidinyl group N1, which is assumed to be protonated, and the O3 hydroxyl to Asp167 side chain (the ‘DFG' aspartate) (Figure 5C). Similar contacts were made for deschloro-flavopiridol bound to CDK2. The chloro-phenyl ring has a different orientation to accommodate the chlorine, which makes a long contact to the main chain oxygen of Ile25 (4.2 Å) and an internal contact to the oxygen of the adjacent ring (2.8 Å). The pocket that accommodates the chloro-phenyl ring is more open in CDK9. Gly112 takes the place of CDK2 Lys89 and creates a less crowded and possibly more favourable electrostatic environment. In the CDK2/flavopiridol complex, Lys89 shifted to accommodate flavopiridol.
Flavopiridol binding induces major conformational changes in CDK9 compared with the CDK9 apo-enzyme and the AMPPNP-complexed structures. The glycine-rich loop (G-loop) between β1 and β2 (residues 17–36) closes and folds over the active site bringing Ile25 and Val33 into van der Waals contact with the inhibitor (similar to the corresponding residues in the closed conformation of inactive CDK2) (Figure 5B and C). Phe30 in the G-loop rotates and makes additional van der Waals contacts (edge to face) with the piperidinyl group of the inhibitor. The β3/αC loop (residues 51–55), which in the AMPPNP complex packs against the G-loop, changes its conformation so that Leu51 occupies the space of Phe30 in the AMPPNP structure and locks the G-loop in its closed conformation. The new position of the G-loop would exclude ATP binding. Although the resulting G-loop conformation is ‘closed', the position of Phe30 is different from that of the equivalent residue (Tyr15) in closed inactive CDK2. Tyr15 is external in the CDK2/flavopiridol complex and the conformation of the β3/αC loop is different.
Autophosphorylation on CDK9/CycT
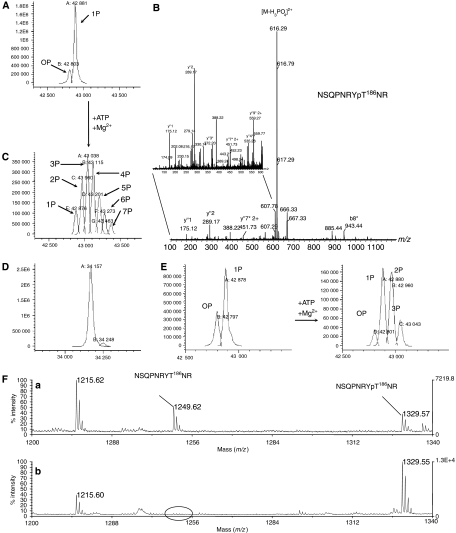
To understand the role of autophosphorylation in the regulation of CDK9, we used the full-length CDK9 (residues 1–372) (in contrast to the truncated CDK9(1–330) used in the crystallographic studies) in complex with CycT1 (residues 1–298) (CDK9-FL/CycT1-298). LC-ESMS showed that CycT1-298 was unphosphorylated in the purified complex (results not shown). CDK9-FL was present as mono-phosphorylated (∼80%) and unphosphorylated species (Figure 6A). Tryptic digestion and peptide analysis by MALDI-MS showed that phosphorylation is homogeneous on the CDK9 activation segment Thr186 (Figure 6B), consistent with the electron density seen in the crystal structure of CDK9(1–330)/CycT1 (Figure 3B).
Figure 6.
Mass spectrometry analysis of CDK9-FL/CycT1-298 phosphorylation state. (A) CDK9, in the purified CDK9/CycT-298 complex, is ∼80% mono-phosphorylated. The expected MW value of the non-phosphorylated protein is 42800.5. The observed masses are: 0P, 42 803; 1P, 42 881. (B) Phosphorylation is homogeneous on Thr186. MS/MS spectrum of the m/z 665.272+ ion corresponding to CDK9-phosphorylated peptide 179–188. A phosphate loss is observed (m/z 616.292+). Peaks marked with an asterisk correspond to dehydroamino-2-butyric acid containing y″- and b-ions and indicate phosphate location on Thr186 (zoom panel). (C) Distribution of phosphorylation states for CDK9 following autophosphorylation reaction. The observed masses are: 1P, 42 876; 2P, 42 960; 3P, 43 038; 4P, 43 115; 5P, 43 201; 6P, 43 273; 7P, 43 433. (D) Autophosphorylation reaction does not affect CycT1-298 phosphorylation state. The expected MW value for CycT1 34 157 corresponds to the observed mass 34 157. (E) Analysis of autophosphorylated CDK9/CycT1-298 by LC/ESI-MS. Left: starting preparation as purified from insect cells. Right: same preparation following limited in vitro autophosphorylation reaction. The expected MW value of the non-phosphorylated protein is 42 800.5. The observed masses are: 0P, 42 797; 1P, 42 878 (left panel) and 0P, 42 801; 1P, 42 880; 2P, 42 960; 3P, 43 043 (right panel). (F) m/z region (1200–1340) of the MALDI mass spectra from tryptic digests of CDK9 showing the peaks corresponding to peptide 179–188 both in parent and in phosphorylated forms. (a) Digest of CDK9 starting preparation as purified from insect cells. (b) Digest of the same preparation following in vitro autophosphorylation reaction.
Incubation of the CDK9-FL/CycT1-298 complex with ATP (10 mM for 2 h at 37°C) resulted in a Gaussian distribution of autophosphorylated forms ranging from 1P to 7P CDK9, centred on the 3P and 4P states, and the disappearance of the unphosphorylated form (Figure 6C). CycT1-298 was not phosphorylated (Figure 6D). Limited autophosphorylation was performed (0.1 mM ATP for 30 min at 20°C) to identify the major and the most rapidly phosphorylated sites. CDK9 phosphorylation shifted to a mixture of unphosphorylated, mono-, di- and tri-phosphorylated forms at 30 min (Figure 6E). Tandem MS identified, in addition to Thr186, the new autophosphorylation sites as Ser347, Thr362 and Thr363 (Supplementary Figure S5). For these last two sites, autophosphorylation was mutually exclusive so that tri-phosphorylated CDK9 had either pThr186, pSer347 and pThr362 or pThr186, pSer347 and pThr363. The amount of peptide corresponding to the pThr186 activation segment increased after autophosphorylation, indicating that Thr186 is also a site of autophosphorylation (Figure 6F) consistent with the observation that shortened CDK9(1–330), which lacks all C-terminal phosphorylation sites, is still capable of incorporating radioactive phosphate in the autophosphorylation reaction to Thr186 (Supplementary Figure S6). We estimate that after 30 min Ser347 is phosphorylated about 50% (based on the sum of 2P and 3P forms; Figure 6E), whereas autophosphorylation of Thr362 and Thr363 (present only in the 3P form) is only 10–15%.
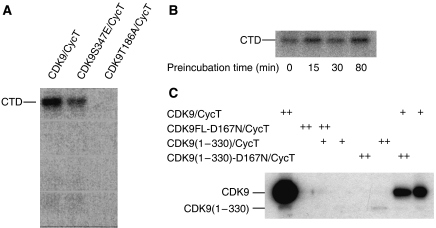
To explore the role of Thr186 phosphorylation in catalysis, we generated the T186A mutant. The CDK9-FL-T186A/CycT complex was unable to phosphorylate the CTD substrate as compared with the WT-CDK9-FL/CycT (Figure 7A). We conclude that Thr186 phosphorylation is essential for CDK9-FL/CycT1-298 activity against the CTD substrate. Comparison of CDK9 sequences (Supplementary Figure S7) shows that Thr186 is conserved in human, mouse, chicken, Xenopus and worm, indicating that activation by phosphorylation of Thr186 is likely to be universal in eukaryotes but other phosphorylation sites are not conserved in lower eukaryotes.
Figure 7.
Catalytic activity of CDK9/CycT1 on RNA Pol II CTD substrate. (A) CDK9-T186A/CycT1-298 showed no activity on CTD in contrast to CDK9-WT/CycT1-298 or the mutant CDK9-S347E/CycT1-298. (B) CDK9/CycT1-288 activity towards the CTD after preincubation with ATP and removal of excess ATP/ADP. (C) Autophosphorylation reactions for active and kinase-dead CDK9-D167N/CycT1-288 complexes. CDK9-D167N/CycT1-288 or CDK9/CycT1-288 variants as substrates (0.6 μg, (++)) were incubated with or without catalytic amounts (50 ng, (+)) of active CDK 9/CycT1-288.
To explore the effect of C-terminal phosphorylations on kinase activity, we used prephosphorylated CDK9/CycT. Autophosphorylation was carried out for 0–80 min, after which ADP was removed and the enzymes were assayed. C-terminally phosphorylated CDK9 shows catalytic activities similar to non-phosphorylated CDK9 in in vitro kinase assays (Figure 7B). We conclude that C-terminal phosphorylation does not affect CDK9 activity against the CTD substrate.
To explore if autophosphorylation occurs in cis or trans, we generated the kinase-dead mutants D167N for CDK9-FL and for the truncated CDK9(1–330), which lacks the C-terminal phosphorylation sites. CDK9-FL/CycT autophosphorylates (Figure 7C, lane 1), but kinase-dead CDK9-FL-D167N/CycT as expected does not (lane 2). We then used catalytic amounts of active CDK9(1–330)/CycT to phosphorylate CDK9-FL-D167N/CycT in trans. Surprisingly CDK9-FL-D167N/CycT was not able to exert an effect as a substrate (Figure 7C, lane 3), indicating that phosphorylation occurs in cis. To explore if Thr186 phosphorylation also occurs in cis, we analysed the kinase-dead mutant CDK9(1–330)-D167N/CycT for its ability to exert an effect as a substrate for active CDK9/CycT. Thr186 in wild-type CDK9/CycT is already 80% phosphorylated when purified from insect cells and incorporation of radioactive phosphate is more difficult to detect. At concentrations equivalent to substrate concentrations autophosphorylation for CDK9(1–330)/CycT is observed (lane 6) corresponding to the increase from ∼80 to 100% phosphorylation of Thr186. Kinase-dead CDK9(1–330)-D167N/CycT does not autophosphorylate, as expected (lane 5), but surprisingly incubation with catalytic amounts of active CDK9-FL/CycT gave rise to no phosphorylation of CDK9(1–330)-D167N/CycT and only autophosphorylation of the active CDK9-FL/CycT (lanes 7 and 8) is observed. We conclude that neither the full-length nor the C-terminally truncated CDK9-D167N mutant is able to exert an effect as a substrate for phosphorylation in trans and hence that autophosphorylation on Thr186 and the C-terminal sites occurs in cis.
Discussion
The most remarkable observation from the CDK9/CycT1 structure is the rotation of the cyclin relative to the CDK compared with the cell cycle CDK/cyclin complexes. Relative rotation between cognate pairs within large protein families has proven to be an effective mechanism for evolving specificity, as demonstrated by competition between bacterial colicins and their immunity proteins (Kuhlmann et al, 2000). Several of the core contacts in CDK9/CycT1 and CDK2/CycA are conserved, but of the extensive contacts from the cyclin motif at the end of H3 KΦEEΦ (where Φ is a hydrophobic residue), which in CDK2/CycA and other cell cycle CDK/cyclin complexes form a link to the CDK activatory phospho-threonine, only that from the lysine is made in the CDK9/CycT1. The N-terminal helix HN is required for the cell cycle CDK/cyclin complexes to form stable active complexes with cognate CDKs. In the transcriptional CDK9/CycT1 complex, HN adopts an orientation that is almost 180° opposite to that of the cell cycle cyclins HN and is only superficially involved in recognition of the CDK. Alternative orientations of the N-terminal helices are also observed in the structures of the other transcriptional cyclins, CycC and CycH, suggesting that their interactions with their CDKs may be similar to the CDK9/CycT1 structure. Transcriptional CDK/cyclin complexes are generally part of multi-molecular complexes. CDK9/CycT complexes are associated with inhibitors (HEXIM1 and 7SK RNA) or activators (Brd4) and also larger complexes (Jeronimo et al, 2007). These multi-molecular assemblies may assist in promoting stability, regulation and recruitment with a pliable transcriptional CDK/cyclin complex.
The conformation of the activation segment of CDK9 generates the specificity for a Ser/Thr-Pro motif present in the major substrates of P-TEFb, namely the CTD heptad repeat of RNA Pol II and the C-terminal repeats of SPT5 subunit of DSIF. The structure of pCDK9/CycT1 indicates that the activating role of pThr186 is likely to be the localization of the activation segment in its correct position for substrate recognition.
The present study has confirmed Thr186 as an autophosphorylation site and a site whose phosphorylation promotes kinase activity, in agreement with the results of Li et al (2005). We have also shown that autophosphorylation occurs in cis. Cell cycle CDK/cyclin complexes are activated by CAK (CDK7/CycH/Mat1) and are not able to autophosphorylate. CAK does not phosphorylate CDK9 (Kim and Sharp, 2001), nor has any activatory CDK9 kinase been identified. The sequence around Thr186 (184YTNRVVT) has none of the hallmarks of a consensus CDK Ser-Pro phosphorylation site. The predicted more open conformation and the additional length of the activation segment in CDK9 may be the key features that make Thr186 more readily available for autophosphorylation in cis. The other sites of CDK9 autophosphorylation, Ser347 (346GSQITQQ) and Thr362 or Thr363 (361ATTNQTE), also lack a consensus sequence motif. The results with the kinase-dead mutants show that these sites are only autophosphorylated in cis, suggesting that additional recognition features within CDK9/CycT1 coupled with localization within the active enzyme are needed to enable them to become substrate sites for autophosphorylation.
Even though the C-terminal CDK9 phosphorylation does not affect CDK9 kinase activity in in vitro kinase assays, these phosphorylation sites may become relevant for the regulation of CDK9 activity in vivo. Garber et al (2000) had previously shown that CDK9 C-terminal phosphorylation is required for high-affinity binding of Tat-CDK9/CycT1 to TAR RNA. Additionally, CDK9 phosphorylation is required for CDK9/CycT association with the regulatory molecules HEXIM1/7SK RNA (Li et al, 2005). Garber et al found that the sequence around Ser347 is a PKA consensus sequence (343RRKGS347QI) and showed that the promotion of C-terminal phosphorylated CDK9/CycT1 binding to HIV TAR could be reproduced by phosphorylation of Ser347 by PKA. Thus, in vivo Ser347 may be an autophosphorylation site or a PKA site. However, CDK9 purified from insect cells is not phosphorylated on its C terminus by the insect cell PKA.
Flavopiridol shows its strongest CDK inhibitory effects with CDK9 and this is reflected in the downregulation of RNA synthesis after flavopiridol treatment of cells (Lam et al, 2001). The regulation of gene expression is exploited in cancer treatment, for example, in gastrointestinal stromal tumour cells, where flavopiridol mediates downregulation of c-Kit and antiapoptotic proteins and thereby renders the cells sensitive to apoptosis (Sambol et al, 2006). The activity of flavopiridol in CLL has also been attributed to the resulting reduction of levels of antiapoptotic proteins (reviewed in Shapiro, 2006). These results provide a rationale for combination of flavopiridol as a transcriptional inhibitor together with other agents in cancer treatment. The crystallographic structure reveals the molecular basis for CDK9 inhibition by flavopiridol. Although the mode of binding to the active site of CDK9 was expected from structural studies of inactive CDK2/deschloro-flavopiridol complex, the conformational changes in the G-loop and the β3/αC loop of active CDK9 could not have been predicted. It seems likely that the greater potency against CDK9 arises from the ability of flavopiridol to induce conformational changes, which lock CDK9 in a conformation that cannot bind ATP. In Abl kinase in complex with imatinib-related compounds, the aromatic residue (Tyr253) from the G-loop has a similar function in selectivity and affinity (Cowan-Jacob et al, 2007).
Flavopiridol targets other CDKs. The inhibitor and ATP complex structures we present here show that the ‘Lys89 pocket' in CDK2 is more open in CDK9. This and the G-loop dynamics might provide a starting point for the rational development of flavopiridol analogues, which maintain this potency but have increased selectivity against other kinases or reduced binding to human serum albumin. Such a specific CDK9 inhibitor would maintain inhibition of transcriptional elongation while minimizing potential side effects. In addition, a specific CDK9 inhibitor could find application as therapy for HIV infection (Salerno et al, 2007).
Materials and methods
Expression, purification and crystallization
CycT2 (residues 1–273) was expressed in Escherichia coli cells and purified on a Ni-Sepharose column, followed by cleavage of the hexa-His tag, and further purification on a size exclusion column. Crystals were obtained using 0.2 M sodium formate, 0.1 M BisTrisPropane, 20% PEG 3350 and 10% EtGly as the precipitant. Data were collected at the Swiss Light Source X10 beam line to 1.8-Å resolution for native crystals and the structure was solved by SeMet SAD phasing with data collected at 0.979-Å wavelength (Table I).
Table 1.
Crystallographic parameters for CDK9/cyclin T1 and cyclin T2 structures
| Cyclin T2 | CDK9/CycT1 | CDK9/CycT1/AMPPNP | CDK9/CycT1/ flavopiridol | ||
|---|---|---|---|---|---|
| Native | SeMet | ||||
| Data collection | |||||
| Beam line | SLS X10 | SLS X10 | ESRF ID-29 | ESRF ID-14–2 | ESRF ID-14–4 |
| Space group and unit cell | P1 a=43.03 Å, b=58.64 Å, c=65.63 Å, α=110.6°, β=102.4°, γ=90.1° | P1 a=42.92 Å, b=58.75 Å, c=65.32 Å, α=110.8°, β=102.3°, γ=90.4° | H3 a=b=172.92 Å, c=95.76 Å | H3 a=b=173.13 Å, c=96.64 Å | H3 a=b=173.92 Å, c=97.17 Å |
| Resolution (highest resolution shell) (Å) | 59.76–1.80 (1.847–1.80) | 59.76–12.50 (2.60–2.50) | 48.7–2.48 (2.61–2.48) | 32.7–2.9 (3.06–2.9) | 38.4–2.80 (2.95–2.8) |
| Total observations | 193 211 | 194 590 | 103 359 | 123 733 | 154 163 |
| Unique | 53 530 | 91 090 | 37 491 | 23 641 | 26 960 |
| Rmerge | 0.11 | 0.11 | 0.063 (0.472) | 0.085 (0.512) | 0.093 (0.585) |
| Multiplicity | 3.57 | 4.48 | 2.8 (2.8) | 5.2 (5.3) | 5.7 (5.8) |
| Mean I/σI | 14.31 (5.36) | 14.43 (3.93) | 12.0 (2.0) | 16.7 (2.4) | 10.8 (3.0) |
| Completeness | 99.08 (90.07) | 99.1 (100) | 99.8 (100) | 99.8 (100) | |
| Refinement | |||||
| Total number of atoms | 4322 | 4603 | 4527 | 4553 | |
| Number of water molecules | 271 | 139 | 30 | 20 | |
| R (highest resolution shell) | 0.191 (0.254) | 0.179 (0.299) | 0.174 (0.252) | 0.176 (0.243) | |
| Rfree (highest resolution shell) | 0.223 (0.334) | 0.221 (0.364) | 0.234 (0.309) | 0.228 (0.286) | |
| r.m.s. bonds (Å) | 0.011 | 0.005 | 0.006 | 0.004 | |
| r.m.s. angles (deg) | 1.131 | 0.646 | 0.745 | 0.666 | |
The pFast-BAC dicistronic vector containing full-length CDK9 and CycT1 (kindly provided by Dr Simona Rizzo, Nerviano Medical Science) was used as template for PCR reactions. CDK9 and CycT1 constructs were cloned separately into the vector pVL1393 (Pharmingen BD) modified with the insertion of MBP or GST fusions and 3C protease cleavage sites. After transfection of Sf21 or Sf9 insect cells and amplification, expression was obtained by co-infecting insect cells with GST–CycT1 baculovirus (MOI 0.3) and MBP-CDK9 baculovirus (MOI 0.5) at 27°C for 3 days. The CDK9/CycT1 complex was purified using a GSH-Sepharose column, cleavage of GST and MBP fusions and further purification by size exclusion chromatography. Over 25 different constructs were evaluated. Diffracting crystals were obtained from the constructs CDK9 (residues 1–330)/CycT1 (residues 1–259) using 4.5 mg/ml CDK9/CycT1 with the precipitant solution 100 mM Tris, pH 8.5, 20% PEG 1000, 500 mM NaCl and 4 mM TCEP at 4°C. Mass spectrometry showed that CDK9 was mono-phosphorylated on Thr186 (approximately 85%). The best diffracting crystals were obtained after in vitro autophosphorylation by incubating with ATP to achieve 100% phosphorylation on Thr186 (Supplementary Figure S6). AMPPNP crystals were obtained in similar conditions by co-crystallizing CDK9/CycT1 in the presence of 2 mM AMPPNP and 2.4 mM MgCl2. The complex with flavopiridol was obtained by co-crystallization with 100 μM flavopiridol in 100 mM NaK-phosphate pH 6.2, 20% PEG 1000, 200 mM NaCl and 4 mM TCEP. Data were collected at ESRF beam lines (Table I) and the CDK9/CycT1 structure was solved by molecular replacement using as independent search objects a truncated CDK2 model and the structure of free CycT1 (PDB: 2PK2), derived from a fusion complex with EIAV Tat. The CycT1 structure had been solved (Anand et al, 2007) by molecular replacement from the CycT2 structure (2IVX) reported here.
Kinase assay
CDK9 (residues 1–372)/CycT1-298 (residues 1–298) activity was measured by following the incorporation of radiolabelled phosphate into substrate GST–CTD. Substrate (5 μg) was incubated with 50 ng of CDK9-FL/CycT1-298 in 10 μl kinase buffer (0.1 mM ATP, 10 mM MgCl2, 50 mM Tris/HCl pH 7.5, 1 μCi γ-32P-labelled ATP; MP Biomedicals). The reaction mixture was incubated for 5 min at 20°C and terminated by the addition of SDS sample buffer. CDK9 autophosphorylation was measured by incubating inactive CDK9-D167N/CycT1-288 and CDK9(1–330)-D167N/CycT1-288 (0.6 μg) with 50 ng of the respective active CDK9 complex in kinase buffer as above. The reaction mixture was incubated for 15 min at 30°C and terminated by the addition of SDS sample buffer. Samples were analysed on 12% SDS–PAGE gels subjected to autoradiography.
Limited autophosphorylated CDK9-FL/CycT1-298 samples were obtained by incubating each complex in 50 mM Tris/HCl pH 7.5, 0.1 mM ATP and 10 mM MgCl2 at 20°C for 30 min and analysed by mass spectrometry. Prephosphorylated samples were prepared in the same way and residual ATP/ADP was subsequently removed by desalting spin columns (Pierce) before kinase assays were performed as described above.
Accession numbers
Coordinates have been deposited in the Protein Data bank for CycT2 (2IVX), CDK9/CycT1 (3BLH), CDK9/CycT1/AMPPNP (3BLQ) and CDK9/CycT1/flavopiridol (3BLR).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Data
Acknowledgments
We thank the beam line scientists on stations ID 29, ID 14-4 and ID 14-2 at ESRF and those on station X10 at SLS for their help with synchrotron data collection. This study has been funded by the Medical Research Council and the Wellcome Trust. SB is supported by a Swiss National Foundation Fellowship. GL was supported by a BBSRC CASE award and by a FEBS Fellowship. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from a number of organizations (Supplementary data).
References
- Anand K, Schulte A, Fujinaga K, Scheffzek K, Geyer M (2007) Cyclin box structure of the P-TEFb subunit cyclin T1 derived from a fusion complex with EIAV tat. J Mol Biol 307: 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR (1999) Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci USA 96: 7791–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Lowe ED, Petri E, Skamnaki V, Antrobus R, Johnson LN (2007) Cyclin B and cyclin A confer different substrate recognition properties on CDK2. Cell Cycle 6: 1350–1359 [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN (1995) The crystal structure of cyclin A. Structure 3: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN (1999) The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol 1: 438–443 [DOI] [PubMed] [Google Scholar]
- Byrd JC, Lin TS, Dalton JT, Wu D, Phelps MA, Fischer B, Moran M, Blum KA, Rovin B, Brooker-McEldowney M, Broering S, Schaaf LJ, Johnson AJ, Lucas DM, Heerema NA, Lozanski G, Young DC, Suarez JR, Colevas AD, Grever MR (2007) Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood 109: 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH (2000) Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem 275: 28345–28348 [DOI] [PubMed] [Google Scholar]
- Chen R, Yang Z, Zhou Q (2004) Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem 279: 4153–4160 [DOI] [PubMed] [Google Scholar]
- Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, Shen K, Cole PA, Siligardi G, Johnson LN (2006) The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem 281: 23167–23179 [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, Rheinberger P, Centeleghe M, Fabbro D, Manley PW (2007) Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr 63: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Azevedo WF Jr, Mueller-Dieckmann HJ, Schulze-Gahmen U, Worland PJ, Sausville E, Kim SH (1996) Structural basis for specificity and potency of a flavonoid inhibitor of human CDK2, a cell cycle kinase. Proc Natl Acad Sci USA 93: 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM (1999) Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA 96: 1285–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA (2000) CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P–TEFb complex to TAR RNA. Mol Cell Biol 20: 6958–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA (1998) The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 12: 3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz F, Morris MC, Fesquet D, Cavadore JC, Doree M, Divita G (1997) Interactions of cyclins with cyclin-dependent kinases: a common interactive mechanism. Biochemistry 36: 4995–5003 [DOI] [PubMed] [Google Scholar]
- Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, Johnson LN (2005) The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J 24: 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Ferrin-O'Connell I, Zhang W, Leonard GA, O'Shea EK, Quiocho FA (2007) Structure of the Pho85-Pho80 CDK–cyclin complex of the phosphate-responsive signal transduction pathway. Mol Cell 28: 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP (1995) Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature 376: 313–320 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Sharp PA (2001) Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem 276: 12317–12323 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T (1992) Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell 3: 1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C (2000) Specificity in protein–protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol 301: 1163–1178 [DOI] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, Wahl LM, Harris ED, Mikovits JA, Monks AP, Hollingshead MG, Sausville EA, Staudt LM (2001) Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2, Research0041.0041–0041.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH (2005) Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem 280: 28819–28826 [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285: 2177–2198 [DOI] [PubMed] [Google Scholar]
- Lolli G, Lowe ED, Brown NR, Johnson LN (2004) The crystal structure of human CDK7 and its protein recognition properties. Structure 12: 2067–2079 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH (1996) Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O (2003) MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol 23: 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G, Majello B, Lania L (2003) Catalytic activity of Cdk9 is required for nuclear co-localization of the Cdk9/cyclin T1 (P-TEFb) complex. J Cell Physiol 197: 1–7 [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O (2001) 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325 [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 12: 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH (2006) Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP (1996a) Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A–Cdk2 complex. Nature 382: 325–331 [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP (1996b) Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol 3: 696–700 [DOI] [PubMed] [Google Scholar]
- Salerno D, Hasham MG, Marshall R, Garriga J, Tsygankov AY, Grana X (2007) Direct inhibition of CDK9 blocks HIV-1 replication without preventing T-cell activation in primary human peripheral blood lymphocytes. Gene 405: 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambol EB, Ambrosini G, Geha RC, Kennealey PT, Decarolis P, O'Connor R, Wu YV, Motwani M, Chen JH, Schwartz GK, Singer S (2006) Flavopiridol targets c-KIT transcription and induces apoptosis in gastrointestinal stromal tumour cells. Cancer Res 66: 5858–5866 [DOI] [PubMed] [Google Scholar]
- Shapiro GI (2006) Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 24: 1770–1783 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Tarricone C, Dhavan R, Peng J, Areces LB, Tsai LH, Musacchio A (2001) Structure and regulation of the CDK5–p25(nck5a) complex. Mol Cell 8: 657–669 [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fisher WH, Jones KA (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–237 [DOI] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q (2008) Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q (2001) The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322 [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q (2003) Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell 12: 971–982 [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125: 1137–1149 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH (2006) The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Data