Abstract
Genome-wide studies of steady-state mRNA levels revealed common principles underlying transcriptional changes in response to external stimuli. To uncover principles that govern other stages of the gene-expression response, we analyzed the translational response and its coordination with transcriptome changes following exposure to severe stress. Yeast cells were grown for 1 h in medium containing 1 M NaCl, which elicits a maximal but transient translation inhibition, and nonpolysomal or polysomal mRNA pools were subjected to DNA-microarray analyses. We observed a strong repression in polysomal association for most mRNAs, with no simple correlation with the changes in transcript levels. This led to an apparent accumulation of many mRNAs as a nontranslating pool, presumably waiting for recovery from the stress. However, some mRNAs demonstrated a correlated change in their polysomal association and their transcript levels (i.e., potentiation). This group was enriched with targets of the transcription factors Msn2/Msn4, and the translational induction of several tested mRNAs was diminished in an Msn2/Msn4 deletion strain. Genome-wide analysis of a strain lacking the high salinity response kinase Hog1p revealed that the group of translationally affected genes is significantly enriched with motifs that were shown to be associated with the ARE-binding protein Pub1. Since a relatively small number of genes was affected by Hog1p deletion, additional signaling pathways are likely to be involved in coordinating the translational response to severe salinity stress.
Keywords: mRNA, translation, microarray, yeast, salinity stress, potentiation
INTRODUCTION
Eukaryotic cells have the ability to respond to various changes in their surrounding environment by changing their protein repertoire. These responses are often elicited by signal transduction pathways and involve both transcriptional and post-transcriptional regulatory mechanisms. One of the most important regulatory mechanisms at the post-transcriptional level involves the control of protein synthesis. Regulation at this step is usually manifested by a global effect in which the synthesis of almost all proteins is affected similarly and involves a specific effect in which a few key factors demonstrate a unique response. For example, the translational response of yeast cells to various stress situations was shown to consist of a global inhibition effect on the synthesis of most proteins (Tzamarias et al. 1989; Fuge et al. 1994; Barbet et al. 1996; Ashe et al. 2000, 2001; Kuhn et al. 2001; Uesono and Toh 2002; Shenton et al. 2006) and a specific effect in which the translation of key proteins was maintained and even induced (Kuhn et al. 2001; Smirnova et al. 2005; Shenton et al. 2006). Reducing global translation in response to different stress situations usually involves pathways that regulate the activity of general factors (such as eIF2α or eIF4E). However, the cellular mechanisms underlying the differential translation response, where some specific mRNAs do not follow the general trend, are poorly understood. One suggested regulatory mechanism stems from the observed correlation between changes in translation and changes in transcription of particular genes (Preiss et al. 2003; Smirnova et al. 2005; Shenton et al. 2006). In this model, gene-specific transcription potentiates the mRNA and makes it prone to a similar translational regulation. Such potentiation may occur if, for example, regulatory proteins are deposited on the mRNA by the transcription machinery (Lotan et al. 2005) or if an alternative transcription start site is selected (Law et al. 2005).
High salinity stress affects cellular physiology at multiple levels: it creates a hyperosmotic force that leads to water efflux and consequently to loss of internal pressure (Hohmann 2002); it imbalances membrane potential and thereby affects the activity of membrane transporters (Norbeck and Blomberg 1998); it disrupts ion homeostasis within cells and intracellular pH equilibrium, which result in misfolding of proteins and the generation of reactive oxygen species (ROS) (Mendoza et al. 1994; Lahav et al. 2004; Koziol et al. 2005; Mortensen et al. 2006); finally, it causes a solute-specific toxic effect. For example, sodium or lithium ions, but not potassium, inhibit the phosphatase activity of Hal2p and, thus, reduce the ability of cells to resist high salinity (Murguia et al. 1995, 1996).
The cellular response to high salinity was studied extensively and serves as a model for gene-expression changes in response to external stimuli. The response is mediated by several stress responsive signaling pathways, of which the high osmolarity glycerol (HOG) mitogen activated protein kinase (MAPK) pathway plays a major role. This pathway is involved in sensing the increase in turgor pressure and transducing the appropriate signals to the gene-expression program (Hohmann 2002). Changes at the transcriptional level usually occur following phosphorylation of the terminal MAPK in this pathway (Hog1p), which regulates the activity of several transcription factors, including Msn2/Msn4, Msn1, Hot1, and Sko1 (Proft and Serrano 1999; Rep et al. 1999b, 2000).
While the transcriptional response to high salinity is well studied (Posas et al. 2000; Rep et al. 2000; O'Rourke and Herskowitz 2004; Gat-Viks and Shamir 2007), changes at the translational level are not fully characterized. In response to high salinity (NaCl), yeast cells transiently inhibit the rate of protein synthesis but resume translation at normal levels following an adaptation period, the duration of which correlates with the severity of the saline stress (Uesono and Toh 2002). The recovery of the translation system, but not the initial inhibition of protein synthesis, requires a functional HOG signaling pathway (Uesono and Toh 2002). Intriguingly, under severe high salt stress Hog1p was shown to retain in the cytosol in an activated form (Van Wuytswinkel et al. 2000), thus suggesting a role in cytosolic functions.
Here, we performed a genome-wide analysis of the translational response of yeast cells to high salinity stress, either in the presence or absence of the Hog1 protein. In the presence of Hog1p, we observed a significant change in the translation of many mRNAs involved in the cellular response to various stresses. Many of these genes were also affected at their mRNA levels, consistent with a potentiation process involving Msn2/Msn4 transcription factors. Interestingly, the translation of only a small fraction of these mRNAs appeared to be dependent on Hog1p. A relatively large fraction of the affected mRNAs contain within their UTRs sequence motifs that were shown previously to be associated with the ARE-binding protein Pub1p.
RESULTS AND DISCUSSION
Ribosomal association following high salinity stress
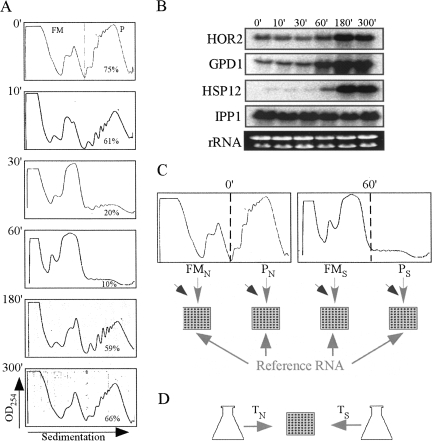
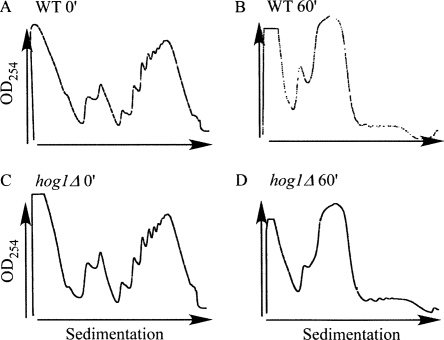
Following exposure to high salinity stress conditions, yeast cells experience a strong yet transient reduction in the number of translating ribosomes (Fig. 1; Uesono and Toh 2002). We performed a time course analysis in which yeast cells grown in optimal conditions were shifted to a medium containing 1 M NaCl for 0, 10, 30, 60, 180, and 300 min. Cells were harvested at these time points, and the sedimentation pattern of polysomal complexes was determined (Fig. 1). Within 1 h of the application of high salinity stress, the percentage of signal in the polysomal region dropped from 75% to 10%. This strong inhibition of protein synthesis is merely temporal, as the cells replenish their polysomal pool to almost normal levels within 5 h after administration of NaCl. Based on these ribosomal association parameters, one can suggest that the translation machinery exhibits the most drastic change at the 1-h time point and is fully adjusted to the new conditions at the 5-h time point. Interestingly, under these conditions, the steady-state mRNA levels for at least three salt-stress related mRNAs (Fig. 1B) started increasing at the 1-h time point and reached a maximal level at the 3-h time point. This is consistent with previous studies that revealed that higher salt concentrations elicit a delayed transcriptional response (Rep et al. 1999a; Posas et al. 2000). Thus, at the 1-h time point genes are likely to be strongly affected at the translation level and to a much lesser extent at their transcript levels. Since our main interest herein is the effect at the translation level, we used this time point in all further experiments. Moreover, at high NaCl concentrations the main regulator of the salinity response (Hog1p) was shown to be retained in the cytosol in its active form for at least 45 min (Van Wuytswinkel et al. 2000). Thus, if the HOG pathway plays a direct role in translation through the Hog1p, it is likely to be apparent in these conditions, in which Hog1p is cytosolic.
FIGURE 1.
Time course analysis and experimental strategy used to study the translational response to high salinity. (A) Cells grown in optimal medium (YPD) to the midlogarithmic phase were subjected to 1 M NaCl stress for the indicated times. Cells were harvested and complexes were separated by velocity sedimentation in sucrose gradients. The sedimentation profile of ribosomal complexes was determined by measuring the OD254 along the gradient. The dashed line indicates the partition line between mRNAs free of ribosomes or associated with monosome (FM) and mRNAs associated with polyribosomes (P). The percent of signal obtained in the P region is indicated in each panel. (B) Northern analysis of changes in transcript levels following exposure to 1 M NaCl. Cells were harvested at the indicated time points (identical to the time points in the polysomal analysis, panel A). RNA was extracted from the samples and subjected to Northern analysis using the indicated probes. (C) Scheme of the procedure for translational profiling. Sucrose density gradients from time points 0 min (normal, N) and 60 min (salt, S) were fractionated to free and monosomal (FM) or polysomal (P) mRNA. Each pool was converted to Cy5-labeled cDNA, mixed with Cy3 labeled unfractionated RNA from an unrelated sample (reference RNA), and hybridized to a DNA microarray. Slanted arrows represent the spike-in RNA that was added to each sample in equal amounts. (D) Scheme of the transcriptome analysis. Unfractionated RNA was extracted from these two time points (TN, TS), labeled differentially, and hybridized together to a DNA microarray. Note that the same cell culture was used for both C and D in order to reduce technical variation.
Changes in the yeast transcriptome following high salinity stress
Large-scale analyses of changes in steady-state mRNA levels following high salinity stress have demonstrated that the transcriptome response to this environmental stress is highly dynamic and varies greatly with time, salt type and concentration, and yeast strain used (Garcia et al. 1997; Posas et al. 2000; Yale and Bohnert 2001; O'Rourke and Herskowitz 2004; Hirasawa et al. 2006; Gat-Viks and Shamir 2007). To establish the salt response of our strain (BY4741), we isolated RNA samples from cells at time zero or after 1 h of stress and used them for microarray analysis. The samples were differentially labeled and hybridized together to a DNA microarray containing ∼6000 known and predicted yeast ORFs. The experiment was repeated three times, and the results per gene are the average of at least two out of three independent experiments (4789 genes) (Supplemental Table. 1). Following 1 h in high salinity growth conditions, the relative abundance of transcripts representing 124 genes was induced by more than 1.5-fold. Of these, 58 genes (∼47%) are of an unknown biological process according to the Saccharomyces Genome Database (SGD). Only nine genes (YGR243W, GPD1, BTN2, HSP42, YDL023C, HSP12, CUP1-2, SPI1, and STL1) showed a relative increase of more than twofold in their steady-state mRNA levels, and all were previously reported to be induced in response to high salinity (Posas et al. 2000; Yale and Bohnert 2001; Hirasawa et al. 2006). The fact that only a few genes showed more than a twofold increase in mRNA levels is in agreement with reports describing a much reduced transcriptional changes in response to severe osmotic stress compared to mild stress conditions (Fig. 1B; Rep et al. 1999a; Van Wuytswinkel et al. 2000). A decrease in relative transcript abundances was more significant, and 205 ORFs showed more than a twofold reduction. Analysis of the biological process they are involved in by SGD gene ontology (GO) term analysis revealed that ∼40% are involved in the translation process (P value = 1.06 e−37) and many encode for ribosomal proteins (RP). A similar decrease in RP mRNA levels following salt stress was previously observed (Gasch et al. 2000; Rep et al. 2000; Causton et al. 2001; O'Rourke and Herskowitz 2004).
Translational response to high salinity
To study whether mRNA molecules are translated differentially in response to high salinity, we fractionated yeast cells either before or after 1-h exposure to 1 M NaCl into two fractions: nonpolysomal mRNA, representing mRNA that is free or monosomal (FM), and polysomal mRNA (P) that contained two or more ribosomes (Fig. 1). The monosomal fraction was pooled with the free sample because this population of mRNAs is unlikely to represent efficiently translated mRNAs. RNA was purified from each fraction and converted into Cy5-labeled cDNA. An unfractionated reference RNA sample was converted into Cy3-labeled cDNA and hybridized with all samples, thereby serving as a common reference. This experimental procedure was repeated three times using three independent polysomal preparations. Hybridization results provided the relative abundance of each mRNA within each fraction. Notably, to be able to determine the mRNA distribution across a gradient, we added an equal amount of spike-in mix (normalization mix) composed of five different exogenous mRNAs to each fraction. These exogenous mRNAs had at least 100 complementary spots on the DNA microarray. Since the spike-in mix was added to each fraction immediately after collection, any variation in RNA processing between the FM and P fractions (e.g., RNA purification, reverse transcription, Cy labeling and microarray hybridization, and scanning efficiencies) was represented and corrected by differences in the spike-in signals between the two fractions.
The normalized results were used to study the overall translational trend and to identify genes with significant changes in their polysomal status in response to high salinity. We first describe the global trends observed and then discuss the specific groups of genes that deviated from these global trends.
Global changes in yeast ribosomal association following high salinity stress
The experimental design for ribosomal association results in two values per gene: the signal in the nonpolysomal fraction (free and monosomal [FM]) and the signal in the polysomal fraction (P). Both signals were obtained in the format of a ratio between the FM or P fractions and the reference RNA sample. Three criteria were used to analyze the changes in mRNA translation between salt stress (S) and normal (N) conditions.
(1) Change in the polysomal fraction after salt stress (PS/PN). This parameter represents the relative change in mRNAs that are associated with ribosomes and presumably engaged in translation. Low values in this parameter may indicate a strong shift of mRNA from polysomes to the free mRNA fraction. Our experimental data indicate an average decrease of ∼3.5-fold in polysomes for all mRNAs, which is consistent with the strong decrease in the OD254 of the gradient (Fig. 1). This decrease was very similar for most mRNAs, as the range from the 5th to the 95th percentile of PS/PN is 0.2–0.45.
(2) Change in the nonpolysomal fraction (FMS/FMN). This parameter represents the relative change in untranslated mRNAs. Higher values represent accumulation of mRNAs in the FM fraction after salt stress, and therefore may suggest a decrease in translation. Our experimental results indicate accumulation of mRNA in the FM fraction (FMS/FMN > 1) for ∼60% of the genes. In comparison to the small variation in PS/PN, the range of changes in the FM was much larger (5th to 95th percentile range of 0.4–1.7). This difference may be a result of changes in mRNA transcription or degradation that are likely to be apparent in the FM fraction (discussed below). Note that the PS/PN and FMS/FMN parameters do not necessarily complement each other. For example, an mRNA that changed from 99% to 98% in polysomes will have a negligible change in the polysomal fraction (PS/PN), yet the change in FM (FMS/FMN) will seem significant (from 1%–2%, or twofold) (Melamed and Arava 2007).
(3) Change in the ratio of polysomal to free and monosomal mRNA [(P/FM)S/(P/FM)N]. This value is derived from the above two parameters and represents the distribution of mRNAs within the gradient. Using this criterion, we observed that practically all genes (98%) had a lower P/FM after the salt stress [(P/FM)S/(P/FM)N < 1] with the vast majority (86%) presenting a reduction of more than twofold.
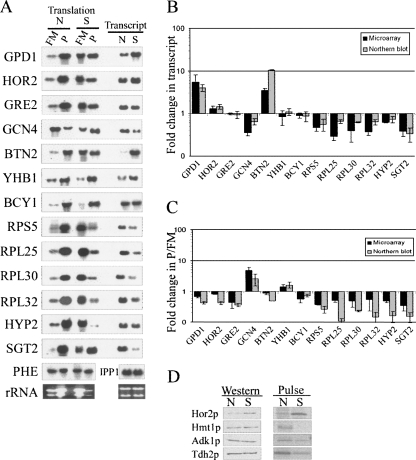
We used Northern blot analysis for a variety of mRNAs to confirm the microarray results in terms of changes in mRNA abundances and mRNA distributions between the P and FM fractions (Fig. 2). Overall, we observed a good correlation between the two methods with respect to changes in total mRNA levels and P/FM values. The slight differences in the extent of changes are probably due to the different normalization strategies used for each method (see Materials and Methods).
FIGURE 2.
Comparison between microarray and Northern results of several candidate genes. (A) Sucrose gradients from cells grown in normal (N) conditions (i.e., YPD) or after 60 min growth in 1 M NaCl (S) were fractionated to two fractions representing free and monosomal (FM) or polysomal (P) mRNA. RNA samples were subjected to Northern analysis using probes recognizing the indicated mRNAs. Transcript levels comparison (right panels) was done by analyzing 5 μg of unfractionated RNA collected from either N or S conditions. (B) Quantified signals from three biological replicates (either microarray [black bars] or Northern [gray bars]) were used to determine the average fold change in steady-state transcript levels for the indicated genes. IPP1 probe was used to normalize for loading variations because it shows no significant change following high salinity stress (Rep et al. 1999a; this study). (C) Quantified signals from three biological replicates (either microarray or Northern) were used to determine the average change in P/FM ratios for the indicated genes. Signals were normalized to an mRNA spike (PHE) that was added in equal amounts to each fraction immediately at collection. (D) Western analysis and pulse labeling of TAP tagged proteins. For Western analyses, the indicated strains were grown in optimal media (YPD) and shifted for 1-h growth to medium containing 1 M NaCl. Protein extracts were prepared from the cells, and equal amounts were subjected to Western analysis using peroxidase anti peroxidase antibody. For pulse labeling, cells were grown overnight in minimal medium with only the necessary amino acids but without methionine. Half of the cells were shifted to a medium supplemented with 1 M NaCl for another 1 h of growth. 35S-methionine was added to the cells 20 min before harvesting. Labeled proteins were immunopercipitated using IgG-sepharose beads, resolved on a gel, and exposed to a PhophorImager. N indicates no salt stress and S indicated salt stress.
To test if the changes in ribosomal association are reflected in changes in protein levels, we performed Western analysis for several representative genes. To avoid effects that are due to changes in transcript levels, we selected genes that displayed insignificant changes at the transcript level. Strains expressing TAP-tagged HOR2 or HMT1 (representing genes with relatively high ribosomal association after the treatment) and TDH2 or ADK1 (low ribosomal association) were subjected to 1-h treatment with NaCl, and protein extracts were taken for Western analysis (Fig. 2D). Western analysis revealed that Hor2p levels increased by 2.6-fold, suggesting an induction of protein synthesis. The seeming discrepancy between the slight decrease in ribosomal association of HOR2 (change in P/FM = 0.8) and the increased protein synthesis rates is attributed to the difference in growth medium; when cells were grown in minimal medium (as in the case of the protein analysis) HOR2 mRNA showed a significant increase in ribosomal association (data not shown). Hmt1p levels, as well as the levels of Adk1p and Tdh2p, were similar before and after the salt treatment. We reasoned that the lack of apparent changes in protein levels as determined from Western analyses might be due to the stability of these proteins, which would obscure changes in protein synthesis rates. We therefore pulse-labeled cells with 35S methionine for the last 20 min of exposure to the stress and utilized the TAP tag to perform immunoprecipitation (IP) of the newly synthesized proteins. As can be seen in Figure 2D, Hor2p has a higher synthesis rate than Adk1p and Tdh2p after the 1-h salt shock, consistent with the differences in polysomal association found for these genes. For Hmt1p, however, there is no detectable signal after the treatment, indicating that it is not synthesized under these conditions. The discrepancy between the changes of HMT1 ribosomal association and its apparent rate of protein synthesis may be due to differences in the growth media used for the two assays. Indeed a difference in growth media was found to affect HOR2 since its ribosomal association was even higher when the salt treatment was performed with minimal medium (data not shown). We were unable to obtain satisfactory Northern data for the tagged HMT1 when cells were grown in minimal medium; therefore, the possibility that changes in transcript levels occurred also for this gene cannot be excluded. Alternatively, these data may indicate that HMT1 is regulated at other stages of translation. For example, the elongation factor EF2 was shown to be highly phosphorylated after salt treatment (Teige et al. 2001), which suggests regulation during the elongation step. Overall, this limited pulse-labeling analysis indicates that differences between mRNAs in their ribosomal association are usually, but not always, reflected at their protein synthesis rates. Proteomic studies are necessary to fully investigate the extent of concordance between ribosomal association and protein synthesis rates, and these may reveal additional layers of expression regulation.
Global relationships between changes in transcript levels and polysomal association
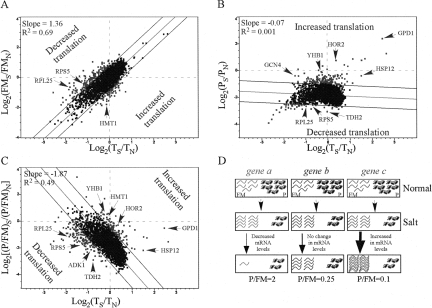
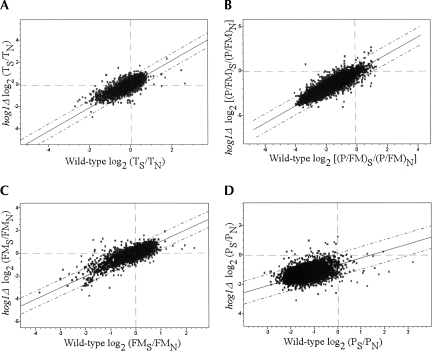
To examine the relationships between changes in translation and transcript levels, the values obtained for each of the above parameters were compared with the data regarding changes in steady-state mRNA levels. For each gene, the PS/PN, FMS/FMN, or (P/FM)S/(P/FM)N values from at least two experimental repeats were averaged (Supplemental Table 2), and each of the averaged values was plotted against the changes in transcript abundances (average of at least two biological replicates) (Fig. 3). Interestingly, changes in the FMS/FMN values appeared to have a strong positive correlation with changes in mRNA transcript abundances (TS/TN) (Fig. 3A). Changes in polysomal association (PS/PN), however, had no simple correlation with TS/TN (Fig. 3B). A reasonable explanation for these observations might be that that the strong reduction in translation affects most of the genes similarly (i.e., their polysomal fraction decreases similarly), and changes in mRNA transcript levels that result from altered mRNA transcription or decaying rates occur mainly in the FM fraction. Consistent with this explanation, the change in P/FM ratio following high salinity stress shows an inverse correlation with the change in mRNA levels (Fig. 3C).
FIGURE 3.
Graphical representations of the relationships between translational changes and changes in the transcriptome. (A–C) Changes in ribosomal association following salt stress, calculated as [log2(FMS/FMN)](A), [log2(PS/PN)] (B), and (log2[(P/FM)S/(P/FM)N]) (C), were plotted against their respective changes in transcript levels (TS/TN). The best-fit linear trend lines are marked, and their slope and R 2 values are indicated. Dashed lines represent two standard deviations (2 SD) from the trend line. Genes that were tested by Northern analysis and disussed in the text are indicated by arrows. (D) A model explaining the inverse relationship between the global translational response and the transcriptome response. (Top) The translational status of three representative genes. For simplicity, at optimal growth conditions, five mRNA molecules of each gene are free of ribosomes (FM) and five are associated with translating ribosomes (P), thereby yielding a P/FM of one. (Middle) Following 1 h of high salinity stress, mRNA molecules are shifted from the P pool to the FM pool in an identical manner for all genes. (Bottom) Concomitant with the translational change or soon after, changes in mRNA transcript levels take place only within the FM fraction. For some genes, a fast degradation (left) occurs, leading to an apparent low P/FM. For other mRNAs (middle), no change occurs in transcript levels. Finally, transcription induced for some genes may lead to an increase in mRNA levels and a decrease in P/FM (right). Thus, although the shift from P to FM was similar to all genes, the changes in mRNA levels, which are reflected mainly in the FM fraction, lead to the apparent inverse correlation between changes in T and changes in P/FM.
Based on these observations, we propose a model that links the general changes in the transcriptome to the changes in translation following high salinity stress (Fig. 3D). According to this model, high salinity stress results in a transfer of mRNA molecules from the polysomal fraction to the FM fraction. This strong shift is similar for most genes (three molecules per gene in this example). Concurrent with or soon after the drop in polysomal mRNA, changes in steady-state mRNA levels that include changes in both transcription and decay rates occur mainly in the FM fraction. Accordingly, a decrease in mRNA levels will be manifested mainly in the FM pool (Fig. 3D, gene a), while those mRNAs still engaged with translating ribosomes will be less accessible for the mRNA decaying machinery (Parker and Sheth 2007). An increase in mRNA abundance (Fig. 3D, gene c) results in accumulation in the FM fraction with little or no increase in its translation. Thus, as illustrated in Figure 3D, changes in steady-state mRNA levels are inversely correlated with changes in P/FM values.
The main implication of these results is that, in general, an increase in mRNA abundance (either from lower decay rates or increased transcription) does not necessarily lead to an immediate increase in protein synthesis levels. Rather, many of these mRNAs are accumulating in the FM fraction, probably in P-bodies, and serving as an mRNA pool to be translated during the subsequent recovery of the translation apparatus (Brengues et al. 2005). This conclusion is relevant for most genes in yeast responding to high salinity stress. Yet, as will be elaborated below, some genes do not follow this general trend and the increase in their transcript levels do correlate with changes in their translatability.
Genes that deviate from the global translational trend
As illustrated in Figure 3, most of the genes analyzed followed a common translational response with respect to changes in mRNA transcript levels. To identify gene candidates having a different translational response, we selected genes that deviated by more than two standard deviations from the overall linear trend lines (Fig. 3A,B,C, dashed lines) in at least one of the three criteria used. Since any biased change in FM or P necessarily affects the change in P/FM, we increased the stringency of selecting genes by the FMS/FMN or PS/PN criterion by filtering in only those genes that showed a change also in (P/FM)S/(P/FM)N. Note that genes were selected as translationally regulated if they deviated from the global translational response. Thus, some genes might show an increase in their P/FM ratio (e.g., Fig. 2A, GCN4,YHB1; Fig. 3C, genes above zero with respect to the Y-axis) and others might display a decrease in their P/FM values (e.g., Fig. 2A, GPD1,HOR2,BTN2; Fig. 3C, genes below zero with respect to the Y-axis), yet all will be considered as translationally up-regulated since they all show a significant increase in translation with respect to the general trend line.
Translationally upregulated genes
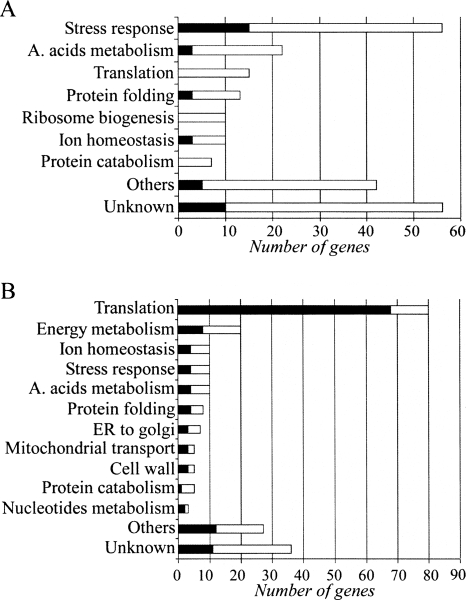
A total of 201 genes were assigned as translationally up-regulated following high salinity stress (the full list appears in Supplemental Table 3). These genes were further grouped into functional categories using the SGD GO Slim Mapper tool, the phenotypic profiles database (Prophecy) (Fernandez-Ricaud et al. 2007), and the literature search (Fig. 4A; Supplemental Table 3).
FIGURE 4.
Bar diagrams of translationally regulated genes grouped by their common processes. The bars indicate the total number of translationally induced genes (A) or translationally repressed genes (B). The black section in each bar indicates the proportion of genes that are within the top 5% of genes having the highest change in transcript levels.
Stress responsive genes
Fifty-six genes (∼27%) are involved in different stress responses. Of these, 27 genes were previously reported to be involved in the cellular response to high salinity. These include genes whose contribution to high salt resistance is known, such as GPD1, HOR2, RHR2, and STL1 that are involved in the biosynthesis and transport of glycerol to counterbalance the high osmotic pressure (Albertyn et al. 1994; Pahlman et al. 2001; Ferreira et al. 2005), and genes whose contribution to high salt resistance is unclear but their expression was shown to be required for resistance (such as HMT1, ERG6, IMD4, NCL1, PMP1, and YNL168C) (Fernandez-Ricaud et al. 2007). The other stress genes identified were shown to be involved in stress responses such as oxidative stress (GRX2, NCE103, PST2, and YHB1), pH tolerance (BTN2, CHS7, CYS4, KRE1, LYP1, SBE22, VMA6, SLT2, and ORM2), the unfolded protein response (HSP104, HSP42, SEC61, FES1, ZUO1, SSA4, and SIS2), and cellular detoxification (AQR1, PDR15, and YGR035C), which are probably secondary outcomes of high salinity.
Signaling
Four genes, BCY1, TIP41, DBF2, and CBK1, which are involved in cell signaling, appeared to be up-regulated by salt stress. Bcy1p is the regulatory subunit of the cAMP-dependent protein kinase (PKA). Binding of cAMP to Bcy1p releases it from the catalytic subunit, thereby increasing its kinase activity. PKA signaling acts as a negative regulator in most types of stress. Consistent with this, a deletion mutation in BCY1 or overexpression of the PKA catalytic subunit reduces yeast viability following high salinity shock (Norbeck and Blomberg 2000). The observed up-regulation of Bcy1p (Fig. 2) might promote yeast survival by increasing its association with PKA catalytic subunits and, therefore, inhibit its activity.
Tip41p is involved in the TOR (target of rapamycin) signaling pathway, and its expression was recently shown to be required for the accumulation of the transcription factor Msn2 in the nucleus in response to different stress conditions (Santhanam et al. 2004). Therefore, induction of TIP41 mRNA translation might be required for optimal accumulation of Msn2p in the nucleus.
Dbf2p and Cbk1p Ser/Thr kinases are two (out of three) members of the S. cerevisiae NDR (nuclear Dbf2-related) family. Both proteins regulate processes that are linked to the exit from mitosis (Hergovich et al. 2006). Indeed, high salinity stress causes a shift from mitosis to the G1 stage, and this transition is partly controlled by the HOG signaling pathway (Reiser et al. 2006). The importance of Dbf2p for salt resistance is emphasized by the strong sensitivity of the dbf2-deletion mutant to high salinity (Lee et al. 1999; Warringer et al. 2003). Thus, these proteins may provide the molecular link between salt sensitivity and the cell cycle effect.
Amino acids
Our analysis indicates that high salinity activates the translation of GCN4, a transcription factor that mediates the cellular response to amino acid starvation, together with its coactivator MBF1. Induced translation of GCN4 following NaCl stress was previously observed and shown to cause sodium sensitivity (Goossens et al. 2001). Surprisingly, though GCN4 translation was induced following NaCl treatment, its mRNA level appeared to be strongly reduced (Fig. 2). These opposite responses might reinforce the suggestion by Alain Goossens et al. that GCN4-induced translation following high salinity stress results from the toxic effect of sodium ions that activate the Gcn2p-dependent phosphorylation of eIF2α. Therefore, reducing GCN4 mRNA steady-state levels might be a cellular response used to counterbalance the negative effect of its induced translation. Indeed, the effect of GCN4-induced translation on the transcriptional activation of genes involved in amino acid biosynthesis was moderate, since, of the top 20% of genes that demonstrated increased mRNA levels (958 genes), only 24 were associated with amino acid metabolic processes. Interestingly, one third of these genes are linked to arginine metabolism (ARG2, ARG3, ARG5, ARG6, ARG80, ARG81, ARG82, ECM40, CAR1, BTN2). Accumulation of arginine, a positively charged amino acid, was previously shown to render cells more tolerant to freezing (Takagi et al. 1997; Morita et al. 2002), and resistance to these stress conditions was shown to be related to hyperosmotic adaptation (Panadero et al. 2006). Only two of these genes (CAR1 and BTN2) were induced also at the translation level. These two genes act to reduce the levels of arginine within cells, while the other genes are involved in arginine biosynthesis. Therefore, regulation of arginine levels in response to high salinity may play a defensive role and probably relies on differential expression of genes involved in arginine metabolism.
Genes with induction of transcript levels and of polysomal association
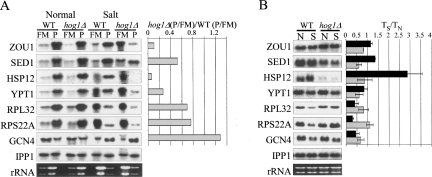
A unidirectional change in transcript levels and in protein synthesis (termed potentiation) (Preiss et al. 2003) was reported to occur following heat shock, oxidative stress, rapamicin treatment, and amino acid starvation (Preiss et al. 2003; Smirnova et al. 2005; Shenton et al. 2006). We found that 12.5% (25 genes) of the genes that were assigned as translationally induced also showed a significant increase in their mRNA levels (ranked in the top 5% of genes), i.e., have a positive potentiation trend (Fig. 4A). Interestingly, this group was enriched with genes that contain binding sites to the stress-response transcription factors Msn2/Msn4 (ALD3, AST1, DDR2, GPD1, HSP12, FMP12, HSP42, HSP104, MBF1, MSC1, and STL1) (P value = 5 e−8) (Shamir et al. 2005) and with other experimentally verified Msn2/Msn4 targets (BTN2, NCE103, MAS1, SPI1, and YHR033w) (Teixeira et al. 2006). We, therefore, hypothesized that the molecular basis underlying the common effects at the transcriptional and translational levels involves these factors. To further explore this possibility, we tested the steady-state levels and polysomal distributions of several of these Msn2/Msn4 targets (GPD1, MBF1, HSP12, and BTN2) in a strain containing deletions of these factors (Fig. 5). Indeed, after high salinity stress, both the steady-state levels (Fig. 5B) and the polysomal association (Fig. 5A) of all four mRNAs showed a clear decrease in comparison to the parental strain. This effect was not apparent for GCN4 mRNA, which is not regulated by Msn2/Msn4. These results reveal the involvement of Msn2/Msn4 in coupling changes in transcription and translation of specific genes following high salinity stress.
FIGURE 5.
Effect of deletion of MSN2/MSN4 genes. (A) Translational response. Yeast strains either carrying (WT) or deleted (Δ) of MSN2/MSN4 genes were grown for 60 min in YPD supplemented with 1 M NaCl, and then subjected to polysomal analysis. RNA samples from FM and P fractions were used for Northern analyses using the indicated probes. The histograms present the ratio between the P/FM of the deletion strain and the normal strain after normalization according to the signals of the PHE mRNA. (B) Transcript changes. RNA samples (5 μg each) from wild-type (WT) or Msn2Δ/Msn4Δ (Δ) cells grown either in normal (N) or salt (S) conditions were subjected to Northern analysis. Black and gray bars represent the ratio of S to N signals in the WT and deletion strain, respectively. Loading variations were corrected according to the IPP1 signals.
Translationally down-regulated genes
Two hundred and twenty-five genes showed a significant decrease in mRNA translation. Of these, ∼36% encode for ribosomal proteins or are associated with other aspects of the translation machinery and ∼9% are involved in energy metabolism (Fig. 4). Additional repressed targets include genes involved in cell-wall metabolism (CWP2, GUK1, PSA1, PIR1, and CCW14), ion homeostasis (VMA4, VMA13, MIR1, PHO84, CUP5, and PDR5), and nucleotide metabolism (ADK1, URA5, and RNR2). These genes are closely linked to cell growth and proliferation processes, and their transcripts are highly abundant in cells grown at optimal conditions (Wang et al. 2002). Since yeast cells experience growth arrest following high salinity stress, it is conceivable that the synthesis of proteins involved in cell growth and metabolism will be reduced.
Genes with a reduction of transcript levels and of polysomal association.
The potentiation effect also occurs in order to repress gene expression under high salinity stress (“negative potentiation”). About 55% (123 genes) of the genes found to be translationally reduced also had a significant decrease in their mRNA abundance, i.e., were found in the top 10% of genes with reduced mRNA levels. Importantly, almost all of the RP genes presented this effect (Fig. 4B), namely, a significant decrease in their polysomal pool and in their transcript levels. The fact that relatively large amounts of these mRNAs are found in the FM pool even after considerable reduction in their levels might suggest that these transcripts serve as a reservoir of mRNAs waiting to be translated following recovery from the shock. A possible means for retaining these transcripts as a translationally inactive pool is through interactions with P-bodies. These cellular compartments were shown to accumulate nontranslating mRNAs in cells undergoing various stresses and to allow their movement back into polysomes once stress is relieved (Brengues et al. 2005). Alternatively, since many of these mRNAs are found in many copies per cell, this negative potentiation effect might not be a regulated response of the cells to stress. Rather, it might represent the result of an incomplete mRNA degradation process, as a large amount of these mRNAs shifted from the P to the FM fraction.
Transcriptome response of hog1 deleted cells
The Hog1 MAPK pathway is a major modulator of the gene expression response to salt stress. Activation of this pathway leads to transcriptional induction of many genes involved in the cellular response to high salinity (Hohmann 2002). However, while following a mild osmotic shock, phosphorylated Hog1p rapidly translocates to the nucleus and exerts its function on transcription, severe osmotic stress results in delayed nuclear accumulation of Hog1p and, consequently, a decreased transcriptional response after 60 min (Rep et al. 1999a; Van Wuytswinkel et al. 2000).
To identify the genes that may be transcriptionally affected by Hog1p, we compared changes in the transcriptome of hog1Δ and wild-type cells (Fig. 7A; Supplemental Tables 4, 5). Almost all of the 126 genes that showed induced mRNA levels in wild-type cells (more than 1.5-fold induction) were induced also in the deletion strain. Therefore, under this harsh stress condition, the increase in transcript levels appears to be controlled by pathways other than the HOG signaling, as was suggested previously (O'Rourke and Herskowitz 2004).
FIGURE 7.
Comparisons of the transcriptome and translational response of hog1Δ and wild-type strains to high salinity. (A) Average values of changes due to salt stress in the hog1Δ transcriptome are plotted against the wild-type average change. The best-fit linear trend line and two SD lines (dashed lines) are shown. (B–D) Comparison of hog1Δ and wild-type changes in P/FM (B), FM (C), and P (D) due to salt stress. The results from one representative experiment are shown. Genes that deviated by more than two SD from the trend line were considered as translationally misregulated in hog1Δ cells. Note that for the analysis presented in the Results, genes were further filtered based on the results from the other two experiments (see Materials and Methods for filtration details).
In contrast to the similar induction profile, the reduction in mRNA levels of many genes was not as strong as in the wild-type strain; about 30% of the 205 genes that showed reduced mRNA levels (greater than twofold reduction) in the wild-type strain were less affected in the hog1Δ. Many of the nonreduced genes encode for ribosomal proteins (e.g., , see below, Fig. 8B, RPL32,RPS22A); this is in agreement with a previous observation demonstrating that Hog1p strongly affects the reduction in mRNA levels of RPL27B and RPL28 (CYH2) (Uesono and Toh 2002). Thus, under severe salt stress conditions Hog1p down-regulates the mRNA levels of ribosomal proteins by reducing their transcription levels or accelerating their decay. We speculate that the latter is more likely because Hog1p is mostly cytosolic under this condition.
FIGURE 8.
Validation of hog1Δ translationally misregulated genes by Northern analysis. (A) Sucrose gradients from wild-type (WT) or hog1Δ cells grown in rich medium (normal) or after 60 min growth in 1 M NaCl (salt) were fractionated to free and monosomal (FM) or polysomal (P) mRNA. RNA samples were subjected to Northern analysis using the indicated probes. The histogram presents the ratio between the P/FM of the deletion strain and the normal strain after normalization according to the signals of the PHE mRNA. (B) Transcript changes. RNA samples (5 μg each) from wild-type or hog1Δ cells grown either in normal or salt conditions were subjected to Northern analysis. Black and gray bars represent the ratio of S to N signals in the WT and deletion strain, respectively. Loading variations were corrected according to the IPP1 signals.
Hog1p-dependent translation
To examine whether the HOG pathway is involved also in translation regulation following salt stress, we compared the mRNA polysomal distributions of hog1Δ cells that were subjected to identical stress conditions. Following 1 h in the presence of 1 M NaCl, the polysomal profile of the hog1 deletion mutant was essentially the same as that of the wild-type strain (Fig. 6), though a longer incubation time resulted in a more severe reduction of its polysomes compared to wild-type cells (data not shown). DNA microarray analyses for FM or polysomal fractions collected from the gradients were performed essentially as described for the wild-type strain. To select candidates for translational regulation by Hog1p, values representing changes in hog1Δ FM, P and P/FM ratios from each biological replicate were plotted against their equivalent changes in wild-type cells (Fig. 7B,C,D). Genes were considered to be translationally misregulated if their change in hog1Δ P/FM ratio deviated from the change in wild-type P/FM ratio by more than two standard deviations (SDs) in one experiment and more than one SD in at least one of the other two biological replicates. We further enhanced the stringency of filtration by considering only genes that showed also significant deviations from the wild-type strain in FMS/FMN or PS/PN values. Using this approach, we identified 99 genes that showed reduced translation levels in hog1Δ cells when compared to the wild-type strain response (Supplemental Table 7). We verified the reduced translation of several mRNAs (SED1, ZUO1, YPT1, HSP12, RPL32, and RPS22A) using Northern blot analysis (Fig. 8). Overall, the Northern data confirmed that these mRNAs are affected by hog1 deletion at the translational level.
FIGURE 6.
Polysomal profiles for wild-type and hog1Δ cells. (A) Wild-type cells grown under normal conditions (before shift to salt conditions). (B) Wild-type cells after 60 min of 1 M NaCl stress. (C) hog1Δ cells grown at normal conditions. (D) hog1Δ cells after 60 min of 1 M NaCl stress.
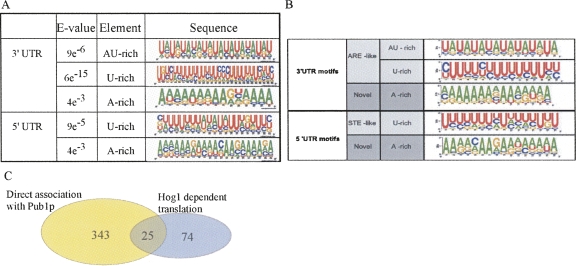
Sequence analysis (Bailey et al. 2006) of the 5′ and 3′ UTRs of the group of genes that showed stronger translation repression compared to the wild-type strain response revealed several statistically enriched motifs (Fig. 9A), that were not detected in a group of nonaffected genes. These motifs contain A-rich, U-rich, or A:U-rich (ARE) sequences and are highly similar to elements that were found as binding sites for the ARE-binding protein Pub1 (Fig. 9B; Duttagupta et al. 2005). Application of the Tomtom algorithm (Gupta et al. 2007) to measure the statistical similarity between a Pub1 motif and its corresponding Hog1 motif revealed for all pairs a significant similarity (E values were <10−4 for all pairs and the lowest was for the U-rich motifs in the 3′ UTRs [E = 10−8]). Moreover, a significant fraction of the mRNAs that were found to be translationally affected by deletion of the HOG1 gene were shown previously to be associated with Pub1p (Fig. 9C; Duttagupta et al. 2005). These data may suggest that Hog1-dependent regulation of many mRNAs is mediated by Pub1p binding to specific sequence motifs in the UTRs. Consistent with this hypothesis, Hog1p has previously been shown to regulate the translation of an ARE-containing mRNA in a manner that necessitates Pub1p (Vasudevan et al. 2005).
FIGURE 9.
Sequence analysis of genes that are translationally regulated by Hog1. (A) Sequences upstream (100 nt) or downstream (200 nt) of each of the affected ORFs were used for classification by the MEME algorithm (Bailey et al. 2006). Shown are motifs that were significantly enriched in either upstream (5′ UTR) or downstream (3′ UTR) regions of the ORF. (B) Sequence motifs of Pub1 targets. Analysis was performed as in A. The figure is reprinted with permission from Duttagupta et al. (2005); © 2005 the American Society for Microbiology. (C) Venn diagram representing the number of mRNAs that were shown to be associated with Pub1p (Duttagupta et al. 2005) and the mRNAs that showed a Hog1-dependent translation. The P-value for the overlap is 2.6e−10 (Fisher test).
Some of the translationally repressed genes in hog1Δ were also significantly down-regulated in wild-type cells, but to a lesser extent. This group mainly includes mRNAs encoding ribosomal proteins. One probable explanation for the more drastic response of these genes is that loss of Hog1p results in stronger stress sensing by the mutated strain, and therefore a stronger translation inhibition of these mRNAs is apparent. Another possible explanation is that large amounts of specific mRNAs accumulate in the FM pool of hog1Δ cells, either from increased transcription or slower decay rates, as discussed above. This will lead to an apparent reduction in the P/FM value as compared to the wild-type strain.
The genes that showed reduced translation levels in the hog1Δ mutant compared to the wild-type strain include several stress responsive proteins, some with a known role in response to high salinity (HSP12, GPD1, PMC1, and SNA3). HSP12 and GPD1 were not induced also at their transcript levels (see Supplemental Table 5). Interestingly, also MSN2/MSN4 deletion mutant failed to induce the translation and transcript levels of HSP12 and GPD1 (Figs. 5, 8). This might indicate that Hog1p potentiates HSP12 and GPD1 expression by the activation of MSN2/MSN4.
Nine of the translationally reduced genes (GRX2, TSA1, TRX1, YHB1, SOD1, SED1, YDL124W, VMA4, and CYS4) play a role in the adaptive response to oxidative stress. The translation response to oxidative stress is partially mediated by the protein kinase Rck2 (Swaminathan et al. 2006), which was shown to be phosphorylated by Hog1p (Teige et al. 2001). Thus, it is conceivable that Hog1p regulates the translation of these genes through activation of Rck2.
Interestingly, a group of 32 genes was found to be translationally up-regulated in hog1Δ compared to the wild-type strain. About one third of this group is related to mitochondrial function. Functional mitochondria have been shown to be required for oxidative stress protection (Grant et al. 1997). Since hog1Δ cells fail to activate the translation of oxidative stress responsive genes, the translation of mitochondrial genes might be indirectly induced to protect cells against oxidative stress.
Hog1p-independent translation
Most of the genes that were translationally regulated in the wild-type strain following high salinity shock were not affected by the loss of Hog1p. This finding implies that the translational adaptation phase to a high saline environment also requires some Hog1-independent mechanisms. One possible mechanism is the potentiation effect we postulate to be promoted by the Msn2/Msn4 general stress responsive transcription factor. As discussed above, many of the potentiated genes were found to be transcriptionally regulated by Msn2/Msn4. Indeed, while Hog1p regulates gene expression under low and modest osmotic challenge, transcriptional regulation by these stress responsive factors is favored under severe osmotic stress (O'Rourke and Herskowitz 2004). Nuclear localization of Msn2/Msn4 in response to stress requires decreased activity of the PKA signaling pathway (Gorner et al. 1998) and inhibition of the TOR signaling pathway (Beck and Hall 1999). Our results show that following high salinity, translation of inhibitors of either pathway (BCY1 and TIP41, respectively) is induced. Therefore, the increased translation levels of these two proteins probably serve as an additional mode of regulation for inhibiting the two signaling pathways and, eventually, to promote the nuclear migration of Msn2/Msn4 following high salinity stress. Since none of these genes (BCY1 or TIP41) was translationally affected by the loss of Hog1p, these modes of PKA and TOR signaling regulation are probably Hog1p independent. Interestingly, the TOR, but not the HOG, pathway was shown to be required for resistance to cation toxicity (Crespo et al. 2001; Ye et al. 2006). Thus, it is possible that some of the HOG-independent regulation includes a response to the cation toxic effect imposed by the sodium ions.
CONCLUSIONS
The work presented here describes the translational response of yeast cells to severe salinity stress and its relationship to the transcriptome response. We identified the genes that follow the global trend of translation repression and revealed that this repression may be associated with accumulation of transcripts as nontranslating pool. We also identified specific groups of differentially translated mRNAs and provide evidence that the regulation of some of them occurs through a potentiation effect involving the stress-response transcription factors MSN2/MSN4. Analysis of the hog1Δ mutant reveals that translation regulation for a small number of mRNAs occurs through the HOG pathway and suggested a mediation of Pub1p in this regulation. Whether this is through direct association of cytosolic Hog1p with the translation machinery is yet to be determined. Finally, translational regulation through the HOG pathway represents only a minor fraction of the regulation that occurs under high salinity stress. The results presented herein provide data regarding the translational response to a single solute concentration in a single time point. Future studies will reveal the dynamics of the translational response following other salt stress conditions.
MATERIALS AND METHODS
Yeast strains and growth conditions
The following Saccharomyces cerevisiae strains were used: BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0), hog1Δ (BY4741, Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 and YLR113w∷kanMX4) (Euroscarf), msn2Δ/msn4Δ (MATα, ura3-52, trp1Δ, leu2-3,112, his3∷hisG, ade2-R8, GAL+, CANS, gal80∷hisG, LEU2∷IME1 IREu-his4-lacZ, msn2∷HIS3, msn4∷URA3) and its isogenic parental strain (kindly provided by Dr. Yona Kassir, Technion-—Israel Institute of Technology), and TAP-tagged strains (Mat a ade2 arg4 leu2-3,112 trp1-289 ura3-52) (Euroscarf numbers SC0520, SC3114, SC1684, SC0877). Cells were grown at 30°C to mid-log phase (OD600 = 0.6) in YPD (1% yeast extract, 2% peptone, 2% glucose). For salt stress, cultures were harvested, centrifuged at 3000g for 5 min at room temperature, washed once with doubly distilled water, resuspended in YPD media supplemented with 1 M NaCl, and incubated at 30°C for the time points specified in the Results section.
Polysomal analysis
Polysomes were isolated as previously described with minor modifications (Arava et al. 2003). Briefly, a 50 mL culture of yeast was harvested by centrifugation (3000g, 4 min, 4°C) in the presence of 100 μg/mL cycloheximide. Following two washes in 4 mL of lysis buffer (20 mM Tris-HCl at pH 7.4, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg/mL cycloheximide, 1 mg/mL heparin, 1% Triton X-100), cells were resuspended in 400 μL lysis buffer, transferred to a screw capped microfuge tube supplemented with 1 mL chilled glass beads, and lysed in a bead beater by two rounds of 90 sec pulses). The lysate was transferred to a clean microfuge tube and centrifuged for 5 min at 8000g at 4°C. The supernatant was transferred to a clean microfuge tube, supplemented with lysis buffer to 800 μL, and loaded onto an 11 mL 10%–50% sucrose gradient (containing 20 mM Tris-HCl at pH 7.4, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg/mL cycloheximide, 500 μg/mL heparin). The gradient was centrifuged in a SW41 rotor (Beckman) at 35,000 rpm for 2.5 h, and polysomal profiles were determined by monitoring RNA absorbance at 254 nm. Concurrently, the gradient was fractionated into two fractions representing free and monosomal RNA (FM) and polysomal RNA (P).
RNA preparation for microarray analysis
RNA from FM and P fractions was precipitated and purified as previously described (Arava et al. 2003). Amino-allyl cDNA synthesis was performed by standard procedures utilizing Improm II Reverse Transcriptase (Promega) in the presence of 2 mM amino-allyl dUTP and oligo-dT primer followed by Cy5 fluorescent dye (Amersham) coupling to the amino-allyl group. RNA extracted from exponentially growing BY4741 strain and labeled with Cy3-dUTP was used as a reference sample. Each cDNA labeled from the fractionated RNA (FM or P) was mixed with labeled reference RNA and hybridized to a DNA microarray containing PCR products of all known and predicted S. cerevisiae open reading frames. To study changes in mRNA abundances, RNA was extracted from the same yeast cultures used for polysomal analysis according to the hot phenol procedure (Schmitt et al. 1990) and labeled with Cy3-dUTP (before salt stress) or Cy5-dUTP (after salt stress), as described above.
Microarray scanning and data filtration
The DNA microarrays were scanned with a GenePix 4000B apparatus (Axon Instruments) at two wavelengths to detect emissions from both Cy5 and Cy3. The images were acquired with GenePix Pro 5.1 software (Axon Instruments) and analyzed with Acuity 4.0 software (Axon Instruments). To minimize artifacts that can arise from low expression values, genes were filtered out if less than 80% of their Cy3 or Cy5 raw intensity values exceeded two standard deviations above background levels. Genes with Cy5/Cy3 inconsistent raw intensities ratios (Rgn R2 < 0.6) or gene features that were smaller then 55 μm were also removed.
Microarray data normalization
The polysomal data were normalized using Bacillus subtilis mRNA spike-in mix derived from lys (ATCC no. 87,482), trp (ATCC no. 87,485), dap (ATCC no. 87,486), thr (ATCC no. 87,484), and phe (ATCC no. 87,483) clones. The spike-in mix (80 pg/μL lys, 160 pg/μL trp, 200 pg/μL dap, 240 pg/μL thr, and 320 pg/μL phe) was added in equal amounts (40 μL) to either FM or P fractions immediately after gradient fractionation and to the reference RNA samples (5 μL). About 20 spots per spike were spotted across the complete grid of the microarray in a spatially even manner. Since the spikes were added in equal amounts, any variation in their signals between the two fractions was the result of differences in preparation steps (e.g., RNA precipitation, reverse transcription, and dye labeling) and could, therefore, be easily corrected. Changes in mRNA abundances were normalized to the median of the signal intensities ratios of all data points in the microarray.
Microarray data analysis
The data are based on three independent biological repeats for the wild-type and hog1Δ strains under normal (N) or stress (S) conditions. The PS/PN, FMS/FMN, and (P/FM)S/(P/FM)N values of each biological repeat were multiplied by a constant value in order to equalize their median values. Next, for each gene, the average values for PS/PN, FMS/FMN, (P/FM)S/(P/FM)N, and TS/TN were calculated from at least two experiments. Respectively, 80%, 80%, 66%, and 90% of the wild-type genes and 83%, 74%, 62%, and 80% of the hog1Δ genes showed coefficient of variation (CV) values that were lower than 0.25. To identify gene candidates for translation regulation under salt stress in the wild-type strain, the PS/PN, FMS /FMN, and (P/FM)S/(P/FM)N average values were plotted against the average values of changes in mRNA abundances (TS/TN). Genes that deviated from the general linear trend line by more than two standard deviations were selected. To select for hog1Δ translationally misregulated genes, hog1Δ PS/PN, FMS/FMN, and (P/FM)S/(P/FM)N values of one biological repeat were plotted against their wild-type equivalents. Gene values that deviated by more than two standard deviations in their (P/FM)S/(P/FM)N ratio of one biological replicate and by more than one standard deviation in at least one of the other two repeats were collected. Of these, only those genes that showed a similar deviation pattern in one of the other two criteria (PS/PN, FMS/FMN) were selected.
Northern blot analysis
Total or polysomal RNA was isolated as described for the microarray experiments from three independent samples. Unfractionated RNA (5 μg) and equal volumes of FM and P RNA were separated by electrophoresis, blotted, and hybridized, essentially as described by Alwine et al. (1977).
Pulse labeling
TAP-tagged strains were grown in SD with the necessary amino acids and without methionine to mid-logarithmic phase, and half of the cells (3 mL) were shifted to the same medium supplemented with 1 M NaCl. Cells were grown for 40 min and then labeled with 35S-methionine (∼100 μCi) for another 20 min. Translation was stopped by addition of cycloheximide to a final concentration of 0.1 mg/mL and, cells were immediately cooled and spun down. Cell pellets were resuspended in 350 μL IP buffer (50 mM HEPES KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Tritone, 0.1% Na-deoxycholate, 1 mM PMSF, 0.5 μg/mL leupeptin, and 0.7 μg/mL pepstatin) and lysed in a bead beater. Lysates were cleared by centrifugation for 10 min at 10,000 rpm and subjected to IP by mixing with 40 μL of IgG sepharose beads (GE Healthcare Cat number 17-0969-01, prewashed three times with IP buffer) for 2 h at 4°C. Beads with their bound complexes were precipitated and washed three times with 1 mL IP buffer. Labeled proteins were then eluted from the beads by cleavage with 5 units of TEV protease (Invitrogen Cat. number 12575-015) for 2 h at 16°C, in 40 μL of TEV buffer (50 mM Tris at pH 8, 0.5 mM EDTA, and 1 mM DTT). Beads were removed by centrifugation at 3000 rpm for 1 min and the supernatant was mixed with 2× Laemmli buffer and a fifth was analyzed on SDS-PAGE.
Data deposition
Microarray data have been deposited in the GEO database at NCBI.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Erez Eliyahu for help in the production of the DNA microarrays and Dr. Yona Kassir for the msn2/msn4 deletion strain. We also thank Drs. Yael Mandel-Gutfreund for help with the Tomtom algorithm and Irit Gat-Viks for fruitful discussions. This work was supported by grants from the Israel Science Foundation and from the Israeli Ministry of Science and Technology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.864908.
REFERENCES
- Albertyn J., Hohmann S., Thevelein J.M., Prior B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J.C., Kemp D.J., Stark G.R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., Slaven J.W., De Long S.K., Ibrahimo S., Sachs A.B. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 2001;20:6464–6474. doi: 10.1093/emboj/20.22.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N.C., Schneider U., Helliwell S.B., Stansfield I., Tuite M.F., Hall M.N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hall M.N. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J.L., Daicho K., Ushimaru T., Hall M.N. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae . J. Biol. Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- Duttagupta R., Tian B., Wilusz C.J., Khounh D.T., Soteropoulos P., Ouyang M., Dougherty J.P., Peltz S.W. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ricaud L., Warringer J., Ericson E., Glaab K., Davidsson P., Nilsson F., Kemp G.J., Nerman O., Blomberg A. PROPHECY—A yeast phenome database, update 2006. Nucleic Acids Res. 2007;35:D463–D467. doi: 10.1093/nar/gkl1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C., van Voorst F., Martins A., Neves L., Oliveira R., Kielland-Brandt M.C., Lucas C., Brandt A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae . Mol. Biol. Cell. 2005;16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge E.K., Braun E.L., Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae . J. Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.J., Rios G., Ali R., Belles J.M., Serrano R. Comparative physiology of salt tolerance in Candida tropicalis and Saccharomyces cerevisiae . Microbiol. 1997;143:1125–1131. doi: 10.1099/00221287-143-4-1125. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat-Viks I., Shamir R. Refinement and expansion of signaling pathways: The osmotic response network in yeast. Genome Res. 2007;17:358–367. doi: 10.1101/gr.5750507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens A., Dever T.E., Pascual-Ahuir A., Serrano R. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 2001;276:30753–30760. doi: 10.1074/jbc.M102960200. [DOI] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Martinez-Pastor M.T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes & Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.M., MacIver F.H., Dawes I.W. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae . FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Stegert M.R., Schmitz D., Hemmings B.A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hirasawa T., Nakakura Y., Yoshikawa K., Ashitani K., Nagahisa K., Furusawa C., Katakura Y., Shimizu H., Shioya S. Comparative analysis of transcriptional responses to saline stress in the laboratory and brewing strains of Saccharomyces cerevisiae with DNA microarray. Appl. Microbiol. Biotechnol. 2006;70:346–357. doi: 10.1007/s00253-005-0192-6. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol S., Zagulski M., Bilinski T., Bartosz G. Antioxidants protect the yeast Saccharomyces cerevisiae against hypertonic stress. Free Radic. Res. 2005;39:365–371. doi: 10.1080/10715760500045855. [DOI] [PubMed] [Google Scholar]
- Kuhn K.M., DeRisi J.L., Brown P.O., Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav R., Nejidat A., Abeliovich A. Alterations in protein synthesis and levels of heat shock 70 proteins in response to salt stress of the halotolerant yeast Rhodotorula mucilaginosa . Antonie Van Leeuwenhoek. 2004;85:259–269. doi: 10.1023/B:ANTO.0000020361.81006.2b. [DOI] [PubMed] [Google Scholar]
- Law G.L., Bickel K.S., MacKay V.L., Morris D.R. The undertranslated transcriptome reveals widespread translational silencing by alternative 5′ transcript leaders. Genome Biol. 2005;6:R111. doi: 10.1186/gb-2005-6-13-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Van Montagu M., Verbruggen N. A highly conserved kinase is an essential component for stress tolerance in yeast and plant cells. Proc. Natl. Acad. Sci. 1999;96:5873–5877. doi: 10.1073/pnas.96.10.5873. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lotan R., Bar-On V.G., Harel-Sharvit L., Duek L., Melamed D., Choder M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes & Dev. 2005;19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D., Arava Y. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. In: Lorsch J.R., editor. Translation initiation: Cell biology, high-throughput and chemical-based approaches. Academic Press; San Diego, CA: 2007. [DOI] [PubMed] [Google Scholar]
- Mendoza I., Rubio F., Rodriguez-Navarro A., Pardo J.M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae . J. Biol. Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Morita Y., Nakamori S., Takagi H. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae . J. Biosci. Bioeng. 2002;94:390–394. doi: 10.1016/s1389-1723(02)80214-6. [DOI] [PubMed] [Google Scholar]
- Mortensen H.D., Gori K., Siegumfeldt H., Nissen P., Jespersen L., Arneborg N. Intracellular pH homeostasis plays a role in the NaCl tolerance of Debaryomyces hansenii strains. Appl. Microbiol. Biotechnol. 2006;71:713–719. doi: 10.1007/s00253-005-0196-2. [DOI] [PubMed] [Google Scholar]
- Murguia J.R., Belles J.M., Serrano R. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- Murguia J.R., Belles J.M., Serrano R. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 1996;271:29029–29033. doi: 10.1074/jbc.271.46.29029. [DOI] [PubMed] [Google Scholar]
- Norbeck J., Blomberg A. Amino acid uptake is strongly affected during exponential growth of Saccharomyces cerevisiae in 0.7 M NaCl medium. FEMS Microbiol. Lett. 1998;158:121–126. doi: 10.1111/j.1574-6968.1998.tb12810.x. [DOI] [PubMed] [Google Scholar]
- Norbeck J., Blomberg A. The level of cAMP-dependent protein kinase A activity strongly affects osmotolerance and osmo-instigated gene expression changes in Saccharomyces cerevisiae . Yeast. 2000;16:121–137. doi: 10.1002/(SICI)1097-0061(20000130)16:2<121::AID-YEA511>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- O'Rourke S.M., Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlman A.K., Granath K., Ansell R., Hohmann S., Adler L. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 2001;276:3555–3563. doi: 10.1074/jbc.M007164200. [DOI] [PubMed] [Google Scholar]
- Panadero J., Pallotti C., Rodriguez-Vargas S., Randez-Gil F., Prieto J.A. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae . J. Biol. Chem. 2006;281:4638–4645. doi: 10.1074/jbc.M512736200. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Posas F., Chambers J.R., Heyman J.A., Hoeffler J.P., de Nadal E., Arino J. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Preiss T., Baron-Benhamou J., Ansorge W., Hentze M.W. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- Proft M., Serrano R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell. Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., D'Aquino K.E., Ee L.S., Amon A. The stress-activated mitogen-activated protein kinase signaling cascade promotes exit from mitosis. Mol. Biol. Cell. 2006;17:3136–3146. doi: 10.1091/mbc.E05-12-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Albertyn J., Thevelein J.M., Prior B.A., Hohmann S. Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae . Microbiol. 1999a;145:715–727. doi: 10.1099/13500872-145-3-715. [DOI] [PubMed] [Google Scholar]
- Rep M., Reiser V., Gartner U., Thevelein J.M., Hohmann S., Ammerer G., Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 1999b;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Krantz M., Thevelein J.M., Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Santhanam A., Hartley A., Duvel K., Broach J.R., Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae . Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir R., Maron-Katz A., Tanay A., Linhart C., Steinfeld I., Sharan R., Shiloh Y., Elkon R. EXPANDER—An integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D., Smirnova J.B., Selley J.N., Carroll K., Hubbard S.J., Pavitt G.D., Ashe M.P., Grant C.M. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Smirnova J.B., Selley J.N., Sanchez-Cabo F., Carroll K., Eddy A.A., McCarthy J.E., Hubbard S.J., Pavitt G.D., Grant C.M., Ashe M.P. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol. Cell. Biol. 2005;25:9340–9349. doi: 10.1128/MCB.25.21.9340-9349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Masek T., Molin C., Pospisek M., Sunnerhagen P. Rck2 is required for reprogramming of ribosomes during oxidative stress. Mol. Biol. Cell. 2006;17:1472–1482. doi: 10.1091/mbc.E05-07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., Iwamoto F., Nakamori S. Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 1997;47:405–411. doi: 10.1007/s002530050948. [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Reiser V., Ruis H., Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.C., Monteiro P., Jain P., Tenreiro S., Fernandes A.R., Mira N.P., Alenquer M., Freitas A.T., Oliveira A.L., Sa-Correia I. The YEASTRACT database: A tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae . Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh E.A. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel O., Reiser V., Siderius M., Kelders M.C., Ammerer G., Ruis H., Mager W.H. Response of Saccharomyces cerevisiae to severe osmotic stress: Evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 2000;37:382–397. doi: 10.1046/j.1365-2958.2000.02002.x. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Garneau N., Tu Khounh D., Peltz S.W. p38 mitogen-activated protein kinase/Hog1p regulates translation of the AU-rich-element-bearing MFA2 transcript. Mol. Cell. Biol. 2005;25:9753–9763. doi: 10.1128/MCB.25.22.9753-9763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Ericson E., Fernandez L., Nerman O., Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale J., Bohnert H.J. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
- Ye T., Garcia-Salcedo R., Ramos J., Hohmann S. Gis4, a new component of the ion homeostasis system in the yeast Saccharomyces cerevisiae . Eukaryot. Cell. 2006;5:1611–1621. doi: 10.1128/EC.00215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]