Abstract
Many viruses avoid immune surveillance during latent infection through reduction in the synthesis of virally encoded proteins. Although antigen presentation critically depends on the level of viral protein synthesis, the precise mechanism used to regulate the generation of antigenic peptide precursors remains elusive. Here, we demonstrate that a purine overloaded virally encoded mRNA lacking secondary structure significantly impacts the efficiency of protein translation and prevents endogenous antigen presentation. Reducing this purine bias through the generation of constructs expressing codon-modified sequences, while maintaining the encoded protein sequence, increased the stem–loop structure of the corresponding mRNA and dramatically enhanced self-synthesis of the viral protein. As a consequence, a higher number of HLA–peptide complexes were detected on the surface of cells expressing this viral protein. Furthermore, these cells were more efficiently recognized by virus-specific T cells compared with those expressing the same antigen expressed by a purine-biased mRNA. These findings delineate a mechanism by which viruses regulate self-synthesis of proteins and offer an effective strategy to evade CD8+ T cell-mediated immune regulation.
Keywords: antigen processing, EBV-encoded nuclear antigen 1, immune evasion, protein synthesis
Viruses that establish persistent infections or are involved in malignant processes have evolved unique mechanisms to evade the potent antiviral cytotoxic T cell response in the immunocompetent host (1–4). These evasion mechanisms include down-regulated gene expression during latent infection, virus replication in immune-privileged tissues, loss of HLA and adhesion protein expression, and sequence variation affecting peptide binding to HLA class I molecules or recognition by the T cell receptor on CD8+ T cells (5–7). It is now firmly established that activation of CD8+ T cells after viral infection critically depends on the efficient presentation of virally encoded epitopes in complex with HLA class I molecules (reviewed in refs. 8 and 9).
Although the human immune system is highly efficient in rapidly processing and presenting peptide epitopes from foreign proteins, many pathogens have adopted strategies to evade this rapid immune scanning by interfering with the HLA class I processing pathway or limiting the HLA–peptide complexes on the cell surface through cis-acting translational inhibition (10–12). Indeed, the EBV-encoded nuclear antigen, EBNA1, which is ubiquitously expressed in all EBV-associated malignancies, is an example of one such protein, which inhibits its self-synthesis and blocks proteasomal degradation, thereby restricting immune recognition by CD8+ T lymphocytes (11, 12). These effects have been accredited to a glycine-alanine repeat domain (GAr) within EBNA1, and observations that removal of this GAr domain led to increased translational efficiency and enhanced immune recognition (11, 13) suggested that the GAr sequence may be contributing to the inhibition of EBNA1 levels and thereby playing a crucial role in determining the efficiency by which EBNA1 epitopes are endogenously generated in virus-infected cells. Although later studies showed that GAr-mediated inhibition of mRNA translation, but not protein degradation, was essential for blocking endogenous processing of CD8+ T cell epitopes from EBNA1 through the class I pathway, it seems that the GAr domain plays a unique dual role as an inhibitor of both ribosomal and proteasomal activity (11). The precise mechanism by which the GAr domain minimizes self-synthesis while maintaining a functional expression level of EBNA1 remains to be determined.
Previous studies by Cristillo et al. (14) hypothesized that many persistent viruses have evolved to overload the coding sequences of latent proteins with purine codons so as to avoid triggering the host cell dsRNA surveillance mechanism. This “stealth” strategy is used by EBNA1, which is uniquely expressed in the most basic form of EBV latency (latency I). In this study we address the mechanism by which EBNA1 overloads its GAr domain with purine codons so as to inhibit self-synthesis and thereby evade immune recognition. Our results reveal that reduction of purine bias within the GAr domain, while maintaining the encoded protein sequence, dramatically altered the mRNA structure of EBNA1 and reversed the cis-inhibitory effect on EBNA1 synthesis. This evasive mechanism by EBNA1 to purine stack its internal GAr to regulate translational efficiency, demonstrates that EBNA1 translational modulation is occurring at the nucleotide level and not at the protein level as previously proposed.
Results
Purine Bias Within the GAr Domain of EBNA1 Results in an Unstable mRNA Secondary Structure.
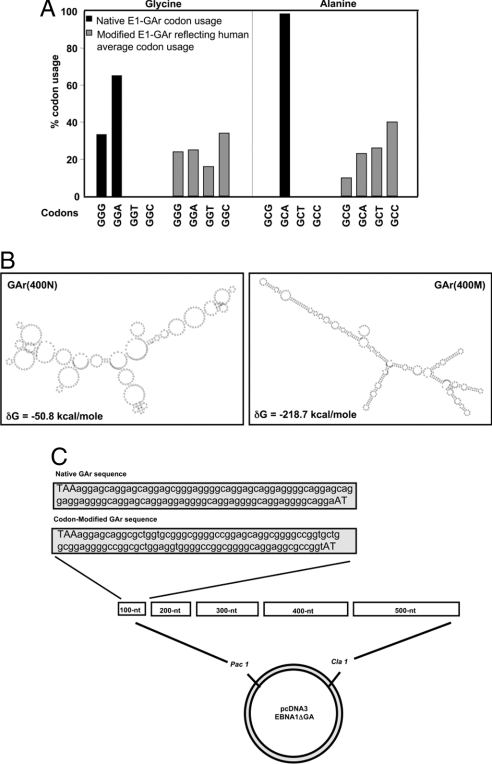
In delineating the mechanism by which the GAr domain exerts its cis-inhibitory effect on self-synthesis, an examination of the EBNA1 sequence revealed a dramatic overrepresentation of purine residues within the GAr domain (14). More than 99% of the glycine residues and 100% of the alanine residues within the GAr domain are comprised of purine codons (GGG, GGA, and GCA) compared with expected human averages for glycine and alanine purine codons of 49.3% and 33.3%, respectively (Fig. 1A). RNA secondary structure analysis (MFOLD) (15, 16) of 400 nt of native EBNA1 GAr sequence revealed a complete absence of secondary structure (Fig. 1B). MFOLD analysis reveals many potential structures. Fig. 1B is a representation of the most stable predicted structure. Similar analysis after modification of this same 400-nt GAr sequence to remove purine bias, while still maintaining the encoded protein sequence, demonstrated an enhanced and more stable mRNA secondary structure, reflected by the significantly more negative Gibbs free energy value, δG, of −218.7 kcal/mol compared with −50.8 kcal/mol for the native form (Fig. 1B). Based on this analysis, we hypothesized that an abnormal mRNA structure caused by unusual codon bias may play a crucial role in regulating the synthesis of EBNA1. To test this hypothesis, a number of glycine (GGN) and alanine (GCN) codons within the GAr domain were modified, by mutating the choice of third bases (N) to reflect the expected human average codon usage (Fig. 1A) (17). A series of sequentially designed GAr oligonucteotides were synthesized and inserted into the coding sequence of EBNA1 devoid of its normal GAr domain (E1-ΔGA) (Fig. 1C). These engineered GAr domains, GAr(N) referring to native EBNA1 GAr sequence or GAr(M) referring to codon-modified EBNA1 GAr sequence, encoded varying lengths of the GAr sequence (100–500 nt) (Fig. 1C).
Fig. 1.
Purine bias within the GAr sequence of EBNA1 affects mRNA secondary structure. (A) The native EBNA1 GAr domain has an overrepresentation of purine codons compared to a synthetically generated modified EBNA1 GAr domain where purine bias has been reduced to reflect human average codon usage. (B) Predicted secondary structure of a 400-nt native or codon-modified GAr domain using the mRNA structure analysis program, MFOLD. Free energy values (δG) are shown for both sequences. (C) A series of GAr oligonucteotides were synthesized and inserted into the coding sequence of EBNA1-ΔGA. These engineered GAr domains encode varying lengths of native or codon-modified GAr sequence (100–500 nt).
A Codon-Modified GAr Influences EBNA1 Expression.
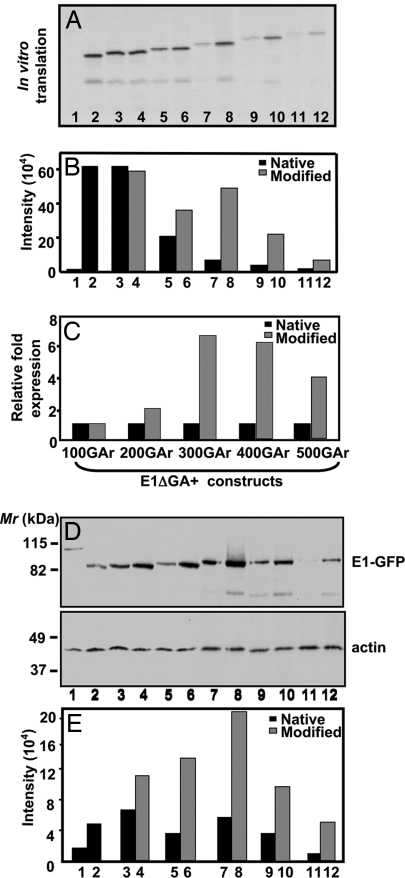
Analysis of in vitro-transcribed EBNA1 mRNA comprised of native or codon-modified GAr sequences by in vitro translation (IVT) assays demonstrated that the translational efficiency of EBNA1 sequences encoding a codon-modified GAr domain was significantly enhanced when compared with its matched pair of EBNA1 sequences encoding a native GAr domain (Fig. 2 A–C). In addition, there was a significant reduction in translational efficiency as the native or modified GAr domain was increased in length (Fig. 2 A and B). Furthermore, in agreement with the IVT assays, Western blot analysis of intracellular expression of EBNA1-GFP sequences in an epithelial cell line (HEK293) also showed a dramatic increase in protein expression for codon-modified EBNA1 constructs (Fig. 2 D and E). Differences in protein expression levels between native and codon-modified EBNA1-GFP sequences were not caused by differential transfection efficiencies because transfectants showed similar percentages (33–36%) of GFP-positive cells. The correlation between GAr size and level of translation varies slightly between the in vitro and in vivo model systems, suggesting that the overall translational efficiency may be related not only to predicted mRNA secondary structure but also to other factors such as tRNA abundance or other fortuitous binding proteins to Gly/Ala-rich proteins. Taken together, these observations demonstrate that purine bias within the EBNA1 GAr domain directly impacts EBNA1 protein expression.
Fig. 2.
A codon-modified GAr sequence influences EBNA1 synthesis. (A) IVT assay of pcDNA3 expression constructs encoding EBNA1 (E1) (lane 1), E1ΔGA (lane 2), E1-GAr(100N) (lane 3), E1-GAr(100M) (lane 4), E1-GAr(200N) (lane 5), E1-GAr(200M) (lane 6), E1-GAr(300N) (lane 7), E1-GAr(300M) (lane 8), E1-GAr(400N) (lane 9), E1-GAr(400M) (lane 10), E1-GAr(500N) (lane 11), or E1-GAr(500M) (lane 12). The constructs were transcribed and translated in vitro with T7 RNA polymerase by using a coupled transcription/translation reticulocyte lysate system. 35S-methionine-labeled proteins were visualized by autoradiography. (B and C) Band intensities from the IVT assay were quantified by densitometric analysis using Imagequant software (Molecular Dynamics) and graphed to demonstrate absolute intensities (B) or relative fold increase of EBNA1 encoded by codon-modified GAr domains compared with EBNA1 encoded by native GAr domains (C). (D) Western blot of EBV-negative HEK293 cells transfected with expression constructs encoding E1-GFP (lane 1), E1ΔGA-GFP (lane 2), E1-GAr(100N)-GFP (lane 3), E1-GAr(100M)-GFP (lane 4), E1-GAr(200N)-GFP (lane 5), E1-GAr(200M)-GFP (lane 6), E1-GAr(300N)-GFP (lane 7), E1-GAr(300M)-GFP (lane 8), E1-GAr(400N)-GFP (lane 9), E1-GAr(400M)-GFP (lane 10), E1-GAr(500N)-GFP (lane 11), or E1-GAr(500M)-GFP (lane 12) with a GFP antibody (Upper) or a monoclonal actin antibody (Lower). Molecular weight markers Mr (kDa) are indicated on the left. (E) Band intensities after immunoblotting were quantified as described for B. Representative data from one of four experiments are presented here.
Regulation of EBNA1 Synthesis Occurs at the Translational Level.
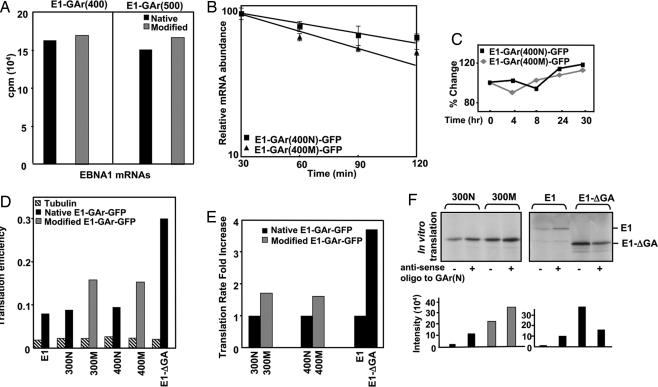
To determine whether the enhanced synthesis of codon-modified EBNA1 was being influenced by transcriptional and/or translational factors, we first assessed mRNA synthesis levels of EBNA1 sequences encoding either native or codon-modified GAr domains. pcDNA3 expression constructs encoding E1-GAr(400N), E1-GAr(400M), E1-GAr(500N), or E1-GAr(500M) were linearised with XbaI and transcribed with T7 RNA polymerase by using a Riboprobe in vitro transcription system supplemented with 50 μCi [α-32P]UTP. Data presented in Fig. 3A demonstrate that modification of the GAr coding sequences had minimal impact on mRNA synthesis levels, consistent with previous studies comparing EBNA1 and EBNA1-ΔGA steady-state mRNA levels (11). To discount the possibility that modification of codons within the GAr domain may have altered EBNA1 mRNA turnover, we next performed mRNA degradation analyses by using quantitative real-time RT-PCR (qRT-PCR). The results presented in Fig. 3B demonstrate that mRNA from EBNA1 sequences encoding a 400-nt modified repeat, GAr(400M), displayed a half-life of 231 ± 142 min, compared with a value of 366 ± 30 min for mRNA from EBNA1 encoding a 400-nt native repeat, GAr(400N). However, after statistical analysis there was no significant difference in the two half-lives (P > 0.05) (Fig. 3B), indicating that the overall rates of decay of mRNAs of the two representative EBNA1 constructs encoding either native or codon-modified GAr sequences are comparable. As expected, protein degradation rates of EBNA1-GFP sequences encoding either GAr(400N) or GAr(400M) domains displayed similar intracellular kinetics after cycloheximide treatment over a 30-h time course (Fig. 3C).
Fig. 3.
Purine bias within the GAr domain decreases the translational efficiency of EBNA1. (A) In vitro transcription assays of EBNA1 encoding either 400 or 500 nt of native or codon-modified GAr sequence demonstrated similar levels of mRNA synthesis. (B) Relative decay of mRNAs for EBNA1-GFP encoding 400 nt of either native or codon-modified GAr sequences, after inhibition of transcription with Actinomycin D. (C) Half-life analysis of E1-GAr(400N)-GFP and E1-GAr(400M)-GFP using FACScan analysis. HEK293 cells, transfected with these expression vectors were treated with 50 μg/ml cycloheximide over a 30-h time course. Expression levels are plotted as a relative percentage of the signal at time 0. (D and E) EBNA1-GFP synthesis rates were measured by transfecting HEK293 cells with either E1-GFP, E1ΔGA-GFP, E1-GAr(300N)-GFP, E1-GAr(300M)-GFP, E1-GAr(400N)-GFP, or E1-GAr(400M)-GFP expression constructs. Transfectants were metabolically labeled overnight in growth medium containing 20 μCi/ml of 3[H]methionine followed by a 30-min pulse with 100 μCi of 35[S]methionine. Cells were lysed, immunoprecipitated with either anti-GFP or anti-tubulin, and subjected to SDS/PAGE. Quantitation of synthesis of these proteins was determined by measuring the 35[S]/3[H] ratio for each protein by liquid scintillation counting (D) and graphed as fold increase in translation rate of modified versus native GAr (E). (F) IVT assay of EBNA1 expression constructs in the presence or absence of a 21-mer antisense oligonucleotide to the native GAr sequence at a molar excess of 10:1 to the GAr. 35S-methionine-labeled proteins were visualized by autoradiography and quantified by densitometric analysis using Imagequant software.
As the observed differences in protein expression of EBNA1-GFP encoded by either native or modified repeat sequences were not caused by significant differences in mRNA transcription or turnover, we assessed absolute rates of EBNA1 synthesis relative to a common protein standard by using a double labeling protocol (18). Although the rate of synthesis for tubulin remained unchanged in HEK293 cells transfected with the different EBNA1 expression vectors, the translation rates for two representative EBNA1 constructs encoding modified GAr sequence, E1-GAr(400M)-GFP and E1-GAr(300M)-GFP, were 60–70% higher, respectively, than their corresponding matched pair of native GAr sequence (Fig. 3 D and E), confirming earlier results that a codon-modified GAr domain of EBNA1 critically modulates translational efficiency (Fig. 2A). Transfection efficiencies for native and codon-modified EBNA1-GFP sequences in the above assays were similar as assessed by the percentage of GFP-positive cells.
A GAr Antisense Oligonucleotide Enhances EBNA1 Synthesis in Vitro.
Having observed an increase in translation efficiency as the mRNA of the codon-modified GAr domain becomes more structured because of a greater propensity to form stem loops after modification of the codons, we reasoned that if we could stabilize the mRNA of native GAr sequences, this modification may override the GAr cis-inhibitory affect on translation. To test this hypothesis, we performed IVT assays of EBNA1 encoded by native or codon-modified GAr sequences in the presence of a 21-mer GAr(N) antisense oligonucleotide. Fig. 3F demonstrates that addition of the antisense oligonucleotide results in an increase in translation product for E1-GAr(300N) and full-length EBNA1, with the EBNA1 expression construct encoding 300 nt of native GAr sequence, E1-GAr(300N), reaching 50% of the translational level observed for the matching codon-modified E1-GAr(300M) construct. An increase in translation product was also observed for E1-GAr(300M) because stretches of the antisense oligo also matched regions of the codon-modied GAr sequence. As the E1-ΔGA expression level was not enhanced after the same antisense oligonucleotide treatment, the data clearly implicate the EBNA1 GAr in translational regulation (Fig. 3F).
Increased Translation Efficiency of EBNA1 Encoded by a Modified GAr Results in Enhanced Antigen Processing.
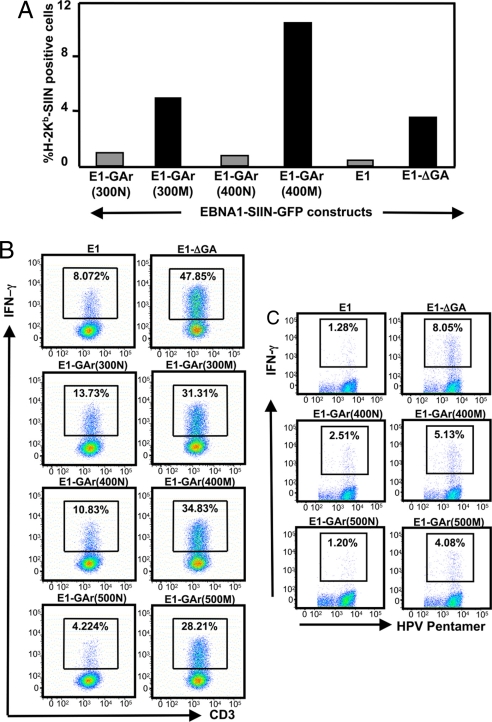
We next investigated the impact of the enhanced translational efficiency of codon-modified EBNA1 on the endogenous presentation of MHC class I-restricted epitopes derived from EBNA1. In the first set of these experiments we assessed the loading of MHC class I molecules with a H-2Kb-restricted epitope from ovalbumin, SIINFEKL (Ser–Ile–Ile–Asn–Phe–Glu–Lys–Leu, residues 257–264), which was inserted into the EBNA1 sequence (13). H-2Kb-expressing HEK293 cells were transiently transfected with either E1-GFP, E1-ΔGA-GFP, E1-GAr(300N)-GFP, E1-GAr(300M)-GFP, E1-GAr(400N)-GFP, or E1-GAr(400M)-GFP expression constructs, with average transfection efficiencies between 75% and 80% (data not shown). For each SIINFEKL containing construct, a separate transfection of the parent construct without SIINFEKL was done to provide a negative control. After an overnight transfection, cells were assessed by flow cytometry for GFP expression and surface expression of H-2Kb-SIIN complexes by using a mAb (25-D1.16) that recognizes the SIIN epitope bound to H-2Kb molecules (19). For each SIINFEKL-containing construct a separate transfection of the parent plasmid without SIINFEKL was done to provide a baseline above which an increase in fluorescence would indicate H-2Kb-SIIN expression. Representative data from one experiment (Fig. 4A) show that the percentage of cells expressing H-2Kb-SIINFEKL complexes was 4- to 11-fold higher when constructs with EBNA1 encoded by codon-modified GAr sequences were transfected compared with the EBNA1 sequences encoding a native GAr domain. We next assessed the endogenous processing and surface presentation of two different CD8+ T cell epitopes. One of these was a HLA B8-restricted epitope derived from the EBNA3 protein, FLRGRAYGL (Phe–Leu–Arg–Gly–Arg–Ala–Tyr–Gly–Leu, residues 345–361), which was inserted into the C terminus of the EBNA1 sequence (13), while the second epitope was encoded within the EBNA1 sequence and restricted through HLA B35, HPVGEADYFEY (His–Pro–Val–Gly–Glu–Ala–Asp–Tyr–Phe–Glu–Tyr, residues 407–417). HLA B8 and HLA B35-positive human cells expressing EBNA1 encoded by either native or codon-modified GAr domains were incubated with the CD8+ T cells specific for FLRGRAYGL or HPVGEADYFEY epitopes and stimulation was assessed by using intracellular cytokine assays. Data presented in Fig. 4 B and C show that human cells expressing EBNA1-GFP protein encoded by a codon-modified GAr domain [E1-GAr(300M)-GFP, E1-GAr(400M)-GFP and/or E1-GAr(500M)-GFP] stimulated 2- to 7-fold more IFN-γ producing EBV-specific T cells compared with cells expressing EBNA1-GFP encoded by a native GAr domain [E1-GAr(300N)-GFP, E1-GAr(400N)-GFP and/or E1-GAr(500N)-GFP]. Interestingly, these experiments demonstrated that as the GAr domain is increased in length within the EBNA1 sequence the number of IFN-γ producing EBV-specific T cells are decreased, closely mimicking the IVT data (Fig. 2 A and B).
Fig. 4.
A codon-modified GAr sequence enhances the presentation of CD8+ T cell epitopes. (A) Expression of surface H-2Kb–SIIN complexes was assessed by flow cytometry on 293KbC2 cells transfected with EBNA1-SIIN-GFP constructs encoding native or codon-modified GAr sequences (as shown). Data are the percent of cells positive for H-2Kb–SIIN when compared with controls cells transfected with relevant parental EBN1-GFP plasmids that lack the SIIN epitope. (B) FLR-specific T cells were incubated overnight in the presence of brefeldin A at a responder to stimulator ratio of 5:1 with fixed EBV-negative SVMR6 cells transfected with selected EBNA1-GFP expression vectors. IFN-γ production by CD3+CD8+ cells was determined by intracellular cytokine staining. (C) HPVGEADYFEY-specific T cells were incubated in duplicate in the presence of brefeldin A for 6 h at a responder-to-stimulator ratio of 5:1 with fixed EBV-negative DG75 cells transfected with selected EBNA1-GFP expression vectors. Cells were incubated with a HPV-specific pentamer followed by anti-CD8 and incubated with anti-IFN-γ and analyzed on a FACSCanto. The top right hand corner of each panel represents the percentage of HPV-specific CD8+ lymphocytes producing IFN-γ. Data shown in C and D are for one representative experiment of three separate experiments.
Discussion
The EBV-encoded EBNA1 protein represents an ideal model for investigating the influence of viral mRNA structure on protein translation and T cell recognition. The use of a series of matched EBNA1 expression constructs differing only within the nucleotide sequence of the internal GAr domain, while maintaining identical protein sequence, provides evidence that purine codon bias of the GAr domain within EBNA1 directly impacts on the translational efficiency of this protein. Possible explanations for why the more structured codon-modified GAr mRNA translates more efficiently may be that the lack of predicted secondary structure in the native purine-rich mRNA leaves this RNA region vulnerable to the binding of proteins that inhibit translation or may simply reflect a difficulty for the ribosome to translate through the purine overloaded sequence within the GAr of the native mRNA. By reducing this purine overload, we were able to override the inhibitory effect of the GAr domain on self-synthesis. The observation that matched pairs of EBNA1 expression vectors, encoding either native or codon-modified GAr sequences, display different translational efficiencies demonstrates that EBNA1 translational modulation is occurring at the nucleotide level and not at the protein level as previously proposed. Because the N- and C-terminal sequences of the matched EBNA1 constructs are identical, it is unlikely that the observed differences in translational efficiency between native and codon-modified EBNA1 are caused by differences in initiation or termination events. Rather our results demonstrate that the overrepresentation of purine codons within the GAr domain promotes an unusual and unstable mRNA secondary structure, which significantly affects the elongation step during the translation of native EBNA1 transcripts.
Having delineated the immune evasive mechanism of codon bias used to inhibit self-synthesis, it will be important to explore strategies for targeting the EBNA1 protein in vivo to enhance its expression in virus-infected cells. This approach should allow for more efficient production of peptide epitopes from the EBNA1 protein and thus more efficient recognition by virus-specific T cells. It is of interest to note that >90% of EBNA1 epitopes mapped to date reside downstream of the EBNA1 GAr sequence, highlighting the significance of the virus's immune evasion strategy of purine stacking its GAr domain so as to impede translation through this unusual RNA structure, thus limiting the level of EBNA1 immunogenic epitopes for immune surveillance. This proposition is strongly supported by the data presented in this study, where we have shown that the increased translation of the EBNA1 protein after modification of codons within the GAr domain dramatically enhanced T cell recognition. Furthermore, the encouraging observation where we demonstrate an enhancement in the translational efficiency of EBNA1 after the addition of a GAr antisense oligonucleotide to IVT assays, may represent a potential therapeutic approach to EBV infection. Here, the antisense oligonucleotide may have the capacity to bind to several sites within the mRNA encoding the GAr region, possibly promoting enhanced and specific secondary structure that increase translation. Future studies should now focus on efficient delivery of GAr antisense oligonucleotides in vivo by using tumor or infectious models. This observation is particularly pertinent for EBV-associated malignancies such as nasopharyngeal carcinoma and Hodgkin's lymphoma where the endogenous processing machinery is intact and EBV gene expression is restricted to only a few viral proteins, including EBNA1. Furthermore, a vaccine formulation based on a codon-modified EBNA1 sequence may be more efficacious in terms of priming EBNA1-specific T cell responses in vivo. This strategy is expected to improve the EBNA1-specific T cell response quantitatively and qualitatively. Indeed, previous studies by Liu et al. (20) have shown that a DNA vaccine based on codon-modified human papillomavirus type 16 E7, which was expressed at higher levels compared with the WT E7, significantly enhanced CTL induction and antitumor activity. Overriding translational repression of EBNA1 should also induce growth inhibition, cell cycle arrest, and cell death in virus-infected normal and malignant cells (21). Indeed, it is now well established that overexpression of the recombinant EBNA1 gene product can be toxic and induces programmed cell death in certain cell types (22).
Materials and Methods
Cell Lines.
Cell lines (HeLa, DG75, and SVMR6) were routinely maintained in RPMI medium 1640 supplemented with 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin plus 10% FCS (referred to as growth medium). HEK293 cells were grown in DMEM/10% FCS.
Generation of EBNA1 Expression Constructs.
Full-length EBV-encoded EBNA1, (E1) and EBNA1-ΔGA, (E1-ΔGA) were cloned into the expression vector pcDNA3 (Invitrogen). To generate EBNA1 expression constructs with varying lengths of native or modified GAr sequences, 105-mer oligonucleotides were synthesized, representing sequential 102-nt increments of the EBNA1 GAr sequence [supporting information (SI) Table S1]. Mutagenesis of the E1-ΔGA expression construct allowed the insertion of PacI and ClaI restriction sites at nucleotide position 250. The first 105-mer sense and antisense oligonucleotides of native or modified GAr sequence were annealed and cloned into the PacI and ClaI restriction sites to generate E1-GAr(100N) or E1-GAr(100M), where N represents the native GAr sequence and M represents the codon-modified GAr sequence. Sequential 105-mer GAr oligonucleotides were cloned into the 3′ ClaI restriction site of each preceding GAr insertion to generate the following EBNA1 expression constructs: E1-GAr(200N), E1-GAr(200M), E1-GAr(300N), E1-GAr(300M), E1-GAr(400N), E1-GAr(400M), E1-GAr(500N), and E1-GAr(500M). The above EBNA1 expression vectors were also subcloned in-frame with a sequence coding for GFP (pEGFP-N1; Clontech). To assess EBNA1 endogenous processing, a sequence encoding the HLA-B8-restricted, EBNA3 epitope FLRGRAYGL (23) was inserted into EBNA1 constructs as described (13). For the assessment of endogenous loading of MHC class I molecules, a H-2Kb-restricted epitope from ovalbumin, SIINFEKL (24), was inserted in-frame between EBNA1 and GFP.
In Vitro Transcription/Translation Assays.
EBNA1/pcDNA3 expression constructs were linearized with XbaI and 1 μg of template transcribed with T7 RNA polymerase by using a Riboprobe in vitro transcription system (Promega) supplemented with 50 μCi [α-32P]UTP (Amersham Biosciences). For translation assays EBNA1/pcDNA3 vectors were transcribed and translated in vitro with T7 RNA polymerase by using a coupled transcription/translation reticulocyte lysate system (Promega) supplemented with 250 μCi 35[S]methionine (Amersham Biosciences). Lysates were subjected to SDS/PAGE and autoradiography as described (13). IVT assays were performed in the presence or absence of a 21-mer antisense oligonucleotide, (5′-TCCCGCTCCTGCTCCTGCTCC-3′) to the native GAr sequence at a molar ratio of 10:1 to the GAr.
Transfection of EBNA1 Constructs and Detection by Immunoblotting.
DG75 cells (5 × 106) were transfected with 10 μg of expression constructs by using the BioRad Gene Pulser (960 μF, 250 V, 0.4-cm gap electrode, 300-μl assay volume, 25°C). For adherent cell lines HEK293 or SVMR6 cells (2 × 105) were transfected with 0.4 μg of EBNA1 expression constructs by using Effectene (Qiagen) according to the manufacturer's instructions. Twenty-four hours posttransfection, cells were harvested and samples were subjected to SDS/PAGE and immuno-blotted with either anti-GFP (1:2,000) or an actin mAb (1:1,000) as described (25). Half-life analysis of EBNA1-GFP fusion proteins were performed as described (25).
Isolation of RNA and RT-PCR.
Total RNA was extracted from HEK293 cells transiently transfected with EBNA1 expression constructs after treatment with 5 μg/ml Actinomycin D (Sigma) over a time course of 2 h. The RNA was purified with an RNeasy Plus Mini Kit (Qiagen) and quantified by spectrophotometric measurements at 260 and 280 nm, and its integrity was verified by the OD260/OD280 absorption ratio (>1.8) and visualization on an agarose gel.
qRT-PCR.
cDNA synthesis of selected EBNA1 sequences was undertaken with 1 μg of isolated RNA per sample by using MMLV SuperScript III reverse transcriptase (Invitrogen) and an anchored oligo(T)18 primer combined with random hexamers. qRT-PCR using the Sybr Green-based fluorescent detection system and the ABI Prism 7900 Sequence Detection System (Applied Biosystems) was used to measure mRNA abundance. Primers were designed by using DS Gene (version 1.5) (Accelrys) software (Table S2). A constant amount of cDNA, corresponding to 10 ng of reverse-transcribed RNA derived from each sample was used for qRT-PCR measurements. Four technical replicates were performed for each gene investigated. This process allowed quantification of the target gene relative to a constant reference gene in each sample by using threshold cycle (Ct) data. Ribosomal protein P0 (RPLP0; GenBank accession no. NM_053275) was used as the reference gene for all samples.
Each qRT-PCR (5 ml total volume) contained 2.5 ml of 2× Sybr Green Master Mix (Applied Biosystems), 0.25 ml of each primer giving a final concentration of 500 nM each, 1.0 ml water, and 1.0 ml of a 1/10 dilution of the stock cDNA template. The cycling conditions consisted of 40 cycles of 95°C for 15 s and 60°C for 1 min. At the completion of each run, a dissociation melt curve analysis was performed. All melt curves for detectable amplicons showed a single peak and were consistent with the presence of a single specific amplicon. Data analyses used Q-gene qRT-PCR analysis software (Gene Quantification; www.gene-quantification.info) and gene expression was normalized relative to RPLP0 expression. A linear least-squares statistical analysis was applied for half-life calculations.
Measurement of EBNA1 Protein Synthesis.
HEK293 cells (2 × 105) were transfected with selected EBNA1-GFP expression constructs. Twenty-four hours posttransfection the cells were metabolically labeled at 37°C for 12–14 h in growth medium containing 20 μCi/ml 3[H]methionine (Amersham Biosciences). Cells were washed in PBS and incubated in methionine-free growth medium for 30 min at 37°C preceding a 30-min pulse with 100 μCi 35[S]methionine. Following the pulse, cells were lysed in Tris-buffered saline with 1% Triton X-100 and protease inhibitors and precleared with Protein A Sepharose, and lysates were immunoprecipitated with anti-GFP or a mAb to β-tubulin (Sigma). Immunoprecipitated samples were added to 10 ml of scintillant fluid, Ultima Gold (PerkinElmer Life and Analytical Sciences), and counted on a Packard liquid scintillation analyzer, Tri-carb 2100TR.
Detection of Cell Surface Kb-SIINFEKL.
293KbC2 cells, which stably express H-2Kb (26), were transfected with E1-SIINFEKL-GFP constructs by using Lipofectamine 2000 (Invitrogen). For each SIINFEKL-containing construct, a separate transfection of the parent construct without SIINFEKL was done to provide a negative control. Cells were harvested after an overnight transfection and stained with mAb 25D1.16 (19) conjugated to Alexa Fluor 647 (Molecular Probes/Invitrogen), a kind gift of M. Princiotta (Upstate Medical University, Syracuse, NY) and J. Yewdell (National Institute of Allergy and Infectious Diseases, Bethesda, MD) (27), on ice for 30 min. Cells were washed and analyzed by flow cytometry (LSR II; BD Biosciences) for GFP expression and 25D1.16 binding. GFP+ events were determined from a population gated tightly on forward scatter (FSC) × side scatter (SSC) (to eliminate cells adversely effected by the transfection), and average transfection efficiencies were 75% or better. As the majority of 25D1.16+ events were contained in the GFPhi population of all SIINFEKL-containing constructs, we restricted further analysis to the brightest half of transfected cells. To determine the level of surface Kb-SIINFEKL for each version of construct, the 25D1.16-Alexa 647 intensity of the brightest 50% of GFP+ events was compared between non-SIINFEKL bearing parent constructs and the relevant test SIINFEKL-containing construct by using the Overton Subtraction method (28) (Flowjo software; Tree Star).
Intracellular Cytokine Staining.
SVMR6 or DG75 cells transfected with EBNA1-GFP constructs were fixed with 2% paraformaldehyde, washed, and resuspended in growth medium. T cells, either an FLRGRAYGL-specific clone (LC13) or a HPVGEADYFEY-specific T cell line, were incubated overnight (FLR) or for 6 h (HPV) at 37°C with fixed EBNA1-GFP transfectants at responder-to-stimulator ratios of 5:1, 10:1, and 20:1 in growth medium supplemented with Brefeldin A (BD Pharmingen). Cells were washed and incubated with either phycoerythrin (PE)-labeled HLA B3508 HPVGEADYFEY pentamer (ProImmune; Oxford) followed by peridinin chlorophyll protein (perCP)-conjugated anti-CD8 for HPV-specific T cells or allophycocyanin-conjugated anti-CD3 and perCP-conjugated anti-CD8 for LC13, rewashed, then fixed and permeabilized with cytofix/cytoperm (BD Pharmingen) at 4°C for 20 min. Cells were washed in perm/wash, incubated with allophycocyanin-conjugated or PE-conjugated anti-IFNγ (BD Pharmingen) at 4°C for 30 min, washed again with perm/wash, resuspended in PBS, and analyzed on a FACSCanto (BD Biosciences).
Supplementary Material
Acknowledgments.
We thank Professor John Shine and Dr. Ross Tellam for critically reading this manuscript and Diem Hoang-Le, Michelle Martinez, and Sarah Wilkins for technical assistance. This work was supported by funding from the National Health and Medical Research Council, Canberra, Australia and the Queensland Cancer Fund. R.K. is supported by a fellowship from the National Health and Medical Research Council. D.C.T. is supported by National Health and Medical Research Council Grant CDA418108.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9135.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801968105/DCSupplemental.
References
- 1.Mocarski ES., Jr Immunomodulation by cytomegaloviruses: Manipulative strategies beyond evasion. Trends Microbiol. 2002;10:332–339. doi: 10.1016/s0966-842x(02)02393-4. [DOI] [PubMed] [Google Scholar]
- 2.Khanna R, et al. EBV peptide epitope sensitization restores human cytotoxic T cell recognition of Burkitt's lymphoma cells. Evidence for a critical role for ICAM-2. J Immunol. 1993;150:5154–5162. [PubMed] [Google Scholar]
- 3.Gandhi MK, Khanna R. Human cytomegalovirus: Clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. 2004;4:725–738. doi: 10.1016/S1473-3099(04)01202-2. [DOI] [PubMed] [Google Scholar]
- 4.Khanna R, Moss DJ, Gandhi M. Applications of emerging immunotherapeutic strategies for Epstein–Barr virus-associated malignancies. Nat Clin Pract Oncol. 2005;2:138–149. doi: 10.1038/ncponc0107. [DOI] [PubMed] [Google Scholar]
- 5.Bauer D, Tampe R. Herpes viral proteins blocking the transporter associated with antigen processing TAP: From genes to function and structure. Curr Top Microbiol Immunol. 2002;269:87–99. [PubMed] [Google Scholar]
- 6.Park B, et al. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity. 2004;20:71–85. doi: 10.1016/s1074-7613(03)00355-8. [DOI] [PubMed] [Google Scholar]
- 7.Kimura Y, et al. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J Virol. 2005;79:4870–4876. doi: 10.1128/JVI.79.8.4870-4876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yewdell JW, Haeryfar SM. Understanding presentation of viral antigens to CD8+ T cells in vivo: The key to rational vaccine design. Annu Rev Immunol. 2005;23:651–682. doi: 10.1146/annurev.immunol.23.021704.115702. [DOI] [PubMed] [Google Scholar]
- 9.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 10.Yewdell JW, Schubert U, Bennink JR. At the crossroads of cell biology and immunology: DRiPs and other sources of peptide ligands for MHC class I molecules. J Cell Sci. 2001;114:845–851. doi: 10.1242/jcs.114.5.845. [DOI] [PubMed] [Google Scholar]
- 11.Yin Y, Manoury B, Fahraeus R. Self-inhibition of synthesis and antigen presentation by Epstein–Barr virus-encoded EBNA1. Science. 2003;301:1371–1374. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- 12.Levitskaya J, et al. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellam J, et al. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J Exp Med. 2007;204:525–532. doi: 10.1084/jem.20062508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristillo AD, et al. Double-stranded RNA as a not-self alarm signal: To evade, most viruses purine-load their RNAs, but some (HTLV-1,Epstein–Barr) pyrimidine-load. J Theor Biol. 2001;208:475–491. doi: 10.1006/jtbi.2000.2233. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson AB, Zuker M. Structural analysis by energy dot plot of a large mRNA. J Mol Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases: Its status 1999. Nucleic Acids Res. 1999;27:292. doi: 10.1093/nar/27.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Garay ML, Cabral F. Overexpression of an epitope-tagged β-tubulin in Chinese hamster ovary cells causes an increase in endogenous α-tubulin synthesis. Cell Motil Cytoskeleton. 1995;31:259–272. doi: 10.1002/cm.970310403. [DOI] [PubMed] [Google Scholar]
- 19.Porgador A, et al. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 20.Liu WJ, et al. Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T lymphocyte induction and antitumor activity. Virology. 2002;301:43–52. doi: 10.1006/viro.2002.1584. [DOI] [PubMed] [Google Scholar]
- 21.Jones RJ, et al. Epstein–Barr virus nuclear antigen 1 (EBNA1)-induced cytotoxicity in epithelial cells is associated with EBNA1 degradation and processing. Virology. 2003;313:663–676. doi: 10.1016/s0042-6822(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 22.Ramage AD, et al. Improved EBV-based shuttle vector system: Dicistronic mRNA couples the synthesis of the Epstein–Barr nuclear antigen-1 protein to neomycin resistance. Gene. 1997;197:83–89. doi: 10.1016/s0378-1119(97)00245-x. [DOI] [PubMed] [Google Scholar]
- 23.Burrows SR, et al. The specificity of recognition of a cytotoxic T lymphocyte epitope. Eur J Immunol. 1992;22:191–195. doi: 10.1002/eji.1830220128. [DOI] [PubMed] [Google Scholar]
- 24.Carbone FR, Bevan MJ. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J Exp Med. 1989;169:603–612. doi: 10.1084/jem.169.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tellam J, et al. Endogenous presentation of CD8+ T cell epitopes from Epstein–Barr virus-encoded nuclear antigen 1. J Exp Med. 2004;199:1421–1431. doi: 10.1084/jem.20040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tscharke DC, et al. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Princiotta MF, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 28.Overton WR. Modified histogram subtraction technique for analysis of flow cytometry data. Cytometry. 1988;9:619–626. doi: 10.1002/cyto.990090617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.