Abstract
Walleye dermal sarcoma virus is a complex retrovirus that is associated with walleye dermal sarcomas that are seasonal in nature. Fall developing tumors contain low levels of spliced accessory gene transcripts A and B, suggesting a role for the encoded proteins, Orf A and Orf B, in oncogenesis. In explanted tumor cells the 35 kDa Orf B accessory protein is localized to the cell periphery in structures similar to focal adhesions and along actin stress fibers. Similar localization was observed in mammalian cells. The cellular protein, receptor for activated C kinase 1 (RACK1), bound Orf B in yeast two-hybrid assays and in cell culture. Sequence analysis of walleye RACK1 demonstrated high conservation to other known RACK1 sequences. RACK1 binds to activated protein kinase C (PKC). Orf B associates with PKCα, which is constitutively activated and localized at the membrane. Activated PKC promoted cell survival, proliferation, and increased cell viability in Orf B-expressing cells.
Keywords: oncogenesis, retrovirus, walleye dermal sarcoma virus, RACK1, PKC
Introduction
Dermal sarcoma in walleye (Sander vitreus) is etiologically associated with the complex retrovirus, walleye dermal sarcoma virus (WDSV) (Martineau et al., 1992; Martineau et al., 1991; Walker, 1969; Yamamoto, Kelly, and Nielsen, 1985; Yamamoto et al., 1976). Walleye dermal sarcoma (WDS) is exceptional in its seasonal nature. Tumors first appear on fish in the fall and are present throughout the winter. During the spawning period, the following spring, the tumors naturally regress (Bowser et al., 1988; Bowser and Wooster, 1991). WDS is efficiently transmitted experimentally to walleye fingerlings by topical, oral, or intramuscular administration of filtrates prepared from regressing tumors. In contrast, filtrates from developing tumors are unable to transmit disease (Bowser, Martineau, and Wooster, 1990; Bowser et al., 1996; Martineau et al., 1990). Associated with the seasonal nature of disease are differences in viral gene expression (Bowser et al., 1996; Quackenbush et al., 1997). The developing fall tumors only express the spliced accessory gene transcripts, A and B, encoding orf a and orf b, while regressing spring tumors were found to express spliced transcripts, the full-length genomic RNA, and infectious virus (Bowser et al., 1996; Martineau et al., 1992; Quackenbush et al., 1997). The presence of only the A and B transcripts during tumor development led to the hypothesis that the Orf A and Orf B proteins mediate oncogenesis.
orf a encodes a retroviral cyclin (rv-cyclin or Orf A) protein, which localizes to the nucleus and interacts with proteins necessary for transcription (LaPierre, Casey, and Holzschu, 1998; Rovnak, Casey, and Quackenbush, 2001; Rovnak et al., 2005; Rovnak and Quackenbush, 2002; Rovnak and Quackenbush, 2006). Rv-cyclin functions to negatively regulate viral gene expression through the direct interaction of its transcription activation domain with TATA binding protein-associated factor 9 (TAF9) (Rovnak et al., 2005; Rovnak and Quackenbush, 2006). WDSV rv-cyclin can induce cell-cycle progression in cyclin deficient yeast and induce hyperplasia in transgenic mice after wound healing (Lairmore et al., 2000; LaPierre, Casey, and Holzschu, 1998).
orf b encodes a protein of 306 amino acids with a molecular mass of 35 kDa and has limited sequence homology to the rv-cyclin (LaPierre et al., 1999) but no homology with other, known proteins. WDSV Orf B was found to localize in the cytoplasm and at the plasma membrane in explanted tumor cells (Rovnak et al., 2007). Cellular proteins that interact with Orf B were identified in a yeast 2-hybrid assay, one of which is the receptor for activated C kinase (RACK1)(see below).
RACK1 is a 36 kDa protein (Ron et al., 1994) composed of seven WD-repeats, domains first identified in the β subunit of the heterotrimeric G proteins that are important in protein-protein interactions (Fong et al., 1987; Neer et al., 1994). RACK1 binds to activated conventional isoforms of protein kinase C (PKC) and functions as an anchoring protein to stabilize PKC at the membrane in an active conformation (Dorn and Mochly-Rosen, 2002; Ron et al., 1994).
PKC comprises a large family of serine-threonine isoenzymes that contain a regulatory domain and a kinase core. PKCs are classified into three groups based on the domain composition of the regulatory component: conventional (α, βI, βII, and γ), novel (δ, ε, η, and θ), and atypical (ζ and ι/λ) (Newton, 2003). PKCs are involved in a broad array of biological functions such as cell proliferation, differentiation, survival and apoptosis (Nakashima, 2002; Nishizuka, 1988; Nishizuka, 1995). All PKC isoforms associate with phospholipids in the membrane, and each group has additional cofactor requirements for full activation. The conventional PKCs are calcium-dependent (Ca+2) and require binding to diacylglycerol (DAG) for activation (Newton, 2003; Nishizuka, 1986). Novel PKCs are Ca+2-independent but require DAG for activation, and the atypical isoforms only require phosphatidylserine for activation (Newton, 2003; Nishizuka, 1984).
In this study we demonstrate a direct interaction of Orf B with RACK1 and constitutive activation of PKCα in Orf B-expressing cells. Further, activation of the PKC signaling pathway is responsible for the ability of Orf B-expressing cells to survive and proliferate under serum deprived conditions.
RESULTS
Expression of WDSV Orf B in explanted tumor cells and cell lines
Cells from a regressing tumor were established in vitro after explanting tumor fragments in culture dishes (Rovnak et al., 2007). The adherent cells, which include cells of mixed lineages, were transferred to glass microscope slides, and expression of WDSV Orf B was evaluated by an immunofluoresence assay using rabbit anti-Orf B serum. Orf B localized to the plasma membrane in structures consistent with focal adhesions and lamellapodia, and along actin stress fibers (Fig. 1, upper panels). Stable expression of Orf B was established in NIH3T3 cells, Chinese hamster ovary cells (CHO), and canine fibroblast cells (Cf2Th) by transfection with an Orf B expression construct (pKH3-Orf B) and selection for neomycin resistance. Orf B expression in the cell lines is localized to the cytoplasm and associated with the membrane and stress fibers, similar to that observed in the explanted tumor cells (Fig. 1, NIH3T3-Orf B cells-lower panel and data not shown). Orf B is also present in the nucleus of these cells.
Fig. 1.
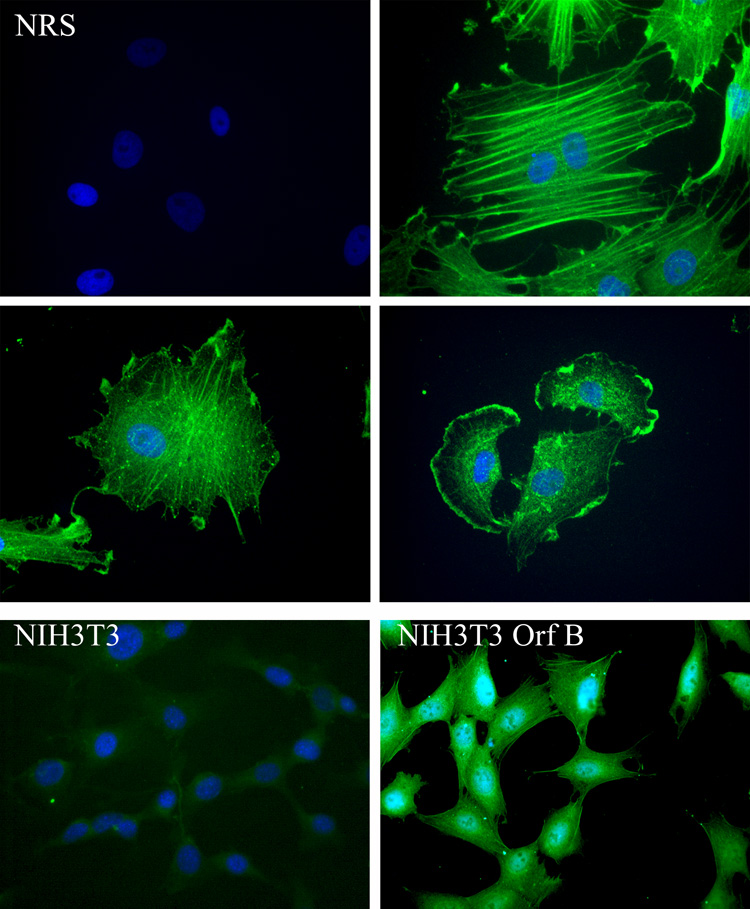
Expression of WDSV Orf B in explanted tumor cells and NIH3T3 cells. (Upper panels) Explanted spring tumor cells were labeled consecutively with rabbit anti-Orf B sera and FITC-conjugated goat anti-rabbit IgG (green). 400 X magnification. (Lower panel) NIH3T3 (left) and NIH3T3-Orf B (right) cells were labeled with anti-HA mAb (HA.11) and FITC-conjugated anti-mouse IgG (green). 400 X magnification. Nuclei were stained with DAPI (blue).
WDSV Orf B interacts with RACK1
A yeast two-hybrid assay was used to identify cellular proteins that interact with WDSV Orf B. WDSV Orf B was fused to the GAL4 DNA binding domain (GAL4 DBD) and used as the bait for screening a human cDNA library. The sequence of the walleye genome is not known, so a human cDNA library was used to facilitate identification of Orf B interacting proteins. Saccharomyces cerevisiae strain Y190, harboring the HIS3 and lacZ reporter genes under the control of the GAL1 UAS, were co-transformed with a HeLa cDNA library and the GAL4 DBD-Orf B construct. Positive HIS3 transformants were screened for β-galactosidase activity by colony-lift filter assay. Sequence analysis of DNA isolated from ten β-galactosidase positive colonies revealed that two clones contained partial cDNAs of the human RACK1 gene encoding amino acids 87–317 or 139–317, limiting potential Orf B interaction to the last 178 amino acids of RACK1. This region of RACK1 encompasses WD repeats 5 through 7.
To verify the interaction of WDSV Orf B with RACK1 in vertebrate cells, immunoprecipitation (IP) assays were used. Cell lysates were subjected to IP with mouse anti-RACK1 antibody or normal mouse serum (NMS) followed by western blot analysis with anti-HA antibody (12CA5) to detect expressed, HA-tagged Orf B. Orf B was co-immunoprecipitated (coIP) with RACK1 from cells that stably express WDSV Orf B (NIH3T3-Orf B) (Fig. 2). In addition, Orf B was coIPed with RACK1 from lysates of Cf2Th and CHO cells that stably express Orf B and HeLa cells transiently transfected with an Orf B expression construct, pKH3-Orf B (data not shown).
Fig. 2.
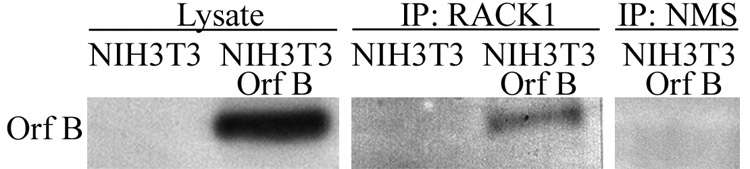
WDSV Orf B interacts with RACK1. Lysates from NIH3T3 cells and 3T3 cells stably expressing Orf B (NIH3T3-Orf B) were immunoprecipitated with mouse anti-RACK1 antibody or normal mouse serum (NMS). HA-tagged Orf B was detected with anti-HA antibody (12CA5). Lysate represents 4% of the total amount used for immunoprecipitation.
The walleye orthologue of human RACK1 was cloned to confirm its interaction with Orf B. RACK1 is highly conserved across species (Chou et al., 1999; Kwak et al., 1997). Therefore, degenerate polymerase chain reaction (PCR) primers, based on highly conserved regions of human, mouse, fugu, and zebrafish RACK1, were used in a reverse transcriptase polymerase chain reaction (RT-PCR) reaction followed by 5’ rapid amplification of cDNA ends (5' RACE) to amplify walleye RACK1. Walleye RACK1 was found to be 317 amino acids in length and predicted to encode a 36 kDa protein. The amino acid sequence of walleye RACK1 is 97% identical to zebrafish and 96% identical to mouse and human RACK1 (Fig. 3). The seven conserved WD repeats were identified in walleye RACK1 by amino acid alignment with human, mouse, and zebrafish RACK1 (Fig. 3). The WD repeats in RACK1 are predicted to form a seven-bladed propeller structure similar to that described for the β subunit of G proteins (Garcia-Higuera et al., 1996). The WD repeats serve as docking sites for binding of multiple proteins (Ron and Mochly-Rosen, 1994; Schechtman and Mochly-Rosen, 2001).
Fig. 3.
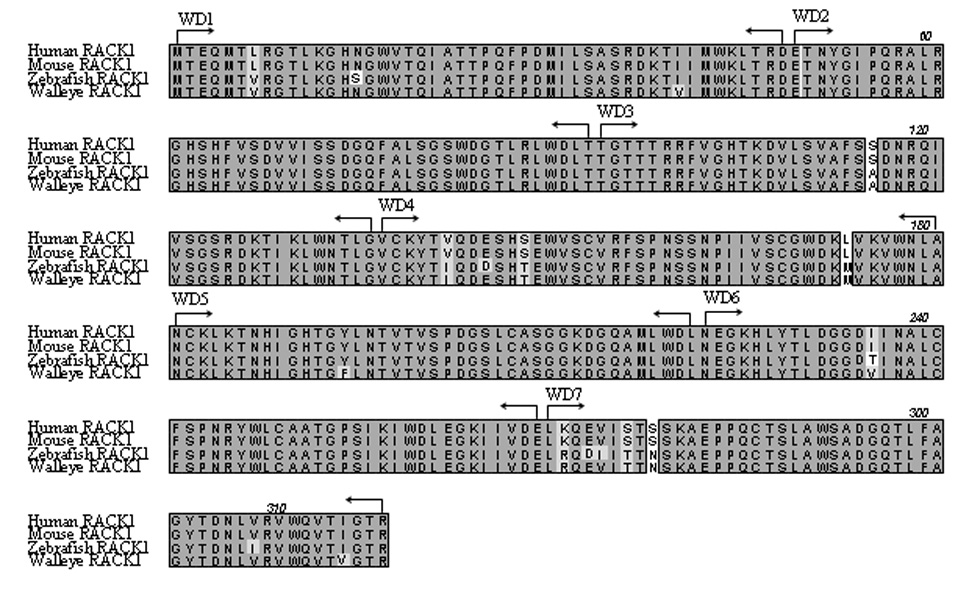
Amino acid sequence alignment of walleye RACK1 with human, mouse, and zebrafish RACK1. Identical residues are darkly shaded, similar residues are lightly shaded, and non-identical residues are unshaded. Positions of the seven WD repeats (WD1-7) are indicated with arrows.
coIP assays were used to test for the interaction of walleye RACK1 with WDSV Orf B. Cell lysates from HeLa cells co-transfected with FLAG-tagged walleye RACK1 (pFLAG-wRACK1) and HA-tagged Orf B (pKH3-Orf B) expression constructs or empty vectors were incubated with anti-HA (12CA5) or anti-FLAG antibodies and protein G sepharose. IP protein complexes were separated by SDS-PAGE and western blots probed with antibodies to HA to detect Orf B or to FLAG to detect walleye RACK1. Results in Fig. 4A demonstrate coIP of Orf B and walleye RACK1 from HeLa cell lysates. Further, an interaction of Orf B with walleye RACK1 was confirmed in walleye fibroblast cells (W12). Cell lysates from W12 cells transiently transfected with pKH3-Orf B or the pKH3 empty vector were subjected to IP with an anti-RACK1 monoclonal antibody. Western blot analysis demonstrates coIP of HA-tagged Orf B with endogenous walleye RACK1 (Fig. 4B). Together, these data show that WDSV Orf B interacts with RACK1 in these cells.
Fig. 4.
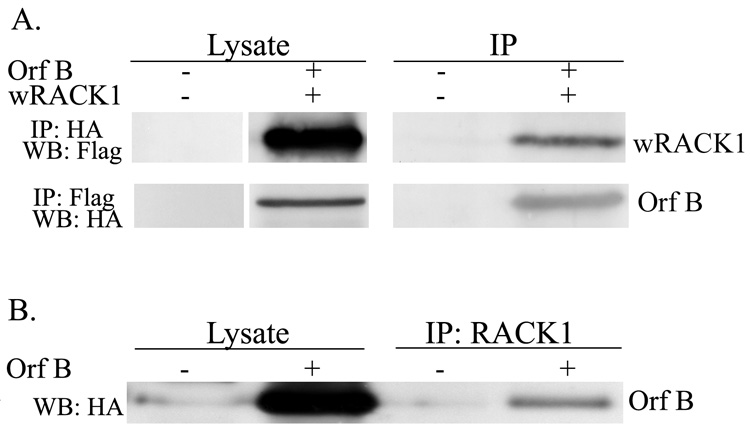
A. Co-immunoprecipitation of Orf B and walleye RACK1 from HeLa cell lysates. Lysates from HeLa cells co-transfected with HA-tagged Orf-B (pKH3-Orf B) and FLAG-tagged walleye RACK1 (pFLAG-wRACK1) or empty vectors pKH3 and pFLAG were immunoprecipitated with anti-HA or anti-FLAG antibodies. HA-tagged Orf B was detected with anti-HA MAb (12CA5) and walleye RACK1 was detected with anti-FLAG MAb (M2). HA-tagged Orf B and FLAG-tagged walleye RACK1 run at approximately 39 kDa and 37 kDa, respectively. Lysates represent 4% of the total amount used for immunoprecipitation. B. Immunoprecipitation of Orf B with endogenous walleye RACK1. Lysates from walleye cells (W12) transfected with pKH3-Orf B or pKH3 empty vector were immunoprecipitated with MAb to human RACK1. Orf B was detected with anti-HA (12CA5).
WDSV Orf B associates with PKCα
Based on the known interaction of RACK1 with conventional PKC isoforms and the data above demonstrating binding of RACK1 to WDSV Orf B, we investigated the possibility that Orf B may be in a complex with PKC in the NIH3T3-Orf B cell line. NIH3T3 mouse fibroblast cells only express PKC isozymes α, δ, ε, and ζ (Mischak et al., 1993; Olivier, 1992). Therefore, we tested possible interaction of Orf B with the conventional isoform, PKCα. Lysates from NIH3T3-Orf B and NIH3T3 control cells were subjected to IP with anti-PKCα antibody and Orf B was detected with anti-HA antibody by western blot analysis of the anti-PKCα IP complexes (Fig. 5). In contrast, antibody that recognizes one of the novel PKC isoforms, PKCδ was unable to coIP Orf B from these lysates (Fig. 5). CoIP of Orf B with another conventional isoform of PKC, PKCβII, has been demonstrated with lysates harvested from Orf B expressing HeLa and CHO cells (data not shown).
Fig. 5.
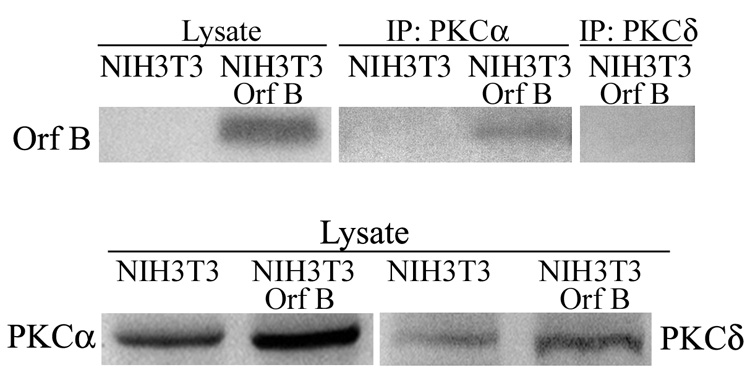
WDSV Orf B interacts with PKCα. Lysates from NIH3T3 cells stably expressing Orf B (NIH3T3-Orf B) and NIH3T3 control cells (NIH3T3) were immunoprecipitated with rabbit anti-PKCα antibody or with rabbit anti-PKCδ antibody. HA-tagged Orf B was detected with anti-HA (12CA5) MAb. Lysate represents 4% of total amount used for immunoprecipitation.
The PKC family of serine/threonine kinases functions to transduce signals that control cell proliferation, differentiation, motility, and apoptosis. Activation of PKCα protects cells from apoptosis (Nakashima, 2002; Ruvolo et al., 1998). The interaction of Orf B with RACK1 and PKCα suggested that PKC function might be altered. To evaluate whether Orf B expression affects PKC function in NIH3T3-Orf B cells, we first tested the ability of cells to survive after exposure to staurosporine, a known PKC inhibitor and inducer of apoptosis. Treatment of cells with concentrations of staurosporine that induce apoptosis significantly reduced the number of viable NIH3T3 cells compared with NIH3T3-Orf B cells (P< 0.004). Exposure to 25, 50, or 100 nM staurosporine for 16 hrs reduced NIH3T3 viability to 81% (mean ±8.22), 79% (±5.07), and 71% (±10.84) of untreated cells, respectively (Fig. 6). In contrast, the viability of staurosporine-treated NIH3T3-Orf B cells was not reduced relative to untreated cells (mean = 104% at 25 nM (±5.4), 102% at 50 nM (±15.3), and 100% at 100nM (±14.6)). Comparable results were obtained when Orf B expressing Cf2Th cells were treated with staurosporine (data not shown). These results suggest that Orf B associates with conventional PKC isoforms and may affect PKC signaling to promote cell survival.
Fig. 6.
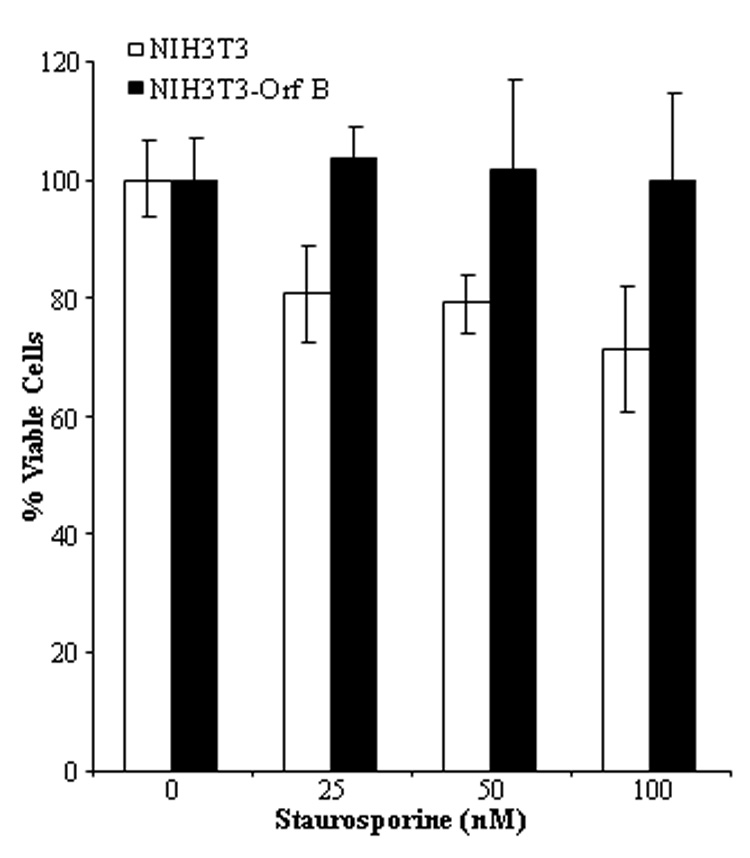
WDSV Orf B expressing cells remain viable after treatment with staurosporine. NIH3T3 and NIH3T3-Orf B cells were treated with 25, 50, or 100 nM staurosporine, and cell viability was measured by MTS assay after 16 hrs of incubation. The mean ± standard deviation of OD490 readings from replicates of six wells was determined and normalized to % viability. The data represent one of three independent experiments. Statistically significant differences (P< 0.004).
PKC is activated in WDSV Orf B expressing cells
Newly synthesized PKC associates with cellular membranes through weak interactions with phospholipids. PKC then undergoes phosphorylation at three sites rendering it mature and catalytically competent but not yet fully active. Phosphorylated PKC is released into the cytosol where it remains in an inactive conformation until elevated levels of Ca+2 and DAG are present, at which time PKC translocates to the membrane. High affinity interactions of PKC with the membrane result in the release of the PKC pseudosubstrate from the substrate binding site and the establishment of an active conformation of PKC (Newton, 2003). Interaction of RACK1 with activated PKC targets PKC to different subcellular sites close to PKC substrates and is thought to maintain PKC in its active conformation.
To determine whether Orf B affects activation of PKCα due to its interaction with RACK1 the activation status and localization of PKCα was investigated. We focused on the NIH3T3 fibroblast cell line for these studies, because, when cultured under serum-deprived conditions, PKC is not activated and the effect of overexpressed RACK1 on cell growth has been extensively investigated (Chang, Chiang, and Cartwright, 2001; Chang et al., 1998; Mamidipudi et al., 2004). Cell lysates were isolated from NIH3T3 and NIH3T3-Orf B cells that were serum deprived for 24 hrs and either treated with 200 nM phorbol myristic acid (PMA) in dimethyl sulfoxide (DMSO), a phorbol ester known to activate PKC, or with an equal volume of DMSO as control. Cytosolic and membrane fractions were separated by ultracentrifugation and subjected to western blot analysis with anti-phospho-PKC α/βII antibody specific for the autophosphorylated sites of PKCα (Thr 638) and PKCβII (Thr641). As expected, full activation and translocation of PKCα to the membrane fraction was only detected in serum-deprived NIH3T3 cells after PMA treatment (Fig. 7A). In contrast, activated PKCα is present in the membrane fractions from serum-deprived NIH3T3-Orf B cells treated with PMA or with DMSO alone (Fig. 7A). The same blot was probed with antibody against the membrane glycoprotein cadherin as a loading control. The blot was then reprobed for total PKCα. The majority of PKCα detected in the membrane fraction from serum-deprived NIH 3T3-Orf B cells appears to be phosphorylated whereas only unphosphorylated PKCα was detected in membranes of NIH3T3 cells. Equivalent levels of phosphorylated PKCα were detected in NIH3T3 and NIH3T3-Orf B cells treated with PMA (Fig. 7A), and phosphorylated PKCα was detected in cytosolic fractions isolated from NIH3T3 and NIH3T3-Orf B cells (Fig. 7B). Treatment of cells with PMA resulted in reduced levels of cytosolic, phosphorylated PKCα, suggesting that PKCα translocated to the membrane. Expression of Orf B and RACK1 in membrane and cytosolic fractions was confirmed by staining the lower portion of the blots with anti-HA and anti-RACK1 and with anti-β actin as a loading control (Fig. 7A and 7B).
Fig. 7.
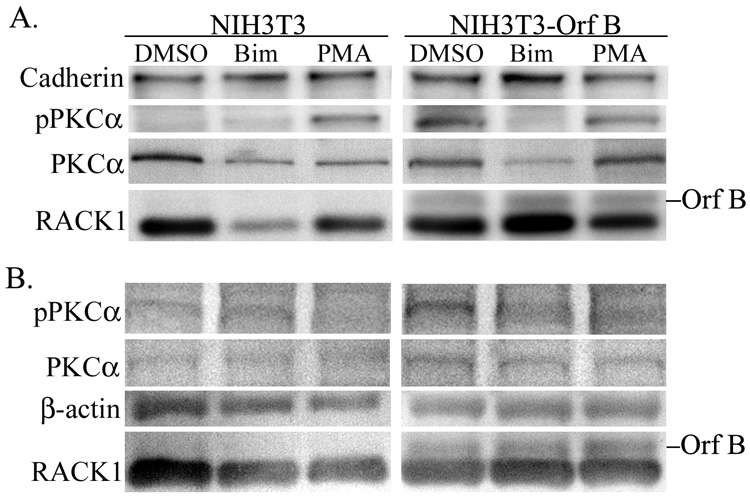
PKCα is constitutively activated in Orf B-expressing cells. NIH3T3 and NIH3T3-Orf B cells were grown without serum for 24 hours and then treated with dimethyl sulfoxide (DMSO), bisindolymaleimide I (Bim), or phorbol ester (PMA). Cell lysates were harvested, separated into membrane (A) and cytosolic (B) fractions by ultracentrifugation, and analyzed by western blot with the indicated antibodies; anti-pPKCα detects PKCα when phosphorylated at threonine 638, anti-PKCα detects total PKCα anti-RACK1 detects endogenous RACK1, and anti-HA (12CA5) detects HA-tagged Orf B. β-actin and cadherin serve as loading controls. The data presented in this figure is representative of four independent experiments.
Next, we assessed the effect of treatment with the PKC inhibitor bisindolymaleimide I hydrochloride (Bim) on PKCα activation in NIH3T3-Orf B cells. Bim binds to the catalytic site of PKC and functions as a competitive inhibitor of ATP (Toullec et al., 1991). Cells were serum deprived for 24 hrs and then treated with Bim for 2 hrs, and lysates collected and processed as above. Treatment with Bim resulted in greatly reduced levels of activated PKCα in membrane preparations from serum-deprived NIH3T3-Orf B cells (Fig. 7A). These data indicate that PKCα is constitutively activated in Orf B-expressing cells.
Activation of PKC in NIH3T3-Orf B cells promotes cell survival
PKC regulates several signaling pathways that mediate cell proliferation and survival. Since PKCα is constitutively activated in Orf B-expressing cells under serum-deprived conditions, we investigated whether activation of PKCα in NIH3T3-Orf B cells influences cell survival. After culture of NIH3T3-Orf B and NIH3T3 control cells for 24 hours in the presence of serum, cells were washed with phosphate buffered saline (PBS) and further cultured in media without serum. An MTS assay was used to measure cell growth. The MTS reagent was added to the culture each day and OD490 readings were taken after 3 hrs of further incubation. Total cell input at day 0, when serum was removed, was normalized to a value of 100% viability and adjusted accordingly each day thereafter for 7 days. The growth of NIH3T3 cells declined progressively through day 7 without serum (Fig. 8A). The phase contrast image presented in Fig. 8B illustrates substantial cell loss by day 3. In contrast, proliferation of NIH3T3-Orf B cells was observed through day 3, during which time 1.7 cell doublings occurred (Fig. 8A). A slight decrease in cell viability was observed at days 4 and 5, after which there was increased proliferation of NIH3T3-Orf B cells through day 7 without serum. NIH3T3-Orf B cell proliferation is shown in the phase contrast images at day 0 and 3 days after serum deprivation (Fig. 8B).
Fig. 8.

Orf B-expressing cells proliferate and survive in the absence of serum. A. NIH3T3 cells and NIH3T3-Orf B cells were grown under serum-free conditions and viable cells measured each day for 7 days by MTS assay. The mean of six replicate wells was determined for each time point. Viable cells at Day 0 were normalized to 100% and ODs from subsequent days were adjusted accordingly. The data presented in this figure is representative of three independent experiments. B. Phase contrast images of NIH3T3 and NIH3T3-Orf B cells illustrate cell morphology at day 0 and day 3 of culture without serum. Magnification was 200X.
To determine whether activated PKC is responsible for the proliferation and survival of NIH3T3-Orf B cells shown in Figure 8, cells were cultured in the presence of the PKC inhibitor, Bim. The growth of serum-deprived NIH3T3-Orf B cells was significantly diminished in the presence of Bim (Fig. 9A, open squares, P< 0.001). After 3 days of treatment there were 60% fewer NIH3T3-Orf B cells than in cultures without Bim. By day 7 the number of viable cells in the Bim treated cultures was reduced to input levels (mean = 90% ±7.9), whereas the cultures without Bim remained elevated (mean = 316% ±39). The decrease in cell growth is evident in the phase contrast images shown in Fig. 9B (compare middle panels vs. right panels). Addition of Bim to NIH3T3-Orf B cells cultured in the presence of serum had no effect on the ability of these cells to proliferate (Fig. 9A and B, left panels). Overall, these results support the conclusion that PKCα is activated in NIH3T3-Orf B cells and that this activation contributes to cell proliferation and survival.
Fig. 9.
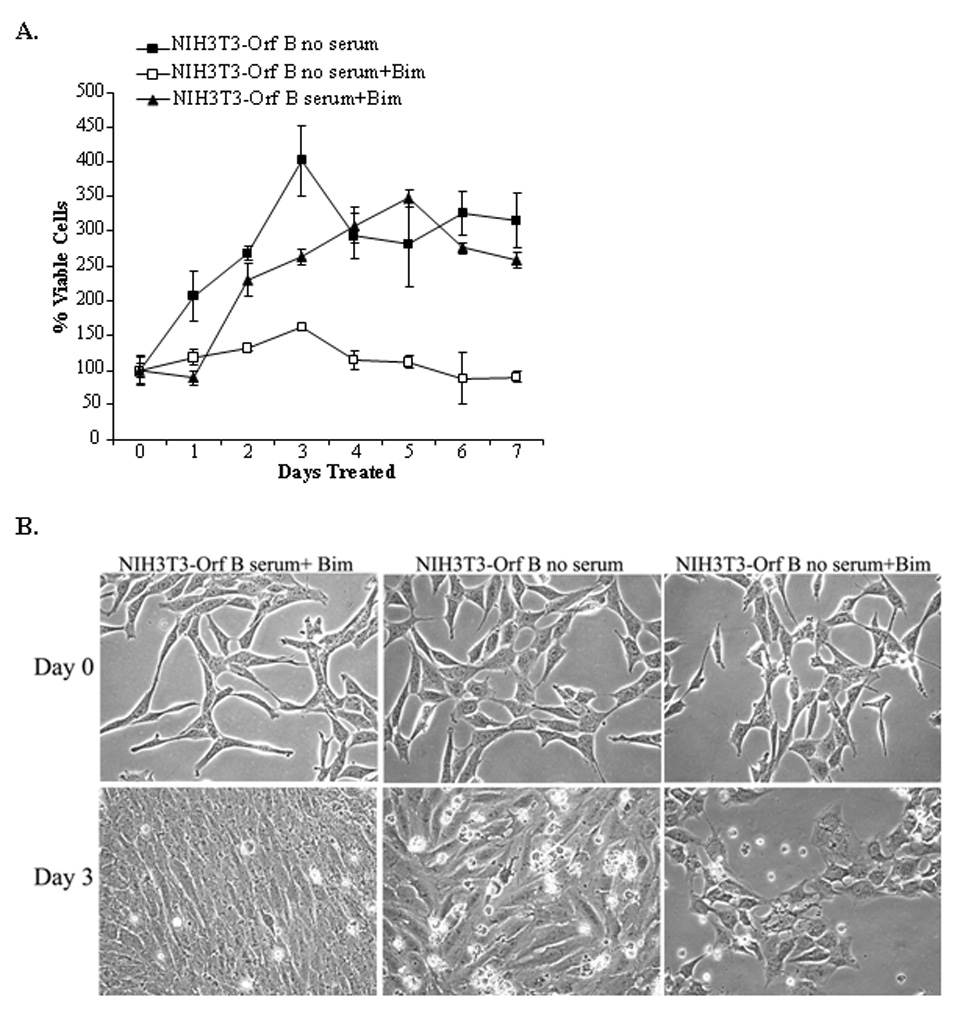
Activated PKC drives proliferation and survival of Orf B-expressing cells. A. NIH3T3-Orf B cells were grown without serum in the presence of 5 µM Bim in DMSO (NIH3T3-Orf B no serum+Bim) or DMSO only (NIH3T3-Orf B no serum). NIH3T3-Orf B cells were also cultured with serum and 5µM Bim (NIH3T3-Orf B serum+Bim). Viable cells were measured each day for 7 days by MTS assay. The mean of six replicate wells was determined for each time point. Viable cells at Day 0 were normalized to 100% viability and the ODs from subsequent days were adjusted accordingly. The data presented in this figure is representative of three independent experiments. B. Phase contrast images illustrate the morphology NIH3T3-Orf B cells cultured with serum and treated with 5 µM Bim (left), cells without serum and treated with DMSO (middle), and cells without serum and with 5 µM Bim (right) at day 0 and day 3. Magnification was 200X.
DISCUSSION
The results from these studies demonstrate the interaction of WDSV Orf B with RACK1 and PKCα. RACK1 was identified as a binding partner of WDSV Orf B by a yeast 2 hybrid screen and this interaction was confirmed by coIP assays. RACK1 is known to interact with over 20 cellular and viral proteins with varying functions (McCahill et al., 2002). Cloned walleye RACK1 exhibits 96% amino acid identity to human and mouse RACK1 and 97% identity to zebrafish RACK1, illustrating the highly conserved nature of this protein. The functional importance of RACK1 is exemplified by this conservation across eukaryotic species (McCahill et al., 2002). RACK1 was first identified as an anchoring protein for the conventional PKCs and activated PKC βII was the preferred binding partner of RACK1 (Csukai and Mochly-Rosen, 1999; Mochly-Rosen, Khaner, and Lopez, 1991; Mochly-Rosen et al., 1991; Mochly-Rosen et al., 1995; Ron et al., 1994; Stebbins and Mochly-Rosen, 2001). One of the novel PKC isoforms, PKCε, is known to interact with RACK2, also known as the coatomer protein β’COP (Csukai et al., 1997). The expression pattern for each PKC isoform varies among different cell types (Jaken, Leach, and Klauck, 1989; Mochly-Rosen, 1990). NIH3T3 cells express abundant levels of only one conventional PKC isoform, PKCα, and low levels of the novel forms, PKCδ and PKCε (McCaffrey et al., 1987; Mischak et al., 1993)
The interaction of Orf B with RACK1 suggested that PKC may be present in an Orf B/RACK1 complex, and coIP assays with lysates from NIH3T3-Orf B cells confirmed an interaction of Orf B with PKCα. Orf B was also coIPed with PKCβII from lysates isolated from cells that express this isoform. An association of Orf B and PKC is further suggested by the localization of Orf B in tumor cells at the plasma membrane and along actin stress fibers. Goodnight et al. (Goodnight et al., 1995) showed that PKCα is diffusely distributed in the cytoplasm in NIH3T3 cells and, upon TPA stimulation, is redistributed to the plasma membrane with marked localization to membrane ruffles and accumulation at focal adhesion contacts. RACK1 is also targeted to focal adhesions where it affects cell motility (Kiely et al., 2006; Vomastek et al., 2007). When over-expressed in NIH3T3 cells, PKCβII associates with actin-rich microfilaments (Goodnight et al., 1995). The localization of Orf B in explanted tumor cells and cultured cell lines exhibited all of these characteristics.
Still unresolved is the question of whether there is direct contact between Orf B and PKCα or whether coIP is dependent on the interaction of Orf B and RACK1. RACK1 holds PKC in an active conformation, and the structural nature of RACK1 indicates that more than one protein may bind to it concomitantly (McCahill et al., 2002). The presence of Orf B in a RACK1/PKC complex may aid in the formation of a more stable, active complex. Another possibility is that Orf B is a substrate for PKC and RACK1 targets PKC to subcellular locations in which Orf B resides. A ProSite Motif analysis (Hulo et al., 2007) identified three predicted PKC phosphorylation sites within Orf B. Binding of viral proteins to RACK1 may be a means to target interaction with PKC. The Epstein-Barr virus protein, ZEBRA, is a transcriptional activator that functions to disrupt latency. ZEBRA binds RACK1 and is phosphorylated by PKC at residue S186. However, this phosphorylation does not appear to be responsible for the disruption of latency (El-Guindy et al., 2002). PKC activity was inhibited in EBV-infected monocytes, likely due to impaired translocation of PKC from the cytosol to the plasma membrane (Tardif et al., 2002). The M1 protein from avian, swine, and human influenza A viruses interacts with RACK1 and is phosphorylated by PKC (Reinhardt and Wolff, 2000). HIV Nef also binds RACK1 and is phosphorylated by PKC, which results in enhanced viral replication (Gallina, Rossi, and Milanesi, 2001; Wolf et al., 2008). The phosphorylation status of Orf B will be the subject of future investigations.
After synthesis, PKCs associate with membranes and are targeted for phosphorylation at the activation loop by PDK-1, followed by autophosphorylation at the turn and hydrophobic motifs, and release into the cytosol in an inactive conformation. Extracellular stimuli elevate the levels of intracellular Ca+2 and DAG, which facilitates translocation of cytosolic PKC to the membrane and results in high affinity interactions (Newton, 2003). Activated PKC was only detected after stimulation of serum-deprived NIH3T3 cells with PMA. PMA binds PKC and recruits it to the membrane. Detection of phosphorylated PKCα in membranes of serum-deprived NIH3T3-Orf B cells, without PMA treatment, indicates that it is in a constitutively active conformation. Activated PKC functions to phosphorylate and activate substrate proteins that affect cell survival and growth.
Tethering of active PKC to the membrane allows binding to substrates, which affect downstream targets important in cell proliferation such as Raf-1 and the mitogen-activated protein kinase kinase-extracellular signal-regulated kinase (MEK-ERK) (Cai et al., 1997). In HL60 cells PKCα co-localizes with and phosphorylates Bcl-2 in mitochondria, resulting in increased cell survival following chemotherapy (Ruvolo et al., 1999). Constitutive expression of Orf B resulted in cell survival after treatment with staurosporine and proliferation and survival of cells cultured under serum-deprived conditions. The constitutive activation of PKCα in these cells suggests that it is responsible for Orf B’s effects. The reversal of these effects by treatment of serum deprived cells with Bim confirmed the requirement for the PKC signaling pathway in this process.
WDSV transcripts encoding the Orf B protein are present during the period of tumor development, and results from these studies suggest that Orf B is tumorigenic. Constitutive activation of PKCα in WDSV infected cells could lead to deregulated cell signaling pathways critical for both apoptosis and cell proliferation and resulting in tumor formation. The evidence presented here demonstrates alterations of cell proliferation and response to apoptotic stimuli, specifically subject to a PKC inhibitor, in cells expressing Orf B. Identification of the downstream targets activated in Orf B-expressing cells will provide valuable insight into the mechanism(s) of WDSV oncogenesis.
Materials and Methods
Cells and transfection
The WDSV orf B coding sequence was cloned into the pKH3 vector (a generous gift from Dr. Jun-Lin Guan, Cornell University) as previously described (Rovnak, Casey, and Quackenbush, 2001). The pKH3 vector contains a CMV promoter and fuses three influenza virus hemagglutinin (HA) tags onto the amimo terminus of the expressed protein (Chen et al., 1996; Mattingly et al., 1994).
NIH3T3 cells (ATCC CRL 1658), Cf2Th cells (ATCC CRL 1430), and HeLa cells (ATCC CCL 2) were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. CHO cells (CHO-K1, ATCC CCL 61) were maintained in Ham's F12 medium supplemented with 10% FBS. Cells were transfected using FuGENE6 (Roche) according to the manufacturer’s instructions. Stable cell lines were established by cotransfection of pKH3-Orf B and a plasmid encoding neomycin resistance pMC1neo (Stratagene). Cells were selected with G418 and single colonies were expanded and assayed for Orf B expression by western blot analysis.
Fluorescence microscopy
Cells cultured from a regressing dermal sarcoma and NIH3T3 cells were grown on glass slides for immunofluorescence assays (Cel-Line; Erie Scientific, Inc) (Rovnak et al., 2007). Slides were fixed in 2% buffered formalin for 15 min, rinsed in phosphate buffered saline (PBS), and fixed for 5 min in cold acetone:methanol (1:1 v/v). Fixed cells were incubated with a 1:100 dilution of affinity purified rabbit anti-Orf B sera (kindly provided by Volker Vogt, Department of Molecular Biology and Genetics, Cornell University) or 1:1000 dilution of mouse monoclonal anti-HA antibody (HA.11, Covance) for 1 hr at 37°C in a humidified chamber. Slides were washed in PBS and incubated with a 1:40 dilution of affinity purified goat anti-rabbit IgG or goat anti-mouse IgG conjugated to fluorescein isothiocyanate (FITC). After washing in PBS, slides were stained for 1 min in 300µM 4',6'-diamidino-2-phenylindole (DAPI). Photomicrographs were captured digitally at a magnification of 200X or 400X and images were assembled with Adobe Photoshop 7.0 software.
Yeast 2-hybrid screening
WDSV orfb was amplified by PCR from the plasmid clone pKH3-Orf B using 5’ and 3’ primers that incorporate EcoRI (5’GCCGAATTCATGTTTTCAGACTCAGATTCCT-3’) and BamHI (5’-GCCGGATCCTTACTCCGTAGGGCTGGGCTCT-3’) restriction sites, respectively. The amplified product was digested and ligated into the GAL4 DNA binding domain vector pAS2-1 and used as the bait to screen a pACT2 GAL4 AD fusion HeLa cDNA library (MATCHMAKER GAL4 Two-Hybrid System, Clontech). Saccharomyces cerevisiae strain Y190 was cotransformed with the bait plasmid, pAS2-1-Orf B and the pACT2 cDNA library. Positive clones were initially selected based on the ability to grow in the absence of histidine, leucine, and tryptophan in the presence of 25 mM 3-amino 1, 2, 4-triazole (3-AT). Positive colonies were further screened for β-galactosidase activity by a colony lift assay. Plasmid DNA was isolated from clones positive for β-galactosidase activity and sequenced. The specificity of the Orf B and RACK1 interaction was shown by transformation of yeast with (1) pAS2-1-Orf B and pACT2 plasmid, (2) pAS2-1 and pACT2RACK1 plasmid, which were negative for b-galactosidase activity. In addition, screening of the same cDNA library with another viral protein, WDSV rv-cyclin did not result in any interaction with RACK1 (Quackenbush, unpublished).
Cloning of walleye RACK1
RNA was isolated from a walleye fibroblast cell line (W12) with RNAzol (Tel-Test, Inc.) and cDNAs were prepared with oligo dT primer and Superscript II according to instructions of the manufacturer (Life Technologies). An internal walleye RACK1 (wRACK1) sequence was amplified from W12 cDNA by PCR using degenerate primers designed from highly conserved regions of zebrafish, pufferfish, human, and mouse RACK1 sequences (5’ primer-ATGACYGAGCARATGACMST; 3’ primer- CKNGTDCCRATRGTBCACCTG). Additional walleye RACK1 5′ sequences were amplified using a walleye specific RACK1 primer A (5′-AGTTGGCCAGATTCCACACCTTCAC-3′) and a primer designed from zebrafish RACK1 (5′-ATGACCGAGCAGATGACAGTAAGGG-3′). The remaining unknown 5’ walleye RACK1 sequence was amplified by RNA ligase-mediated and oligonucleotide capping rapid amplification of cDNA ends (RACE) using the 5’RACE primer and the wRACK1 specific primer B (5'-GTCTGGAAACTGCGGGGTCGTGGC-3') according to manufacturers instructions (GeneRacer Kit, Invitrogen). The 3’ end of wRACK1 was amplified with an oligo dT primer and wRACK1 specific primer C (5′-GATGGACAGGCCATGCTTTGGGAT-3′).
Walleye RACK1 was amplified by PCR from oligo dT primed W12 cDNA using walleye RACK1 specific primer D (5′-GCCGAATTCGATGACCGAGCAGATGACCGTGAGA-3′) and primer E (5′-CGGGGATCCTTATCGGGTTCCGACGGTCACCTG-3′) that incorporate EcoRI and BamHI restriction sites. The amplified product was digested with EcoRI and BamHI and ligated into p3XFLAG-CMV™-10 expression vector (Sigma).
Nucleotide sequence accession number
The final cDNA sequence for walleye RACK1 has been assigned GenBank accession number EU290652.
Immunoprecipitation and western blot analysis
NIH3T3 control cells and NIH3T3 cells stably expressing Orf B (NIH3T3-Orf B) were lysed with immunoprecipitation (IP) buffer (1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0], 1 mM EGTA, 0.2 mM sodium orthovanadate, 0.2 mM PMSF, 2 µg/ml of leupeptin and aprotinin, 1 µg/ml pepstatin). Lysates were centrifuged at 21,000 × g for 10 minutes and protein concentration of supernatants was determined with the Micro BCA Kit (Pierce). 500 µg of cell lysates were diluted to a concentration of 1 µg/ml in IP buffer and precleared for 3 hours with 50 µl of a suspension of Protein G-Sepharose (Pharmacia Biotech). One microgram of antibody was added to the precleared lysates and incubated overnight at 4°C with rotation. Fifty microliters of a Protein G suspension was added and incubated for an additional 3 hours at 4°C with rotation. Protein G pellets were washed three times with cold IP buffer, suspended in 20 µl of SDS-PAGE loading buffer, and heated to 70° for 10 minutes. Samples were separated under denaturing conditions in a 10% polyacrylamide gel. Cell lysates (15 µg of protein) were loaded in control lanes. Denatured proteins were transferred to Immobilon-P-membrane (Millipore) and incubated with primary antibody overnight at 4°C. Blots were washed, incubated with anti-rabbit IgG or anti-mouse IgM antibodies conjugated with horseradish peroxidase, and developed with LumiGLO Chemiluminsescent Substrate Kit (Kirkegaard and Perry Laboratories).
Membrane Protein Fractionation
NIH3T3 and NIH3T3-Orf B cells were plated at 7.5 × 105 cells/ml and grown in DMEM containing 10% FBS supplemented with 4 mM glutamine. Twenty four hours after plating, the growth medium was replaced with DMEM without serum for 24 hours. Cells were then treated with DMSO, for 45 minutes, 200 nM phorbol 12-myristate 13-acetate (PMA) for 45 minutes, or 5 µM Bisindolymaleide I (Bim) for 2 hrs and then cells were immediately placed on ice. Cells were scraped into protein lysis buffer lacking detergent (20 mM Tris [pH7.5], 2 mM EDTA, 2mM EGTA, 1mM PMSF, 2 ug/ml leupeptin, 2 ug/ml aprotinin, 0.1% 2-mercaptoethanol) then sonicated briefly as described in Goodnight et al. (Goodnight et al., 1995). Membrane and cytoplasmic fractions were separated by ultracentrifugation at 100,000 × g for 1 hour at 4°C. The supernatant (cytoplasmic fraction) was collected. The pellet (membrane fraction) was resuspended in protein lysis buffer containing 1.2% Triton X-100, sonicated briefly and centrifuged at 20,000 × g for 10 minutes at 4°C and the supernatant collected and analyzed as the membrane fraction. Protein concentrations were determined using the Micro BCA Kit (Pierce). Fifteen micrograms of cytoplasmic proteins were precipitated in 1/10 volume of trichloroacetic acid (Sigma) and pellets were dissolved in 100 mM NaOH.
Cell viability assays
NIH3T3 and NIH3T3-Orf B cells were plated at 5.0 × 105 cells/ml in 96 well plates and incubated for 24 hours in DMEM containing 10% FBS. Staurosporine was added to final concentrations of 25, 50, and 100 nM, incubated for 16 hours at 37°C, and cell viability was measured using CellTiter 96 AQueous One Solution (Promega) according to manufacturer's instructions. Briefly, 20 µl of CellTiter 96 solution was added to each well, incubated for 3 hours at 37°C, and then OD readings were taken at 490 nm.
NIH3T3 and NIH3T3-Orf B cells were plated at 5.0 × 105 cells/ml in 96 well plates containing DMEM with 10% FBS and incubated at 37°C in 5% CO2. After 24 hours of culture cells were washed with PBS then further cultured in DMEM without serum. Bim was added daily to maintain a final concentration of 5 µM. Cell viability was measured daily for 7 days using the CellTiter 96 solution (Promega) as described above. Student's t test and 95% confidence intervals based on a t distribution were used for statistical analyses. A P value of less than 0.01 was considered significant.
Acknowledgements
The authors thank Volker M. Vogt for providing rabbit antisera and Randall Basaraba for use of the fluorescence microscope. This work was supported by a National Institute of Health grant CA095056, from the National Cancer Institute to S.L.Q. A Ruth L. Kirschstein National Research Service Award F31CA099944, from the National Cancer Institute, supports Ms. Daniels.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bowser PR, Martineau D, Wooster GA. Effects of water temperature on experimental transmission of dermal sarcoma in fingerling walleyes (Stizostedion vitreum) Journal of Aquatic Animal Health. 1990;2:157–161. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- Bowser PR, Wolfe MJ, Forney JL, Wooster GA. Seasonal prevalence of skin tumors from walleye (Stizostedion vitreum) from Oneida Lake, New York. Journal of Wildlife Diseases. 1988;24:292–298. doi: 10.7589/0090-3558-24.2.292. [DOI] [PubMed] [Google Scholar]
- Bowser PR, Wooster GA. Regression of dermal sarcoma in adult walleyes (Stizostedion vitreum) Journal of Aquatic Animal Health. 1991;3:147–150. [Google Scholar]
- Bowser PR, Wooster GA, Quackenbush SL, Casey RN, Casey JW. Comparison of fall and spring tumors as inocula for experimental transmission of walleye dermal sarcoma. Journal of Aquatic Animal Health. 1996;8:78–81. [Google Scholar]
- Cai H, Smola U, Wixler V, Eisenmann-Tappe I, Diaz-Meco MT, Moscat J, Rapp U, Cooper GM. Role of diacylglycerol-regulated protein kinase C isotypes in growth factor activation of the Raf-1 protein kinase. Mol. Cell. Biol. 1997;17(2):732–741. doi: 10.1128/mcb.17.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BY, Chiang M, Cartwright CA. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J Biol Chem. 2001;276(23):20346–20356. doi: 10.1074/jbc.M101375200. [DOI] [PubMed] [Google Scholar]
- Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the b subunit of G proteins, inhibits activity of Src tyrosine kinases and growth of NIH 3T3 cells. Molecular and Cellular Biology. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-C, Appeddu PA, Isoda H, Guan J-L. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. Journal of Biological Chemistry. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- Chou Y-C, Chou C-C, Chen Y-K, Tsai S, Hsieh FMJ, Liu HJ, Hseu T-H. Structure and genomic organization of porcine RACK1 gene. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1999;1489(2–3):315–322. doi: 10.1016/s0167-4781(99)00213-4. [DOI] [PubMed] [Google Scholar]
- Csukai M, Chen C-H, De Matteis MA, Mochly-Rosen D. The Coatomer Protein beta '-COP, a Selective Binding Protein (RACK) for Protein Kinase Cepsilon. J. Biol. Chem. 1997;272(46):29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- Csukai M, Mochly-Rosen D. Pharmacologic modulation of protein kinase C isozymes: the role of RACKs and subcellular localisation. Pharmacological Research. 1999;39(4):253–259. doi: 10.1006/phrs.1998.0418. [DOI] [PubMed] [Google Scholar]
- Dorn Gn, Mochly-Rosen D. Intracellular transport mechnaims of signal transducers. Annu Rev Physiool. 2002;64:407–429. doi: 10.1146/annurev.physiol.64.081501.155903. [DOI] [PubMed] [Google Scholar]
- El-Guindy A, Heston L, Endo Y, Cho M-S, Miller G. Disruption of Epstein-Barr virus latency in the absence of phosphorylation of ZEBRA by protein kinase C. Journal of Virology. 2002;76(22):11199–11208. doi: 10.1128/JVI.76.22.11199-11208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong HKW, Amatruda TT, Birren BW, Simon MI. Distinct Forms of the {beta} Subunit of GTP-Binding Regulatory Proteins Identified by Molecular Cloning. PNAS. 1987;84(11):3792–3796. doi: 10.1073/pnas.84.11.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina A, Rossi F, Milanesi G. Rack1 binds HIV-1 Nef and can act as a Nef- protein kinase C adaptor. Virology. 2001;283:7–18. doi: 10.1006/viro.2001.0855. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko MP, Reiner O, Smith TF, Neer EJ. Folding of Proteins with WD-Repeats: Comparison of Six Members of the WD-Repeat Superfamily to the G Protein β Subunit. Biochemistry. 1996;35(44):13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- Goodnight J, Mischak H, Kolch W, Mushinski JF. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH 3T3 fibroblasts. the Journal of Biological Chemistry. 1995;270(17):9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche B, DeCastro E, Lachaize C, Langendijk-Genevaux P, Sigrist C. The 20 years of Prosite. Nucleic Acids Research. 2007 doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaken S, Leach K, Klauck T. Association of type 3 protein kinase C with focal contacts in rat embryo fibroblasts. J. Cell Biol. 1989;109(2):697–704. doi: 10.1083/jcb.109.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely P, O'Gorman D, Luong K, Ron D, O'Connor R. Insulin-like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and β1 integrin to promote cell migration. Mol. Cell. Biol. 2006;26:4041–4051. doi: 10.1128/MCB.01868-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J, Kim S, Lee S, Oh S, Byoun C, Han J, Nam H. Xenopus oocytes is inhibited by microinjection of a Brassica napus cDNA clone with high similarity to a mamamalian receptor for activated protein kinase C. Planta. 1997;201:245–251. doi: 10.1007/s004250050063. [DOI] [PubMed] [Google Scholar]
- Lairmore MD, Stanley JR, Weber SA, Holzschu DL. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proceedings of the National Academy of Sciences, USA. 2000;97(11):6114–6119. doi: 10.1073/pnas.110024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. Journal of Virology. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Holzschu DL, Bowser PR, Casey JW. Sequence and transcriptional analyses of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: Evidence for a gene duplication. Journal of Virology. 1999;73(11):9393–9403. doi: 10.1128/jvi.73.11.9393-9403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidipudi V, Chang BY, Harte RA, Lee KC, Cartwright CA. RACK1 inhibits the serum- and anchorage-independent growth of v-Src transformed cells. FEBS Lett. 2004;567(2–3):321–326. doi: 10.1016/j.febslet.2004.03.125. [DOI] [PubMed] [Google Scholar]
- Martineau D, Bowser PR, Renshaw RR, Casey JW. Molecular characterization of a unique retrovirus associated with a fish tumor. Journal of Virology. 1992;66(1):596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau D, Bowser PR, Wooster GA, Armstrong GA. Experimental transmission of a dermal sarcoma in fingerling walleyes (Stizostedion vitreum vitreum) Veterinary Pathology. 1990;27:230–234. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- Martineau D, Renshaw R, Williams JR, Casey JW, Bowser PR. A large unintegrated retrovirus DNA species present in a dermal tumor of walleye Stizostedion vitreum. Diseases of Aquatic Organisms. 1991;10:153–158. [Google Scholar]
- Mattingly RR, Sorisky A, Brann MR, Macara IG. Muscarinic receptors transform NIH 3T3 cells through a ras-dependent signalling pathway inhibited by the ras-GTPase-activating protein SH3 domain. Molecular and Cellular Biology. 1994;14(12):7943–7952. doi: 10.1128/mcb.14.12.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey PG, Rosner MR, Kikkawa U, Sekiguchi K, Ogita K, Ase K, Nishizuka Y. Characterization of protein kinase C from normal and transformed cultured murine fibroblasts. Biochemical and Biophysical Research Communications. 1987;146(1):140–146. doi: 10.1016/0006-291x(87)90702-9. [DOI] [PubMed] [Google Scholar]
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62(6):1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- Mischak H, Goodnight J, Kolch W, Martiny-Baron G, Schaechtle C, Kazanietz MG, Blumberg PM, Pierce JH, Mushinski JF. Overexpression of protein kinase C-d and -e in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence, and tumorigenicty. The Journal of Biological Chemistry. 1993;268(9):6090–6096. [PubMed] [Google Scholar]
- Mochly-Rosen D, Henrich CJ, Cheever L, Khaner H, Simpson PC. A protein kinase C isozyme is translocated to cytoskeletal elements on activation. Cell Regulation. 1990;1:693–706. doi: 10.1091/mbc.1.9.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Khaner H, Lopez J. Identification of Intracellular Receptor Proteins for Activated Protein Kinase C. PNAS. 1991;88(9):3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Khaner H, Lopez J, Smith BL. Intracellular receptors for activated protein kinase C. Identification of a binding site for the enzyme. J. Biol. Chem. 1991;266(23):14866–14868. [PubMed] [Google Scholar]
- Mochly-Rosen D, Smith BL, Chen C, Disatnik M, Ron D. Interaction of protein kinase C with RACK1, a receptor for activated C-kinase: a role in beta protein kinase C mediated signal transduction. Biochem Soc Trans. 1995;23(3):596–600. doi: 10.1042/bst0230596. [DOI] [PubMed] [Google Scholar]
- Nakashima S. Protein kinase C α (PKCα): Regulation and biological function. J. Biochem. 2002;132:669–675. doi: 10.1093/oxfordjournals.jbchem.a003272. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371(6495):297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Newton A. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. Turnover of inositol phospholipids and signal transduction. Science. 1984;225(4668):1365–1370. doi: 10.1126/science.6147898. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233(4761):305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334(6184):661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9(7):484–496. [PubMed] [Google Scholar]
- Olivier ARaP, Peter J. Identification of multiple PKC isoforms in Swith 3T3 cells: Differential down-regulation by phorbol ester. Journal of Cellular Physiology. 1992;152:240–244. doi: 10.1002/jcp.1041520204. [DOI] [PubMed] [Google Scholar]
- Quackenbush SL, Holzschu DL, Bowser PR, Casey JW. Transcriptional analysis of walleye dermal sarcoma virus (WDSV) Virology. 1997;237:107–112. doi: 10.1006/viro.1997.8755. [DOI] [PubMed] [Google Scholar]
- Reinhardt J, Wolff T. The influenza A virus M1 protein interacts with the cellular receptor of activated C kianse (RACK) 1 and can be phosphorylated by protein kinase C. Vet. Microbiol. 2000;74:87–100. doi: 10.1016/s0378-1135(00)00169-3. [DOI] [PubMed] [Google Scholar]
- Ron D, Chen C-H, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: A homolog of the b subunit of G proteins. Proc. Natl. Acad. Sci. USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Mochly-Rosen D. Agonists and antagonists of protein kinase C function, derived from its binding proteins. J. Biol. Chem. 1994;269(34):21395–21398. [PubMed] [Google Scholar]
- Rovnak J, Casey JW, Quackenbush SL. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin) Virology. 2001;280:31–40. doi: 10.1006/viro.2000.0731. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Casey RN, Brewster CD, Casey JW, Quackenbush SL. Establishment of productively infected walleye dermal sarcoma explant cells. Journal of General Virology. 2007;88(9):2583–2589. doi: 10.1099/vir.0.82967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Hronek BW, Ryan SO, Cai S, Quackenbush SL. An activation domain within the walleye dermal sarcoma virus retroviral cyclin protein is essential for inhibition of the viral promoter. Virology. 2005;342(2):240–251. doi: 10.1016/j.virol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus cyclin interacts with components of the Mediator complex and the RNA polymerase II holoenzyme. Journal of Virology. 2002;76:8031–8039. doi: 10.1128/JVI.76.16.8031-8039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus retroviral cyclin directly contacts TAF9. Journal of Virology. 2006;80(24):12041–12048. doi: 10.1128/JVI.01425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo PP, Deng X, Carr BK, May WS. A functional role for mitochondrial protein kinase Ca in Bcl2 phosphorylation and suppression of apoptosis. Journal of Biological Chemistry. 1998;273(25):25436–25442. doi: 10.1074/jbc.273.39.25436. [DOI] [PubMed] [Google Scholar]
- Ruvolo PP, Deng X, Ito T, Carr BK, May WS. Ceramide induces Bcl2 dephosphorylation via a mechanism involving mitochondrial PP2A. Journal of Biological Chemistry. 1999;274(29):20296–20300. doi: 10.1074/jbc.274.29.20296. [DOI] [PubMed] [Google Scholar]
- Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20(44):6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- Stebbins EG, Mochly-Rosen D. Binding specificity for RACK1 resides in the V5 region of bII protein kinase C. Journal of Biological Chemistry. 2001;276(32):29644–29650. doi: 10.1074/jbc.M101044200. [DOI] [PubMed] [Google Scholar]
- Tardif M, Savard M, Flamand L, Gosselin J. Impaired protein kinase C activation/translocation in Epstein-Barr virus-infected monocytes. Journal of Biological Chemistry. 2002;277(27):24148–24154. doi: 10.1074/jbc.M109036200. [DOI] [PubMed] [Google Scholar]
- Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991;266(24):15771–15781. [PubMed] [Google Scholar]
- Vomastek T, Iwanicki M, Schaeffer H-J, Tarcsafalvi A, Parsons J, Weber M. RACK1 targets the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway to link integrin engagement with focal adhesion disassembly and cell motility. Mol. Cell. Biol. 2007;27:8296–8305. doi: 10.1128/MCB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. Virus associated with epidermal hyperplasia in fish. National Cancer Institute Monograph. 1969;31:195–207. [PubMed] [Google Scholar]
- Wolf D, Giese S, Witte V, Krautkramer, Trapp S, Sass G, Haller C, Blume K, Fackler O, Baur A. Novel (n) PKC kinases phosphorylate Nef for increased HIV transcription, replication and perinuclear targeting. Virology. 2008;370:45–54. doi: 10.1016/j.virol.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kelly RK, Nielsen O. Morphological differentiation of virus-associated skin tumors of walleye (Stizostedion vitreum vitreum) Fish Pathology. 1985;20:361–372. [Google Scholar]
- Yamamoto T, MacDonald RD, Gillespie DC, Kelly RK. Viruses associated with lymphocystis and dermal sarcoma of walleye (Stizostedion vitreum vitreum) Journal Fish Research Board Canada. 1976;33:2408–2419. [Google Scholar]


