Abstract
The off-rate (koff) of the T cell receptor (TCR)/peptide-major histocompatibility complex class I (pMHCI) interaction, and hence its half-life, is the principal kinetic feature that determines the biological outcome of TCR ligation. However, it is unclear whether the CD8 coreceptor, which binds pMHCI at a distinct site, influences this parameter. Although biophysical studies with soluble proteins show that TCR and CD8 do not bind cooperatively to pMHCI, accumulating evidence suggests that TCR associates with CD8 on the T cell surface. Here, we titrated and quantified the contribution of CD8 to TCR/pMHCI dissociation in membrane-constrained interactions using a panel of engineered pMHCI mutants that retain faithful TCR interactions but exhibit a spectrum of affinities for CD8 of >1,000-fold. Data modeling generates a “stabilization factor” that preferentially increases the predicted TCR triggering rate for low affinity pMHCI ligands, thereby suggesting an important role for CD8 in the phenomenon of T cell cross-reactivity.
CD8+ cytotoxic T lymphocytes (CTL)1 recognize protein antigens in the form of short peptides presented in association with MHCI molecules on the surface of target cells. Antigen specificity is conferred by the TCR, the highly variable complementarity-determining regions of which interact with the peptide-binding platform of the MHC molecule (1, 2). The pMHCI complex also interacts with the T cell surface glycoprotein CD8, which binds to invariable regions of the MHCI molecule (3, 4). CD8 is thought to act as a coreceptor for antigen in concert with TCR during the process of antigen recognition and T cell activation (5).
CD8 plays a key role in signal transduction by recruiting essential signaling components to the cytoplasmic side of the TCR-CD3-ζ complex (6-9). However, there is still disagreement as to whether the interaction between pMHCI and CD8 stabilizes the TCR/pMHCI interaction and whether the pMHCI/CD8 interaction plays a significant adhesive role in T cell activation (10-15). Early experiments indicated that interaction between pMHCI and CD8 might initiate cell-cell adhesion (10). However, equilibrium binding of soluble human CD8αα has been measured with several different HLA A, B, and C gene products and occurs at dissociation constant (KD) values of >100 μm (16). The low solution affinity and fast kinetics compared with conventional cell-cell adhesion molecules that interact with 1:1 stoichiometry suggest that this interaction would be unable to initiate cell-cell adhesion independently (12, 17) except when these molecules are expressed at unphysiologically high surface densities (10). The binding site for CD8 is distinct and spatially separate from the TCR-recognized, peptide-binding domains of MHCI molecules, and it is expected that a single MHCI molecule might bind both TCR and CD8 simultaneously (2, 3). Simultaneous binding of the TCR and CD8 by pMHCI allows the potential for CD8-induced stabilization of the TCR/pMHCI interaction. Despite the low affinity of the pMHCI/CD8 interaction, any such stabilization could be of extreme biological importance, as biophysical measurements (18, 19) support models of TCR triggering (20, 21) in which the off-rate (koff) of the TCR/pMHCI interaction is the principal kinetic feature determining the outcome of TCR ligation. Whether the pMHCI/CD8 interaction could reduce TCR/pMHCI koff has been the subject of intense debate (11-14). Early surface plasmon resonance (SPR) experiments examining the binding of soluble forms of TCR and CD8 to pMHCI were interpreted as showing that the pMHCI/CD8 interaction stabilizes the interaction between the TCR and pMHCI (11). However, more recent studies repeating these experiments have cast doubt on these conclusions by showing that the soluble extracellular domains of the TCR and CD8 molecules bind to pMHCI independently and with distinct kinetics in both human and murine systems (12, 13). Indeed, the data presented by Garcia et al. (11) exhibit characteristics consistent with the presence of multivalent aggregates (12). Even low levels of such aggregates (<2%) can dominate binding if the monomeric interaction is of low affinity and if high concentrations of soluble protein are used (22). In addition, the current view that the extracellular domains of the TCR and CD8 do not cooperate in binding pMHCI (12, 13) is supported by structural studies that indicate that the binding of one molecule is unlikely to alter the affinity for the other (3). However, structural and biophysical studies of soluble TCR and CD8 do not rule out the possibility that CD8 on the CTL surface enhances TCR binding to pMHCI on the antigen-presenting cell surface. Mounting evidence shows that CD8 can interact directly with the TCR (17, 23-29) and appears to have an important role in organizing the TCR on the T cell surface (13, 30). Direct TCR/CD8 association on the CTL surface could therefore enable cooperativity in pMHCI binding that would not be detectable using soluble versions of these molecules.
Here, we use engineered pMHCI mutant proteins that retain faithful interactions with cognate TCR but bind CD8 with either reduced or enhanced affinities to show that the pMHCI/CD8 interaction can significantly affect the decay of soluble fluorescent pMHCI from the CTL surface. By modeling the dissociation rates of these pMHCI mutants with CD8 interactions spanning an affinity range of >1,000-fold, we quantify the contribution that the pMHCI/CD8 interaction makes to TCR/pMHCI stability and dissociation kinetics.
EXPERIMENTAL PROCEDURES
Cell Lines and CTLs
The CTL clone 003 and CTL line 868, both specific for the HLA A2-restricted HIV-1 p17 Gag-derived epitope SLYNTVATL (residues 77–85), have been described previously (31-33). CTLs were grown from cryopreserved stocks in 24-well tissue culture plates in R10 (RPMI medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine) containing 5% T-STIM (BD Biosciences) and 200 units/ml proleukin (Chiron) for 2 weeks following re-stimulation as described previously (27). After this time, cells were maintained in R10 plus 25 ng/ml interleukin-15 (Peprotech). Cells maintained in interleukin-15 remained viable and continued to grow for several months without the need for re-stimulation with antigen and/or mixed lymphocyte reaction. CTL clones maintained in interleukin-15 for three months exhibited an activation profile identical in terms of early tyrosine phosphorylation events and effector function (specific lysis of antigen-bearing target cells and production of macrophage inflammatory protein-1β and interferon-γ) to that of CTL maintained in interleukin-2 with periodic re-stimulation. SLYNTVATL-specific 0400 and SLY-10 were similarly maintained. CTL clone EBV-A is specific for the HLA A2-restricted, EBV-derived, BMLFI-encoded epitope GLCTLVAML.
Inclusion Body Preparation
Biotin-tagged HLA A2 heavy chain was expressed under the control of a T7 promoter as insoluble inclusion bodies in Escherichia coli strain BL21(DE3)pLysS (Novagen). Isopropyl-1-thio-β-d-galactopyranoside-induced E. coli were lysed by repeated freeze/thaw cycles to release inclusion bodies that were subsequently purified by washing with a 0.5% Triton X-100 buffer (Sigma) as described previously (34). The D227K/T228A, A245V, and Q115E HLA A2 heavy chain mutants and the A2/Kb α3 domain fusion protein were produced in a similar fashion.
Production of Soluble pMHCI
Soluble biotinylated pMHCI monomers were produced as described previously (35). Briefly, heavy chain and β2m inclusion body preparations were denatured separately in 8 m urea buffer (Sigma) and mixed at a 1:1 molar ratio. pMHCI was refolded in 2-mercaptoethylamine/cystamine (Sigma) redox buffer with added synthetic peptide (Research Genetics Invitrogen Corp., Huntsville, AL). The HLA A2-restricted peptides used in this study were the HTLV-1 Tax-derived epitope LLFGYPVYV (36), the HIV-derived p17 Gag epitope SLYNTVATL (37), the EBV-derived BMLFI-encoded epitope GLCTLVAML (38), and the CMV-derived pp65 protein epitope NLVPMVATV (39). Following buffer exchange into 10 mm Tris (pH 8.1), the refolded monomers were purified by anion exchange. Purified monomers were biotinylated using d-biotin (Sigma) and BirA enzyme as described previously (34). Excess biotin was removed by gel filtration.
Tetramerization and Flow Cytometry
Fluorescent tetrameric pMHCI complexes were produced by mixing phycoerythrin-conjugated streptavidin (Molecular Probes or Prozyme Inc.) and biotinylated pMHCI monomers at a 1:4 molar ratio, respectively. For ex vivo analysis, 106 peripheral blood mononuclear cells (PBMCs) were stained with the wild type and mutant tetramers shown at the indicated concentrations for 20 min at 37 °C, washed once in fluorescence-activated cell sorter (FACS) buffer (PBS without Ca2+/Mg2+, 1% bovine serum albumin (w/v), and 0.1% NaN3), surface-stained with pre-titered allophycocyanin-conjugated anti-CD8 and peridinin chlorophyll protein-conjugated anti-CD3 monoclonal antibodies (BD Biosciences) for 30 min at 4 °C, and then washed twice and fixed in 1% paraformaldehyde. Data were collected using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (TreeStar Inc., San Carlos, CA). For the in vitro expanded SLYNTVATL-specific CTL line 868, 2 × 105 CTLs were stained as described above with the indicated pMHCI tetramers at the concentrations shown for 20 min at 37 °C, washed, and then stained with allophycocyanin-conjugated anti-CD8 monoclonal antibody (clone SK1; BD Biosciences) and 7-amino actinomycin D (ViaProbe; BD Biosciences) for 30 min on ice prior to two further washes and data collection; analysis was performed with CellQuest software (BD Biosciences). All pMHCI tetramers used in this study were made fresh for the week of use from pMHCI monomers stored at −80 °C to minimize the effect of stability differences (15). Once prepared, tetramers were stored in the dark at 4 °C.
pMHCI Tetramer Decay Assay
Indicated numbers of CTL were stained in 100 μl of azide buffer (PBS, 0.1% NaN3, and 0.5% fetal calf serum) for 20 min on ice with a concentration of tetramer, previously determined by titration, that gave a starting mean fluorescence intensity (MFI) of 200; 7-amino actinomycin D (ViaProbe, BD Biosciences) was included so that dead cells could be gated out of the analysis. After washing twice in ice-cold azide buffer, CTLs were resuspended in azide buffer, split into two separate aliquots, and placed at room temperature. To one sample, an excess of unconjugated anti-HLA A2 monoclonal antibody (clone BB7.2; Serotec) at 100 μg/ml was added to block tetramer rebinding. Cells were then taken at time points 0, 1, 2, 5, 8, 10, 15, 20, 30, 40, and 60 min, resuspended in PBS, and analyzed on a FACSCalibur flow cytometer. The remaining sample was left without BB7.2, and controls were analyzed at the 0-, 10-, 20-, 30-, 40-, and 60-min time points. Decays were repeated for all mutant tetramers in the panel on the same day. Parameter validation for decay assays is shown in Figs. S1 and S2 of the supplemental data, available in the on-line version of this article.
Biophysical Validation of Mutant pMHCI Affinities for CD8 and TCR
The D227K/T228A mutation in the α3 domain of HLA A2 has been shown previously to abrogate CD8 binding (8); the A245V mutation reduces CD8 binding by >4-fold (16). The biophysical properties of the Q115E α2 domain mutant and the A2/Kb hybrid pMHCI proteins were determined by SPR using a BIAcore 3000™ (BIAcore AB, St. Albans, UK) machine. sCD8αα wild type was prepared as described previously (40). The A6 TCR specific for the HLA A*0201-restricted HTLV-1 Tax epitope LLFGYPVYV was refolded as described previously (41). All proteins for analysis were diluted into HBS-EP buffer (BIAcore AB) containing 10 mm HEPES (pH 7.4), 150 nm NaCl, 3.4 mm EDTA, and 0.005% surfactant P20. A standard amine coupling kit (BIAcore AB) was used to activate the surface of a research grade CM5 sensor chip (BIAcore AB). Streptavidin was covalently coupled to the chip surface by primary amines through the injection of a 0.2 mg/ml streptavidin solution (Sigma) diluted in 10 mm sodium acetate (pH 4.5) over the surface. Biotinylated pMHCI monomers were immobilized onto the chip surface at ∼1,000 response units in each flow cell. Serial dilutions of either sCD8αα wild type or soluble A6 Tax TCR in HBS-EP buffer were flowed over the chip to generate kinetic data. Data were analyzed using BIAeval, Excell, and Origin version 6.1 (Microcal software). KD values were calculated both by linear Scatchard plots and non-linear analysis assuming 1:1 Langmuir binding (A + B ⇆ AB) using non-linear curve fitting to the equation AB = B× ABmax/(KD + B). Each batch of protein was validated by SPR prior to experimentation.
Analysis of Tetramer Decay
We assume the following kinetics for x1, x2, and x3, which represent the surface densities of monovalently bound, bivalently bound, and trivalently bound tetramers, respectively, as shown in Equations 1-3,
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
where the prime indicates the derivative with respect to time, μ is the single-site rebinding rate, and koff is the single-site off-rate. Because tetramers do not rebind to CTL (Fig. S1 in the supplemental data), we can safely assume that tetramers in solution are captured by antibody before rebinding can occur. If rebinding is slow (relative to koff) the dissociation curve is sigmoid, with a shape governed by the mixture of monovalently, bivalently, and trivalently bound forms present at the onset of the dissociation experiment with an exponential tail of time scale 1/koff. On the other hand, if rebinding is fast the ratios x1:x2 and x2:x3 rapidly attain a quasi-equilibrium (irrespective of the initial mixture), and the dissociation curve approaches simple exponential decay with a much reduced relaxation rate 3koff3/μ2. Because our data exhibit exponential decay (Fig. 4), we can confidently assume that our experimental system is in this rapid rebinding regime. (It is notable that the sigmoid curves and exponential curves arise as special cases of a single theory.) As shown here,
| (Eq. 4) |
Equation 4 relates the apparent off-rate obtained from non-linear least-squares regression on the dissociation curve, koff,app, to the true (single-site) off-rate. The proportionality constant (μ2/3)⅓ cannot be determined from our data. However, our conclusions depend on ratios of off-rates, which are readily obtained as the cube roots of ratios of the apparent off-rates.
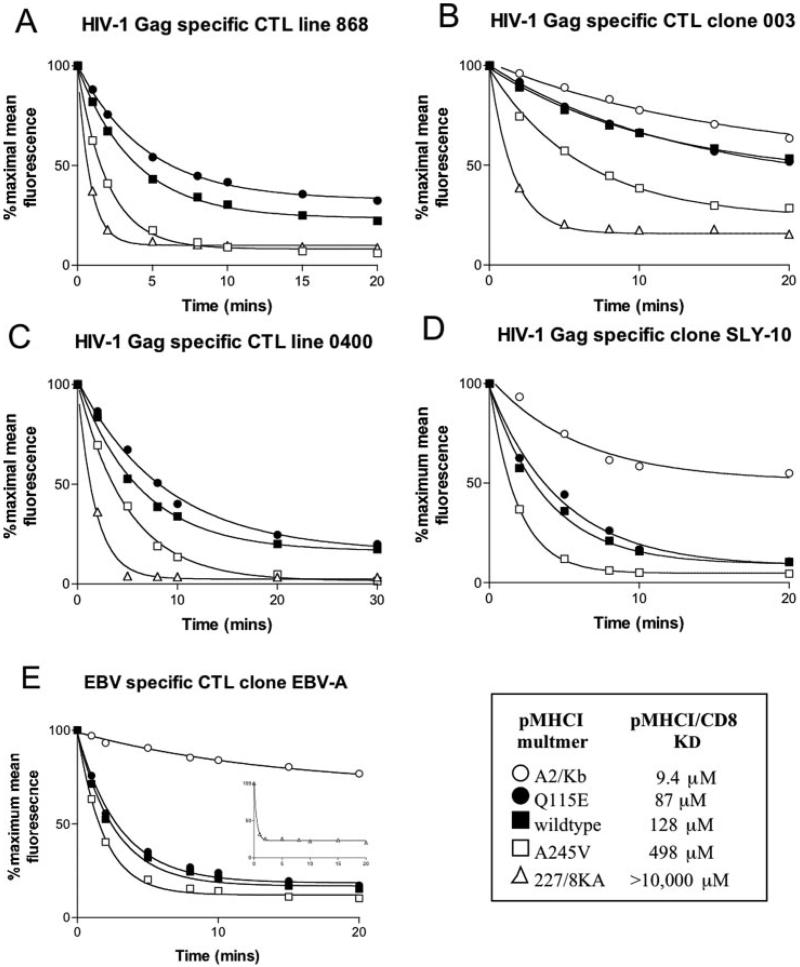
Fig. 4. The effect of the pMHCI/CD8 interaction on TCR/pMHCI dissociation.
1.5 × 106 868 CTL line (A) or 6 × 105 003 CTL clone (B), 0400 CTL (C), and SLY-10 (D) cells per experiment were resuspended in 100 μl of azide buffer (PBS, 0.1% NaN3, and 0.5% fetal calf serum) and stained with either A2/Kb-SLYNTVATL (A2/Kb), Q115E HLA A2-SLYNTVATL (Q115E), HLA A2-SLYNTVATL (wildtype), A245V HLA A2-SLYNTVATL (A245V), or D227K/T228A HLA A2-SLYNTVATL (227/8KA) tetramers conjugated to phycoerythrin and 10 μl of 7-amino actinomycin D (ViaProbe; BD Biosciences) for 20 min on ice. For panel E, 6 × 105 of the EBV-specific CTL clone EBV-A were stained with GLCTLVAML versions of the above tetramers. The concentrations of tetramer used were determined previously by titration to give a starting MFI of 200. The 003 clone stains to a similar level with the D227K/T228A tetramer without the need to adjust the concentration (8). Cells were then washed in ice cold azide buffer and resuspended in azide buffer. An excess of unconjugated anti-HLA A2 monoclonal antibody (clone BB7.2; Serotec) was added at 100 μg/ml, and the sample was moved to room temperature to initiate tetramer decay. Samples were taken at 0, 1, 2, 5, 8, 10, 15, 20, 30, 40, and 60 min and analyzed on a FACSCalibur flow cytometer using CellQuest software. Each panel (A–E) shows data from 0–20 min and the least squares fitting of the dissociation curve (percentage of maximal mean fluorescence = background + (100 − background) × exp{−k off,appt}) from which the apparent off-rate koff,app values shown in Figs. 5A and 6A were calculated. The apparent off-rate koff,app is related to the true (single site) off-rate by Equation 4. Cells stained at the same time were kept without anti-HLA A2 blocking antibody and analyzed at 0, 10, 20, 30, 40, and 60 min to demonstrate that tetramers do not decay significantly in the absence of the blocking antibody. An example of this is shown in Fig. S2 of the supplemental data found in the on-line version of this article. The SLY-10 CTL clone did not stain with HLA A*0201 D227K/T228A-SLYNTVATL at any concentration used, so it was not possible to determine an off-rate for the CD8 null tetramer. The lack of data for tetramer stability in the absence of a pMHCI/CD8 interaction makes it impossible to calculate a stabilization factor for this clone or other CTLs with a very low functional avidity. However, comparison to data obtained on the other CTLs suggests that CD8 provides a similar stabilization factor. To date we have examined >20 tetramer-sorted CTL clones, including clones specific for immunodominant viral antigens. All of these clones are of functional low avidity. Such low avidity clones do not stain with CD8 null tetramers (see Ref. 49 for examples). Our experiments show that tetramer staining rapidly induces cell death of CTLs with high functional avidity (Ref. 61).2 Low avidity CTL grown by tetramer “sort-cloning” are not representative of the types of CTL that function in vivo to clear viral infection. Direct ex vivo staining of CTL specific for the HLA A2 CMV-derived epitope NLVPMVATV shows that these CTL stain well with CD8 null tetramer (Fig. 2). Five further individuals showed a similar pattern (data not shown). CTL specific for the EBV-derived epitope GLCTLVAML can also stain with CD8 null tetramer (data not shown). The decay of the D227K/T228A HLA A2-GLCTLVAML tetramer from EBV-A CTL is shown in the inset of panel E. This CTL clone is of low functional avidity, and it was not possible to stain it to MFI of 200 with this tetramer. The decay shown in the inset, performed at the same time as the other decaying, started at a maximal MFI of 58.
Dependence of the TCR/pMHCI Off-rate on CD8
Let the TCR/pMHCI off-rate be denoted by koff° when the MHC molecule is CD8-bound and by koff* when MHC is not bound to CD8. We assume that these two rates do not depend on the affinity of the MHCI/CD8 interaction. Furthermore, let γ denote the rate at which TCR-bound MHCI molecules associate with CD8; γ is proportional to the surface density of unoccupied (free) CD8 molecules on the T cell. Finally, let ρ denote the rate at which MHCI and CD8 dissociate. Then we attain Equation 5,
| (Eq. 5) |
where [CD8]F is the surface density of free CD8 molecules in the contact area and KD,2D is the two-dimensional dissociation constant of the MHCI/CD8 interaction. We now develop a simple mathematical model that describes how the true single-site off-rate koff is derived from koff° and koff*. Consider a pMHCI molecule that engages a TCR molecule at time t = 0. Let P°(t) denote the probability that dissociation of this TCR/pMHCI does not occur before time t while the MHCI is not associated with CD8 at time t, and let P*(t) denote the probability that TCR/pMHCI dissociation occurs after time t while the MHCI is associated with CD8 at time t. Thus, P°(t) + P*(t) is the probability that TCR/pMHCI dissociation occurs later than t; hence, koff equals the rate of change of −ln{P°(t) + P*(t)}. The rate of change of P° + P* equals the following,
| (Eq. 6) |
as a routine argument shows. Using this equation and the fact that MHCI/CD8 kinetics are much more rapid than the TCR/pMHCI kinetics (12), we obtain P*/(P* + P°) = γ/(ρ + γ), which can be used to find the decay rate of P° + P* as shown here,
| (Eq. 7) |
which we can rewrite as this,
| (Eq. 8) |
by using Equation 5 and κ = [CD8]F KD,3D/KD,2D, where KD,3D denotes the three-dimensional dissociation constant of the MHCI/CD8 interaction. Combining this equation with Equation 4 and κ ≫ KD,3D, an assumption warranted by our data, we obtain Equation 9,
| (Eq. 9) |
which is the linear relationship shown in Figs. 5B and 6B.
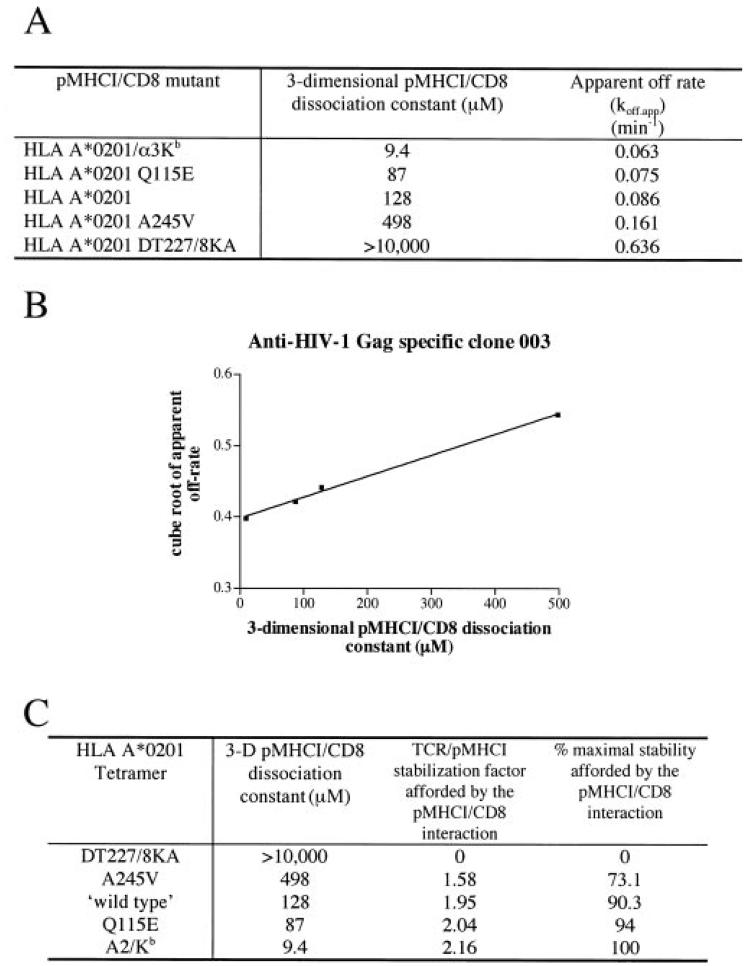
Fig. 5. Modeling the contribution of the pMHCI/CD8 interaction to TCR/pMHCI dissociation of CTL clone 003.
A, dissociation constants for pMHCI/CD8 interactions and apparent off-rates for a spectrum of HLA A2 mutants folded around the HIV-1 Gag-specific epitope SLYNTVATL. The apparent off-rate koff,app was obtained by least squares fitting of the dissociation curve (percentage of maximal mean fluorescence = background + (100 − background) × exp{−koff,appt}) and is related to the true (single-site) off-rate by Equation 4. DT227/8KA, D227K/T228A; α3Kb, A2/Kb. B, the cube root of the apparent off-rate plotted against the KD of pMHCI/CD8; the straight line least squares fit confirms the prediction of the Equation 9. C, summary of the stabilization factor and the percentage of maximal stability that can be afforded by the pMHCI/CD8 interaction for each MHCI mutant. DT227/8KA, D227K/T228A.
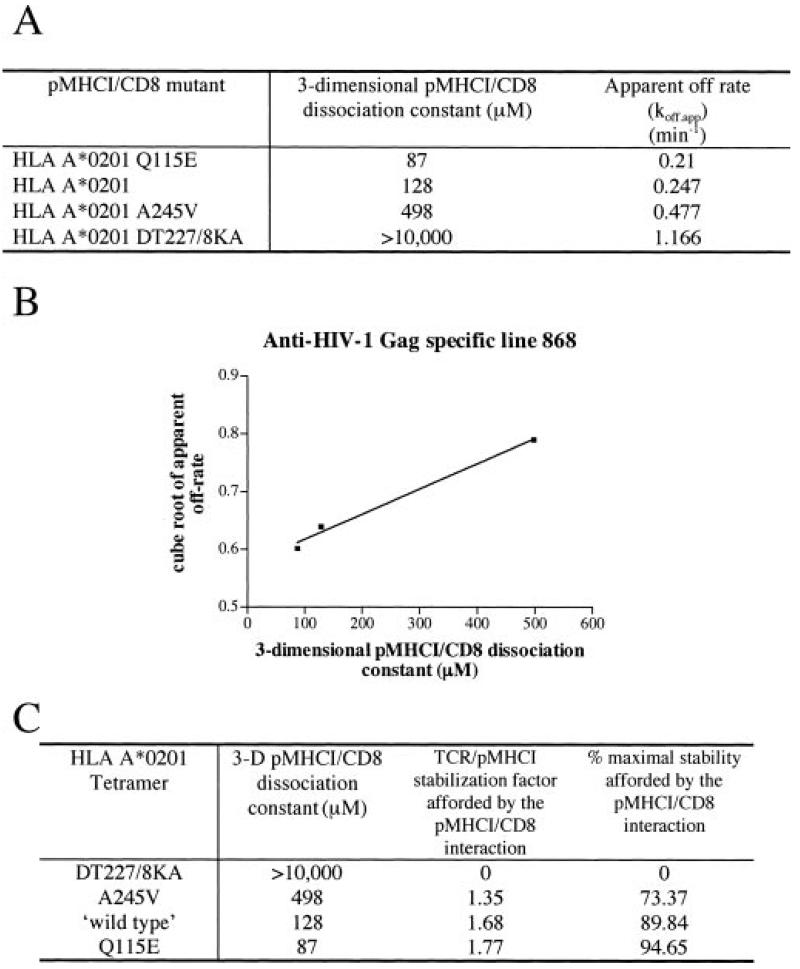
Fig. 6. Modeling the contribution of the pMHCI/CD8 interaction to TCR/pMHCI dissociation of the CTL line 868.
Data as for Fig. 5 but using 868 CTL.
RESULTS
The Design of Mutations to Increase the Interaction between pMHCI and CD8
The D227K/T228A mutation in the α3 domain of HLA A2 abrogates CD8 binding (8); the A245V mutation reduces CD8 binding by >4-fold (16). To develop similar HLA A2-based reagents with enhanced binding affinities for CD8αα, we employed the multi-scale approach used by Glick et al. (42, 43). The algorithm was limited to four feature points. The surface of the CD8αα protein that forms contacts with HLA A2 (3) was systematically searched using amino acid side chains as “probes.” A Gln-115 to Glu mutation in the α2 domain was selected for further molecular dynamics study. The wild type HLA A2 Q115 Oε1 atom forms a weak H-bond interaction with the CD8α1 R4Nη1 atom. (The Oε1 … Nη1 distance in the crystal structure is 3.18Å). This interaction was replaced by a shorter H-bond between HLA A2 Q115E Oε1 and CD8α1 R4 Nη1 atoms; the molecular dynamics simulation showed that the Oε1 … Nη1 average distance was 2.56Å and fluctuated from 2.42 to 2.82 Å. This short distance also indicates that the HLA A2 Q115E carboxylate and CD8α1 R4 guanidinium moieties form a strong electrostatic interaction that is likely to increase the affinity of binding between the two biomolecules. SPR studies supported these predictions (Fig. 1). The human/murine hybrid HLA A2/H2-Kb α3 domain fusion protein (A2/Kb), which has a substantially increased binding affinity for human CD8αα, has been described previously (44).
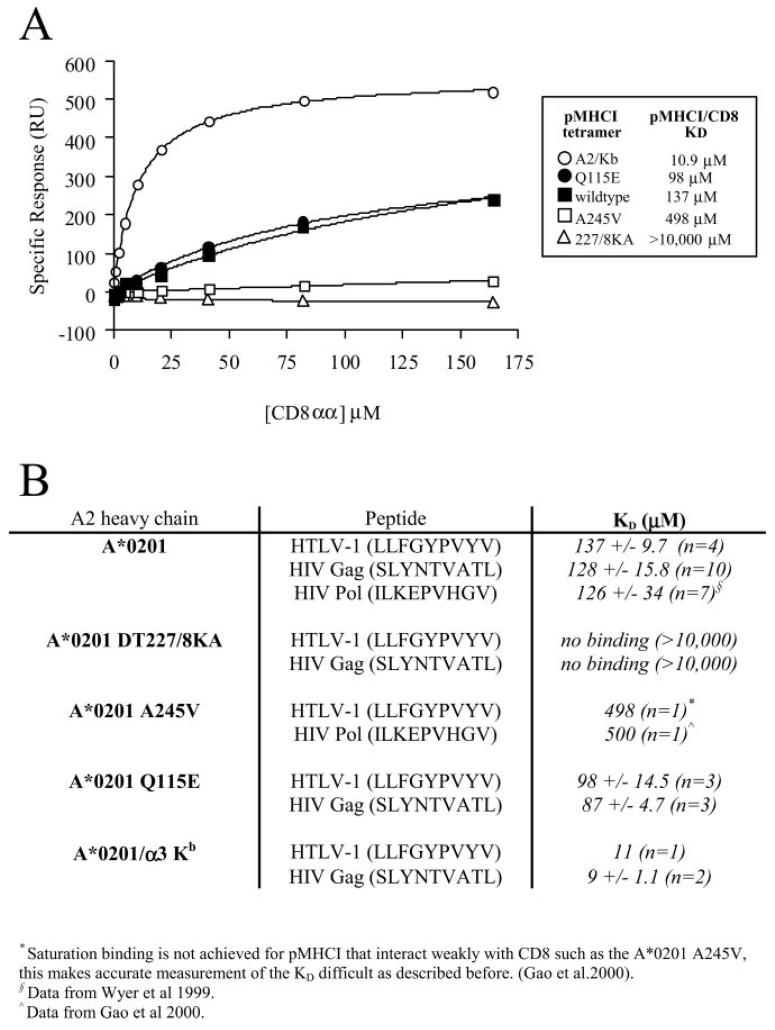
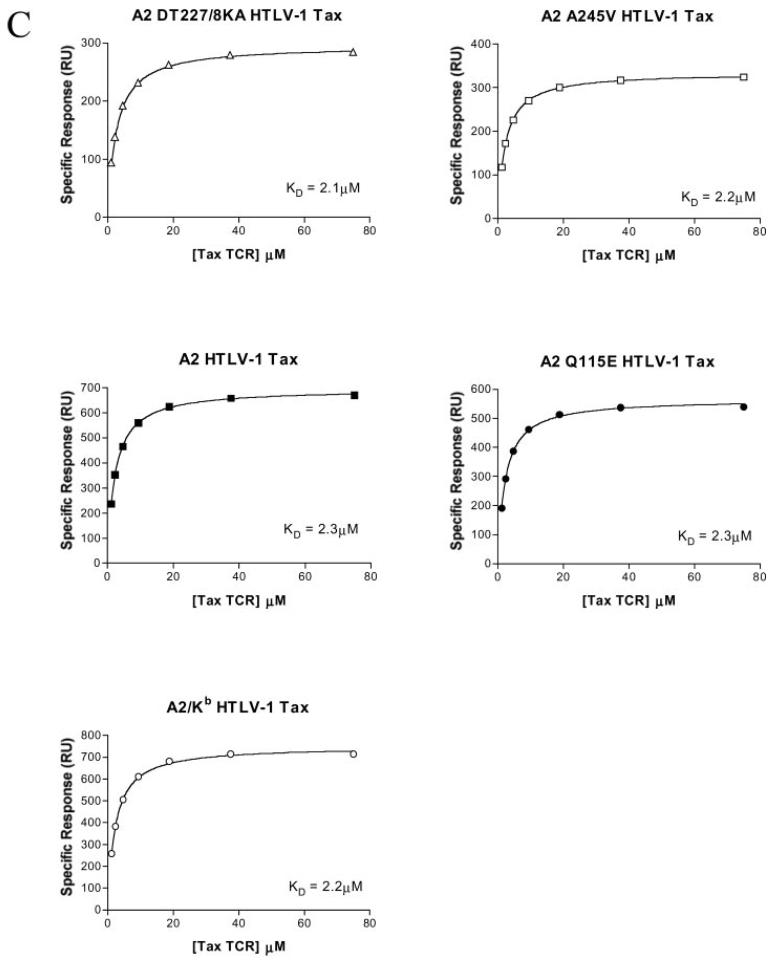
Fig. 1. Mutation of the HLA A2 α2 or α3 domain can abrogate, decrease, or increase the interaction with CD8 without affecting the interaction between the α1/α2 peptide-binding domain and the TCR.
Biotinylated pMHCI monomers were immobilized onto a streptavidin-coated BIAcore chip as described under “Experimental Procedures.” Serial dilutions of either sCD8αα wild type or soluble A6 Tax TCR in HBS-EP buffer were flowed over the chip to generate kinetic data. Data were analyzed using BIAeval, Excell, and Origin version 6.1 (Microcal software). KD values were calculated both by linear Scatchard plots and non-linear analysis assuming 1:1 Langmuir binding (A + B⇆AB) using non-linear curve fitting to the equation AB = B × ABmax/(KD + B). A, equilibrium binding of sCD8αα interacting with either D227K/T228A HLA A2 (227/8KA), A245V HLA A2, wild type HLA A2 (wildtype), Q115E HLA A2, or A2/Kb α3 domain fusion (A2/Kb) folded around the HTLV-1 Tax epitope. B, summary of sCD8αα/pMHCI affinity measurements obtained for pMHCI monomers bearing different mutations and folded around three different epitopes. DT227/8KA, D227K/T228A. C, the KD calculated for the interaction between the A6 Tax TCR and either D227K/T228A HLA A2 (DT227/8KA), A245V HLA A2, wild type HLA A2, Q115E HLA A2, or A2/Kb α3 domain fusion folded around the HTLV-1 Tax epitope was shown to be 2.6 ± 0.4, 2.5 ± 0.3, 2.5 ± 0.3, 2.7 ± 0.4, and 2.5 ± 0.2 μm respectively. In all cases indicated errors are S.D. from the mean of three separate experiments. Data shown are from just one representative experiment (hence the values differ slightly from the averages given above).
Verification That Mutations Affect the pMHCI/CD8 Interaction without Altering the TCR/pMHCI Interaction
SPR was used to determine that the KD values of the pMHCI/CD8 interaction for D227K/T228A HLA A2, A245V HLA A2, wild type HLA A2, Q115E HLA A2, and A2/Kb folded around the HTLV-1 epitope (LLFGYPVYV) were >10,000, 498, 137, 98, and 10.9 μm respectively (Fig. 1A). The KD values of the pMHCI/CD8 interaction for D227K/T228A HLA A2, wild type HLA A2, Q115E HLA A2, and A2/Kb folded around the HIV-1 epitope SLYNTVATL were shown to be >10,000, 128, 87, and 9 μm, respectively (Fig. 1B). These substitutions in the α3 or α2 domain of the pMHCI molecule did not affect TCR binding (Fig. 1C). This spectrum of HLA A2 molecules that have normal TCR/pMHCI interactions but a range of pMHCI/CD8 interactions exceeding 1,000-fold was subsequently used to study the role of the pMHCI/CD8 interaction in the binding of pMHCI antigen.
The CD8 Interaction Determines the Pattern of pMHCI Tetramer Staining at Both Subnormal and Supranormal Affinities
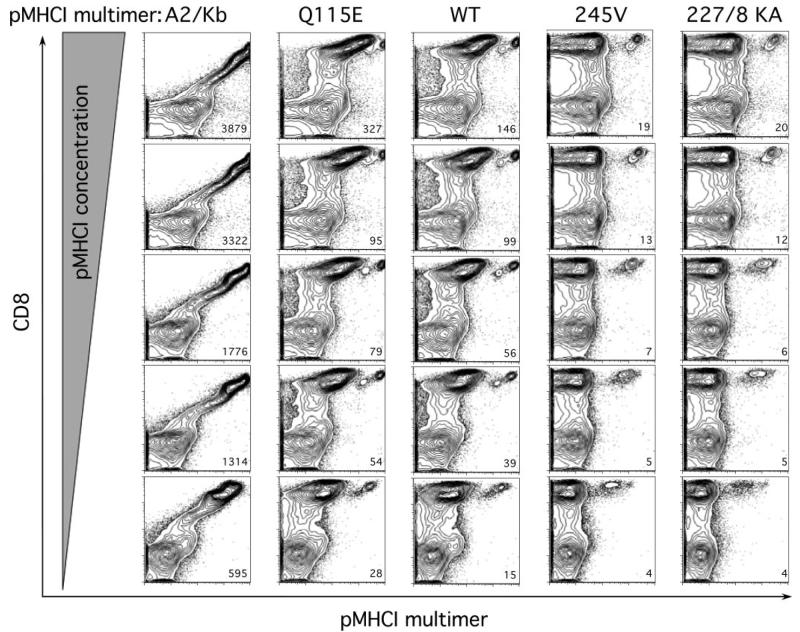
Multimeric pMHC molecules have recently revolutionized T cell immunology by enabling the direct visualization, enumeration, phenotyping, and sorting of T cells based on the antigen specificity of their TCRs (45-47). Emerging evidence suggests that the use of multimerized pMHCI antigens with decreased CD8 binding affinities enhances the specificity of these reagents in relation to background staining (48) and can be used to identify CTLs with a high sensitivity for antigen (49, 50). We used our panel of pMHCI tetrameric complexes with differing affinities for CD8 to examine the staining of human CD8+ T cells specific for the HLA A2-restricted CMV pp65-derived epitope NLVPMVATV across a range of concentrations directly ex vivo (Fig. 2). These data show the utility of these reagents for staining direct ex vivo human PBMCs and eliminate the possibility that the mutations we have engineered into HLA A2 to alter CD8 binding inadvertently introduce new binding properties into the MHC class I molecule. A distinct antigen-specific population was identified with the corresponding pMHCI tetramers of Q115E HLA A2, wild type HLA A2, A245V HLA A2, and D227K/T228A HLA A2; the magnitude of the detected population was equivalent for all of these reagents across all concentrations tested (Fig. 2). Similar results were obtained with PBMCs from two other donors with these CMV-specific reagents and in two further donors with pMHCI tetramers of wild type HLA A2, A245V HLA A2, and D227K/T228A HLA A2 folded around the EBV BMLFI-derived peptide GLCTLVAML (data not shown). Staining intensity declined with decreasing concentrations in all cases as expected. The most notable differentiating feature was background staining. As CD8 affinity increased, the CD8+tetramer− population shifted proportionately toward the CD8+tetramer+ population, as demonstrated by the mean fluorescence intensities of the CD8+tetramer− subset. This trend extended to the extreme case in which all CD8+ T cells were stained by the A2/Kb tetrameric reagent (Fig. 2). Importantly, direct ex vivo staining of PBMC with CMV-specific tetramers showed that wild type and CD8 null reagents could identify a similar population of antigen-specific cells (Fig. 2). This finding suggests that these T cells bear a TCR with a relatively high affinity for cognate antigen (49).
Fig. 2. The effects of differing pMHCI/CD8 interactions on tetramer staining of human PBMC directly ex vivo.
Donor PBMCs were stained with 10, 5, 1, 0.5, and 0.1 μg (with reference to monomer weight) of D227K/T228A HLA A2 (227/8 KA), A245V HLA A2 (245V), wild type HLA A2 (WT), Q115E HLA A2 (Q115E), or A2/Kb α3 domain (A2/Kb) fusion tetramers folded around the CMV pp65-derived peptide epitope NLVPMVATV at 37 °C for 20 min in a final volume of 50 μl. Cells were then washed and stained with anti-CD8 and anti-CD3 monoclonal antibodies for 30 min at 4 °C. Following an additional wash, PBMCs were fixed with 1% paraformaldehyde and analyzed using a FACSCalibur flow cytometer with FlowJo software. At least 300,000 events were collected for each condition. Data plots represent live CD3+ lymphocytes. Background MFI values for the CD8+tetramer− populations are shown (lower right hand corner of each section); for A2/Kb stains, the MFI value is reported for the entire CD8+ T cell population.
The pMHCI/CD8 Interaction Affects pMHCI Tetramer Dissociation
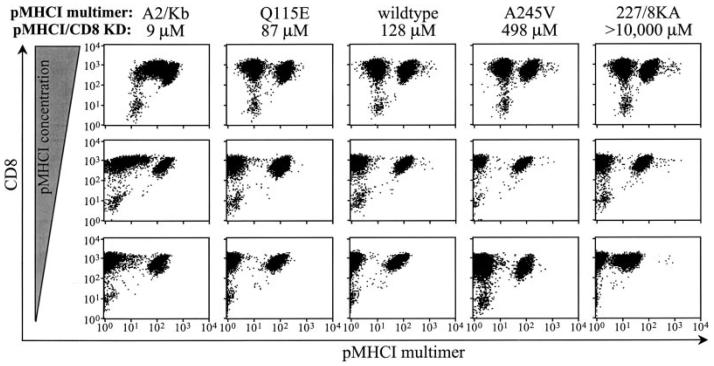
The analysis of interaction kinetics between pMHCI and cell surface TCR has been hindered by the extremely short half-lives of such interactions (1–12 s at 25 °C). Tetrameric pMHCI molecules have considerably longer interaction times and allow a quantitative assessment of dissociation. We examined the decay of pMHCI tetramers with altered pMHCI/CD8 interactions but unaltered TCR/pMHCI interactions from the cell surface of an azide-poisoned CTL line that recognizes the HLA A2-restricted, HIV-1 p17 Gag-derived epitope SLYNTVATL. This epitope is frequently immunodominant in HLA A2+ HIV-infected patients, and the prevalent CTLs in line 868 have been shown previously to be expanded in the patient from whom the line was derived (32). The corresponding pMHCI proteins of A2/Kb, Q115E HLA A2, wild type HLA A2, A245V HLA A2, and D227K/T228A HLA A2 exhibit identical TCR binding but have different CD8 binding (8, 15, 16, 49) (Fig. 1B). All of these reagents stained a population of cells in the 868 CTL line (Fig. 3). The A2/Kb reagent stained all CD8+ cells when used at high concentrations (Fig. 3). This result is consistent with our finding that this reagent stains all CD3+CD8+ human peripheral blood mononuclear cells regardless of their TCR specificity (49) (Fig. 2). When tetrameric A2/Kb is used at a low concentration, the cells with a TCR specific for the SLYNTVATL peptide appeared to out-compete other CD8+ cells for this reagent (Fig. 3; left column). Staining intensity with the D227K/T228A reagent was almost three times lower than with the wild type reagent when 0.5 μg/ml pMHCI was used (Fig. 3), consistent with the 2.5-fold difference in MFI we observed previously with this CTL line (8). Thus, these CTLs exhibit some dependence on the pMHCI/CD8 interaction for staining with pMHCI tetramers in contrast to the 003 CTL clone that recognizes the same antigen (8); these observations suggest that 003 CTL may bear a TCR with higher affinity for the cognate ligand and indicate that there is some variability between SLYNTVATL-specific CTLs in their requirements for the pMHCI/CD8 interaction to aid the binding of pMHCI tetramers.
Fig. 3. The effects of differing pMHCI/CD8 interactions on tetramer staining of 868 CTL.
The CTL line 868 is derived from an HIV-1 infected donor and is specific for the HLA A2-restricted p17 Gag epitope SLYNTVATL. 2 × 105 868 cells per test were resuspended in 20 μl of PBS and stained with D227K/T228A HLA A2 (277/8KA), A245V HLA A2 (A245V), wild type HLA A2 (wildtype), Q115E HLA A2 (Q115E), or A2/Kb α3 domain fusion (A2/Kb) tetramers folded around the SLYNTVATL peptide at either 0.5, 5 or 50 μg/ml for 20 min at 37 °C. Cells were then stained with CD8-allophycocyanin (clone SK1) for 30 min on ice, washed twice, and then resuspended in PBS. Data were acquired using a FACSCalibur flow cytometer and analyzed using CellQuest software. All HLA A2-SLYNTVATL tetramer-positive cells in the 868 CTL line bear a Vα12-2, Vβ5-6 TCR of identical sequence.
The decay of pMHCI tetramers from the cell surface of 868 CTLs clearly correlates with the affinity of the pMHCI/CD8 interaction (Fig. 4A). To study this finding further, we examined CTL clone 003, which expresses a different SLYNTVATL-specific TCR than 868 CTL and is derived from a different patient (31, 32). This clone is believed to be the dominant clone that recognized this epitope in vivo (32). We have shown previously that wild type and D227K/T228A pMHCI tetramers exhibit a similar on-rate and stain these cells within 30 s (8). Using our panel of tetramers with altered CD8 binding properties, we confirmed that a decreased pMHCI/CD8 interaction leads to more rapid tetramer dissociation and that an increased pMHCI/CD8 interaction results in slower tetramer dissociation (Fig. 4B). Similar data were obtained with three further sets of CTLs (Fig. 4, C-E). Thus, the pMHCI/CD8 interaction affects the stability of the TCR/pMHCI interaction on the cell surface. The decay of pMHCI tetramers from 003 CTL (Fig. 4B) was slower than for 868 CTL (Fig. 4A). The former CTL might have a higher affinity TCR, consistent with our finding that 003 CTL are less dependent on CD8 for tetramer binding (8) than 868 CTL (Fig. 3).
The pMHCI/CD8 Interaction Stabilizes the TCR/pMHCI Interaction ∼2-Fold
The apparent off-rates in the tetramer dissociation experiments (Figs. 4-6) exhibit a wide variation; the D227K/T228A HLA A2 decay rate differs from that of wild type HLA A2 by a factor of ∼10. However, these differences in apparent off-rates do not translate directly into single-site off-rates, because tetramer kinetics involves TCR-bound forms of three different valancies (see Equations 1-3). In the regime where tetramer decay curves are virtually exponential, the true ratio of off-rates is the cube root of the ratio of apparent off-rates. In particular, the tetramer dissociation curve fit for D227K/T228A HLA A2 from 003 CTL indicates that (3/μ2)⅓koff° = 0.859 min−⅓ (see Experimental Procedures for notation). The intercept of the straight line fit (Equation 9) shown in Fig. 5B yields (3/μ2)⅓koff* = 0.398 min−⅓. These data suggest that the maximum effect of the pMHCI/CD8 interaction is a prolongation by a factor of 2.16 of the mean TCR/pMHCI binding time. The fit also indicates that (3/μ2)⅓ koff°/κ = 0.29 min−⅓/nm, whence κ = 3 mm, which, in the interaction between a T cell and an antigen-presenting cell, would translate as 6 × 104 free CD8 molecules in the contact area between a T cell and an antigen-presenting cell if we assume σA = 30 × 10−21 m3, where A is the area of the contact interface between the cells and σ is the “confinement length”; the free CD8 count equals Aσκ. Taken together, the tetramer decay experiments indicate that the pMHCI/CD8 interaction stabilizes by ∼2-fold the interaction between the TCR of HIV Gag-specific clone 003 and HLA A2/Kb-SLYNTVATL. The actual effect depends on the fraction of time spent by the MHC molecule in the CD8-bound form, and this fraction increases with increasing affinity. Thus, the wild type pMHCI/CD8 induces a stabilization factor of 1.95, which is close to the maximum of 2.16 by virtue of its low KD of 128 μm. A similar CD8-stabilization effect was observed for 868 CTLs (Fig. 6) and other CTLs with different functional sensitivities (summarized in Table I). We find that pMHCI tetramers rapidly induce the cell death of so-called “high avidity” CTLs (61).2 This may also be true when these reagents are used to sort-clone CTL. The failure of CTL clones with low functional avidity/sensitivity to stain with CD8 null tetramers (44) (Fig. 4)2 makes it impossible to calculate an apparent off-rate for these reagents from these clones. The lack of stability in the absence of a pMHCI/CD8 interaction precludes calculation of a CD8-mediated stabilization factor for such CTLs. Nevertheless, comparison of the off-rates of the other tetramers from the surface of CTL clone SLY-10 to those of other CTLs (Fig. 4) strongly suggests that the pMHCI/CD8 interaction provides a similar stabilization factor of ∼2-fold for CTL clones of very low functional avidity.
TABLE I.
Summary of CTL used and calculated CD8-mediated TCR/pMHCI stabilization factor
| CTL | Specificity | Epitope | Functional avidity | Data figures | Stabilization factor |
|---|---|---|---|---|---|
| 003 | HIV-1 | SLYNTVATL | Very high | 4 and 5 | 2.16 |
| 868 (Line) | HIV-1 | SLYNTVATL | High | 4 and 6 | 1.85 |
| 0400 (Line) | HIV-1 | SLYNTVATL | Intermediate | 4 | 1.7 |
| SLY-10 | HIV-1 | SLYNTVATL | Very low | 4 | NPa |
| EBV-A | EBV | GLCTLVAML | Low | 4 | 2.3 |
It is not possible (NP) to calculate a stabilization factor for CTL clones with a very low functional avidity (see legend to Fig. 4).
DISCUSSION
The T cell surface glycoprotein CD8 interacts with nonpolymorphic regions of MHCI, allowing a single pMHCI to bind both CD8 and TCR simultaneously (3). It has been reported that CD8 can increase the antigen sensitivity of CTL by a factor of >106 and significantly reduce the TCR triggering threshold required for CTL activation (8, 51). Several roles for CD8 in CTL activation have been proposed (reviewed in Refs. 52 and 53). CD8 could do the following: (i) stabilize TCR/pMHCI interactions at the CTL-target cell interface; (ii) play a major role in the topographical organization of cell surface TCR; and, (iii) act to recruit signaling molecules to the TCR-CD3 complex. Dissecting the role of CD8 in antigen recognition has traditionally been tackled using anti-CD8 antibodies. However, such antibodies can have both positive and negative effects on antigen recognition (27, 54) and are able to exert these functions in the absence of any interaction between pMHCI and CD8 (27). In addition, the use of antibodies directed against the CD8 molecule does not allow discrimination between the role of the pMHCI/CD8 interaction, direct coupling of CD8 to the TCR, or other possible roles of CD8 in antigen recognition. The role of CD8 has also been examined by transfecting CD8-negative T cell hybridomas with CD8α and CD8β (51, 55, 56). However, CD8 plays a pivotal role in the organization of cell surface TCRs (13, 27-29, 57-59) and the recruitment of essential signaling components to the cytoplasmic side of the TCR-CD3-ζ complex (6,9), thereby making it impossible to study the role of the pMHCI/CD8 interaction in isolation with such systems. In view of these caveats, we developed the use of MHCI mutations that alter the affinity of CD8 binding without any effect on the TCR/pMHCI interaction in order to study the role of the pMHCI/CD8 interaction, because the precise impact of these mutations can be quantified by SPR (8). Many recent studies have used such molecules (8, 17, 27, 60, 61). These reagents have been used to show that the pMHCI/CD8 interaction enhances sensitivity to antigen by mediating complete phosphorylation of the TCR ζ chain (8). TCR engagement in the absence of a pMHCI/CD8 interaction results in preferential induction of only partially phosphorylated CD3-ζ (p21 phosphoform) and thus cannot effect rapid T cell activation (8). In the present study, we specifically address the role that CD8 plays in the stabilization and kinetics of the TCR/pMHCI interaction at the cell surface. This issue has been the source of much debate but is amenable to precise quantification with the point mutated recombinant proteins used herein.
The use of soluble pMHCI proteins enables TCR/pMHCI interactions and kinetics at the cell surface to be studied without interference from other adhesion or co-stimulatory molecules. In the monomeric form the use of this method is complicated by the extremely short interaction half-life; however, increasing the valency of these reagents by avidin-biotin-based tetramerization significantly increases the cumulative avidity and produces reagents that are valuable tools for the identification and phenotyping of antigen-specific CTLs (45, 46). The effect of the pMHCI/CD8 interaction on the ability of pMHCI tetrameric reagents to form stable interactions with cell surface TCRs has been extensively investigated. Several studies have demonstrated that anti-CD8 antibodies act to block pMHCI tetramer binding to both human and murine CTLs (62-64). In these studies, it was assumed that anti-CD8 antibodies exert their effects by blocking the pMHCI/CD8 interaction, leading to the conclusion that this interaction is “critical” for the stable binding of pMHCI tetrameric reagents to cell surface TCRs. However, we have since shown that anti-CD8 antibodies can reduce tetramer binding even in the absence of a pMHCI/CD8 interaction (27). Anti-CD8 antibodies need not therefore block tetramer binding by interfering with the pMHCI/CD8 interaction and thus cannot be used to define the effect of this interaction on the binding avidity of pMHCI tetramers. We have recently reinforced these findings by showing that some anti-CD4 antibody clones inhibit the binding of pMHCII tetramers in several systems.3
Introducing the α3 domain mutation D227K/T228A into HLA A2 has been shown to abrogate the pMHCI/CD8 interaction (KD of >10,000 μm) without affecting the TCR/pMHCI interaction (8). We have shown that “CD8 null” tetramers bearing this mutation can stain human anti-viral CTL clones efficiently and at an intensity and on-rate similar to that of staining with wild type tetramers (8). For many human anti-viral CTLs, the TCR/pMHCI interaction is almost 100 times stronger than the pMHCI/CD8 interaction (16)2 and of significantly longer duration; therefore, it is not surprising that the requirement for the latter interaction in the stable cell surface binding of pMHCI tetramers is minimal. In this study, we confirm that CD8 null tetramers can efficiently stain human anti-viral CTL in vitro. Furthermore, we show that both wild type and CD8 null pMHCI tetramers efficiently stain similar populations of anti-CMV and anti-EBV CTLs directly ex vivo (Fig. 2, and data not shown). Thus, in accordance with our in vitro studies with human and murine CTL (8, 27, 49), we find that the pMHCI/CD8 interaction is not uniformly essential for multimer binding to cell surface TCRs.
Previous studies have shown that the introduction of α3 domain mutations that reduce the pMHCI/CD8 interaction can significantly decrease the level of pMHCI tetramer binding in human and murine systems and the level of murine pMHCI binding as assessed by photoaffinity labeling (14, 17, 58, 65). Our data demonstrating that the pMHCI/CD8 interaction need not be critical for stable pMHCI multimer binding may at first seem at variance with these studies. However, it is now clear that CTLs exhibit a range of dependence on the pMHCI/CD8 interaction for the stable cell surface binding of pMHCI tetrameric complexes. We have shown previously that D227K/T228A CD8 null tetramers selectively stain only those CTLs with a high sensitivity for antigen (49). Wild type reagents can stain low avidity CTL efficiently; however, CD8 null reagents stain low avidity anti-viral and tumor-specific CTLs poorly or not at all (49). Therefore, the dependence on the pMHCI/CD8 interaction for stable tetramer binding correlates with the functional avidity of the CTL and is thought to reflect the intrinsic affinity of the TCR for the pMHCI ligand, along with other factors such as cell surface organization and density of the TCR (49, 50, 66). In contrast, the pMHCII interaction with the CD4 coreceptor is significantly weaker than the pMHCI/CD8 interaction (67, 68). Early studies in which CD4 and a mutant CD4 without a capacity for cytoplasmic signaling were expressed in T cell hybridomas lacking endogenous CD4 concluded that CD4 has a very minor role as an adhesion molecule in T cell activation (69). This finding has been upheld by more recent reports showing that CD4 does not aid the stabilization of the TCR/pMHCII interaction at the cell surface (70-72).
Our data point to a reconciliation of the apparently disparate findings regarding the requirement for the pMHCI/CD8 interaction in the stable binding of pMHCI tetramers. In attempting such a reconciliation, it is important to treat results gained using anti-CD8 antibodies with caution because these reagents appear to have multiple effects, some of which are independent of the interaction between pMHCI and CD8 (27). It is also important to take the increased affinity of the pMHCI/CD8 interaction in mouse as compared with that in human (8, 73) into account. However, even when such factors are taken into consideration, there appears to be variability within the human (27, 49) and murine (44, 49) systems with regard to the requirement for CD8-mediated stabilization for pMHCI multimer binding. The TCR/pMHCI interaction is of short duration (∼1–12 s at 25 °C). Tetramers derive their high avidity from the large probability that a monovalently bound tetramer will bind bivalently before the single bound site dissociates; this probability is large if the association rate for further sites is much greater than the single-site dissociation rate. This is certainly the case for the tetramers used in this study, as evidenced by the exponential tetramer dissociation curves. Thus, tetramerization of the TCR/pMHCI interaction increases the bound half-life by hundreds of fold (74), as all pMHCI molecules need to be unligated simultaneously for the tetramer to dissociate from the cell surface. As described above, stable cell surface adhesion of pMHCI tetramers has an empirical requirement for the monomeric interaction to be of sufficient duration to allow a further monomer in the complex to interact with another TCR prior to release of the original interaction. Presumably, strong TCR/pMHCI interactions, such as those of immunodominant human anti-viral CTLs, exceed this minimal requirement per se. We have shown that TCR and CD8 cooperate in binding pMHCI at the cell surface (Fig. 4). The pMHCI/CD8 interaction delays the dissociation of the TCR/pMHCI interaction by a factor of ∼2 (Figs. 5 and 6 and Table I) and can thus enable weaker TCR/pMHCI engagements to attain the minimal half-life for stable binding of tetrameric reagents to the CTL surface.
The kinetic proofreading model of T cell activation (20, 21) in which the koff of the TCR/pMHCI interaction and, hence, its half-life, appears to be the principal feature determining the biological outcome of TCR ligation is widely accepted. Recent rigorous testing of this model reinforces its relevance but revises the model to account for how low level undetectable signaling induced by weak TCR ligands can trickle down to affect T cell activation (75) and also to account for how a few ligands can be somewhat more stimulatory than predicted on the basis of the half-life of the TCR/pMHCI interaction alone by the inclusion of a value for heat capacity (76). The potential role of CD8 in stabilizing this interaction is therefore of considerable biological importance and has been the subject of intense debate (11-14). We have shown previously that pMHCI multimers are rapidly internalized by CTL that bear a cognate TCR (77). This process can be prevented by staining on ice in the presence of azide as described under “Experimental Procedures.” If tetrameric pMHCI is prevented from internalizing and re-binding using an anti-MHCI blocking antibody or “cold” unlabeled tetramer, then it is possible to examine tetramer decay from the cell surface (78). Using human anti-viral CTL and pMHCI tetramers with abrogated, reduced, normal, slightly enhanced, and greatly enhanced CD8 binding affinities but unaltered TCR binding, we find that the pMHCI/CD8 interaction can significantly affect the dissociation rate of pMHCI tetramers. Abrogating the pMHCI/CD8 interaction significantly lowers the half-life of pMHCI tetramer binding to cell surface TCR (Fig. 4). Increasing the pMHCI/CD8 interaction without altering the TCR/pMHCI interaction (Fig. 1) results in a pMHCI tetramer that binds to the CTL surface with greater stability than the wild type molecule (Fig. 4). We modeled the contribution of the pMHCI/CD8 interaction to stabilization of the TCR/pMHCI interaction and found that the pMHCI/CD8 interaction stabilizes the monomeric interaction between the TCR of HIV-1 Gag-specific 003 and 868 CTLs and HLA A2-SLYNTVATL by ∼2-fold. A similar value was also determined from the decay from other CTL (summarized in Table I). This factor did not appear to vary with the functional avidity of the CTL or the epitope recognized. It may be shown that the calculated stabilization factor is not dependent on the assumption that koff* does not vary with KD (although the estimate of κ is affected if we allow for such variation).
We would expect the 2-fold stabilization to enhance the ability of the ligand to activate the T cell. Because the rate at which TCRs are triggered depends non-monotonically on the mean TCR/pMHCI binding time (78, 79), the effect of stabilization on the TCR triggering rate may vary, both in size and sense. In particular, a 2-fold increase will change the TCR triggering rate by a factor of exp{1/(2τ)}/2 (80); here τ denotes the average TCR/pMHCI interaction time in the absence of the CD8 stabilization effect divided by the time required to trigger the TCR-CD3 complex. The formula is an upper bound that applies when TCR is present in excess; Ref. 80 shows how to deal with the general case. It follows that the effect can be substantial, arbitrarily >2-fold when τ < 1/ln 8. However, such low τ ligands are very weak agonists, and even a manyfold increase of the triggering rate that they induce will not have a significant impact. For a better agonist (i.e. one such that τ > 1/ln 8), the increase is <2-fold, and for a near optimal agonist such that τ < 1/ln 4 (in the absence of CD8), the effect vanishes altogether. If a ligand is already optimal in the absence of CD8, it becomes 17% less effective in the saturating presence of CD8. Hence, if we rank the strong agonists for a given T cell by potency we conclude that the CD8 stabilization effect can alter the order of that ranking, allowing CTLs to focus their functional avidity on a ligand by adjusting CD8 expression levels.
In summary, we have used a range of pMHCIs with altered CD8 binding but unaltered TCR binding to examine the TCR/pMHCI/CD8 interaction at the cell surface. These experiments allow an assessment of cooperative binding not possible in previous biophysical and structural studies using soluble molecules (3, 12, 13). We show that the TCR and CD8 bind to pMHCI cooperatively at the cell surface. Modeling for the monomeric TCR/pMHCI/CD8 interaction indicates that CD8 provides a stabilization factor of ∼2 that is applicable across all systems tested. The requirement for CD8 to stabilize the TCR/pMHCI interaction beyond a threshold sufficient for TCR triggering or stable binding of multimeric pMHCI to cell surface TCR is minimal with strong TCR ligands (15) but becomes increasingly apparent as the TCR/pMHCI half-life decreases (49, 50), consistent with model predictions (79). The 2-fold stabilization effect provided by the pMHCI/CD8 interaction is expected to enhance T cell activation per se. The TCR triggering rate has been found to depend non-monotonically on the off-rate, with an optimum positioned at a point where 1/koff corresponds to the TCR triggering threshold (79, 80). Reducing koff by a factor of 2 may in fact have a negative impact on ligands with off-rates too slow for optimal stimulation. However, the vast majority of ligands will have off-rates too fast for optimal stimulation of a given TCR, and such ligands will increase in TCR triggering efficacy when koff is reduced by half. Indeed, weak agonist variant peptides cannot be recognized in the absence of pMHCI/CD8 interaction (Fig. S3 in the supplemental data found in the on-line version of this article). Overall, these findings suggest that CD8-mediated stabilization of the TCR/pMHCI interaction contributes to T cell cross-reactivity and promiscuity (81), an effect that might be amenable to therapeutic intervention.
Supplementary Material
Acknowledgments
We are indebted to Bent Jakobsen, Marco Purbhoo, Anton van der Merwe, and Martin Meier-Schellerscheim for helpful comments and constructive criticisms.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental data on the decay of pMHCI tetramers (Figs. S1 and S2) and on pMHCI/CD8 interaction (Fig. S3).
The abbreviations used are: CTL, CD8+ cytotoxic T lymphocyte; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HIV, human immunodeficiency virus; HTLV, human T cell lymphotrophic virus; MFI, mean fluorescence intensity; MHC, major histocompatibility complex; MHCI, MHC class I; pMHCI, peptide-MHCI; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; SPR, surface plasmon resonance; TCR, T cell receptor.
A. K. Sewell, unpublished data.
L. Wooldridge and T. J. Scriba, unpublished data.
REFERENCES
- 1.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph MG, Wilson IA. Curr. Opin. Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 3.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, Jakobsen BK. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 4.Kern PS, Teng MK, Smolyar A, Liu JH, Liu J, Hussey RE, Spoerl R, Chang HC, Reinherz EL, Wang JH. Immunity. 1998;9:519–530. doi: 10.1016/s1074-7613(00)80635-4. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA., Jr. Annu. Rev. Immunol. 1992;10:645–674. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 6.Veillette A, Bookman MA, Horak EM, Bolen JB. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 7.Chalupny NJ, Ledbetter JA, Kavathas P. EMBO J. 1991;10:1201–1207. doi: 10.1002/j.1460-2075.1991.tb08061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purbhoo MA, Boulter JM, Price DA, Vuidepot AL, Hourigan CS, Dunbar PR, Olson K, Dawson SJ, Phillips RE, Jakobsen BK, Bell JI, Sewell AK. J. Biol. Chem. 2001;276:32786–32792. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- 9.Bosselut R, Zhang W, Ashe JM, Kopacz JL, Samelson LE, Singer A. J. Exp. Med. 1999;190:1517–1526. doi: 10.1084/jem.190.10.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norment AM, Salter RD, Parham P, Engelhard VH, Littman DR. Nature. 1988;336:79–81. doi: 10.1038/336079a0. [DOI] [PubMed] [Google Scholar]
- 11.Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 12.Wyer JR, Willcox BE, Gao GF, Gerth UC, Davis SJ, Bell JI, van der Merwe PA, Jakobsen BK. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 13.Arcaro A, Gregoire C, Bakker TR, Baldi L, Jordan M, Goffin L, Boucheron N, Wurm F, van der Merwe PA, Malissen B, Luescher IF. J. Exp. Med. 2001;194:1485–1495. doi: 10.1084/jem.194.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutoit V, Guillaume P, Ayyoub M, Hesdorffer CS, Luescher IF, Valmori D. J. Immunol. 2003;170:5110–5117. doi: 10.4049/jimmunol.170.10.5110. [DOI] [PubMed] [Google Scholar]
- 15.Hutchinson SL, Wooldridge L, Tafuro S, Laugel B, Glick M, Boulter JM, Jakobsen BK, Price DA, Sewell AK. J. Biol. Chem. 2003;278:24285–24293. doi: 10.1074/jbc.M300633200. [DOI] [PubMed] [Google Scholar]
- 16.Gao GF, Willcox BE, Wyer JR, Boulter JM, O'Callaghan CA, Maenaka K, Stuart DI, Jones EY, van der Merwe PA, Bell JI, Jakobsen BK. J. Biol. Chem. 2000;275:15232–15238. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 17.Schott E, Ploegh HL. Eur. J. Immunol. 2002;32:3425–3434. doi: 10.1002/1521-4141(200212)32:12<3425::AID-IMMU3425>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. Nature. 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 19.Lyons DS, Lieberman SA, Hampl J, Boniface JJ, Chien Y, Berg LJ, Davis MM. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- 20.McKeithan TW. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabinowitz JD, Beeson C, Lyons DS, Davis MM, McConnell HM. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1401–1405. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Merwe PA, Brown MH, Davis SJ, Barclay AN. EMBO J. 1993;12:4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki S, Kupsch J, Eichmann K, Saizawa MK. Eur. J. Immunol. 1992;22:2475–2479. doi: 10.1002/eji.1830221002. [DOI] [PubMed] [Google Scholar]
- 24.Naeher D, Luescher IF, Palmer E. J. Immunol. 2002;169:2964–2970. doi: 10.4049/jimmunol.169.6.2964. [DOI] [PubMed] [Google Scholar]
- 25.Doucey MA, Goffin L, Naeher D, Michielin O, Baumgartner P, Guillaume P, Palmer E, Luescher IF. J. Biol. Chem. 2003;278:3257–3264. doi: 10.1074/jbc.M208119200. [DOI] [PubMed] [Google Scholar]
- 26.Osono E, Sato N, Yokomuro K, Saizawa MK. Scand. J. Immunol. 1997;45:487–493. doi: 10.1046/j.1365-3083.1997.d01-427.x. [DOI] [PubMed] [Google Scholar]
- 27.Wooldridge L, Hutchinson SL, Choi EM, Lissina A, Jones E, Mirza F, Dunbar PR, Price DA, Cerundolo V, Sewell AK. J. Immunol. 2003;171:6650–6660. doi: 10.4049/jimmunol.171.12.6650. [DOI] [PubMed] [Google Scholar]
- 28.Kwan Lim GE, McNeill L, Whitley K, Becker DL, Zamoyska R. Eur. J. Immunol. 1998;28:745–754. doi: 10.1002/(SICI)1521-4141(199802)28:02<745::AID-IMMU745>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Cawthon AG, Alexander-Miller MA. J. Immunol. 2002;169:3492–3498. doi: 10.4049/jimmunol.169.7.3492. [DOI] [PubMed] [Google Scholar]
- 30.Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF. J. Immunol. 2000;165:2068–2076. doi: 10.4049/jimmunol.165.4.2068. [DOI] [PubMed] [Google Scholar]
- 31.Sewell AK, Harcourt GC, Goulder PJ, Price DA, Phillips RE. Eur. J. Immunol. 1997;27:2323–2329. doi: 10.1002/eji.1830270929. [DOI] [PubMed] [Google Scholar]
- 32.Wilson JD, Ogg GS, Allen RL, Goulder PJ, Kelleher A, Sewell AK, O'Callaghan CA, Rowland-Jones SL, Callan MF, McMichael AJ. J. Exp. Med. 1998;188:785–790. doi: 10.1084/jem.188.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulder PJ, Sewell AK, Lalloo DG, Price DA, Whelan JA, Evans J, Taylor GP, Luzzi G, Giangrande P, Phillips RE, McMichael AJ. J. Exp. Med. 1997;185:1423–1433. doi: 10.1084/jem.185.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Callaghan C A, Byford MF, Wyer JR, Willcox BE, Jakobsen BK, McMichael AJ, Bell JI. Anal. Biochem. 1999;266:9–15. doi: 10.1006/abio.1998.2930. [DOI] [PubMed] [Google Scholar]
- 35.Glick M, Price DA, Vuidepot AL, Andersen TB, Hutchinson SL, Laugel B, Sewell AK, Boulter JM, Dunbar PR, Cerundolo V, Oxenius A, Bell JI, Richards WG, Jakobsen BK. J. Biol. Chem. 2002;277:20840–20846. doi: 10.1074/jbc.M201819200. [DOI] [PubMed] [Google Scholar]
- 36.Utz U, Koenig S, Coligan JE, Biddison WE. J. Immunol. 1992;149:214–221. [PubMed] [Google Scholar]
- 37.Johnson RP, Trocha A, Yang L, Mazzara GP, Panicali DL, Buchanan TM, Walker BD. J. Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- 38.Scotet E, David-Ameline J, Peyrat MA, Moreau-Aubry A, Pinczon D, Lim A, Even J, Semana G, Berthelot JM, Breathnach R, Bonneville M, Houssaint E. J. Exp. Med. 1996;184:1791–1800. doi: 10.1084/jem.184.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diamond DJ, York J, Sun JY, Wright CL, Forman SJ. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- 40.Gao GF, Gerth UC, Wyer JR, Willcox BE, O'Callaghan CA, Zhang Z, Jones EY, Bell JI, Jakobsen BK. Protein Sci. 1998;7:1245–1249. doi: 10.1002/pro.5560070520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willcox BE, Gao GF, Wyer JR, O'Callaghan CA, Boulter JM, Jones EY, van der Merwe PA, Bell JI, Jakobsen BK. Protein Sci. 1999;8:2418–2423. doi: 10.1110/ps.8.11.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glick M, Robinson DD, Grant GH, Richards WG. J. Am. Chem. Soc. 2002;124:2337–2344. doi: 10.1021/ja016490s. [DOI] [PubMed] [Google Scholar]
- 43.Glick M, Grant GH, Richards WG. J. Med. Chem. 2002;45:4639–4646. doi: 10.1021/jm020830i. [DOI] [PubMed] [Google Scholar]
- 44.Choi EM, Palmowski M, Chen J, Cerundolo V. J. Immunol. Methods. 2002;268:35–41. doi: 10.1016/s0022-1759(02)00198-9. [DOI] [PubMed] [Google Scholar]
- 45.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 46.Klenerman P, Cerundolo V, Dunbar PR. Nat. Rev. Immunol. 2002;2:263–272. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- 47.Kelleher AD, Rowland-Jones SL. Curr. Opin. Immunol. 2000;12:370–374. doi: 10.1016/s0952-7915(00)00102-3. [DOI] [PubMed] [Google Scholar]
- 48.Bodinier M, Peyrat MA, Tournay C, Davodeau F, Romagne F, Bonneville M, Lang F. Nat. Med. 2000;6:707–710. doi: 10.1038/76292. [DOI] [PubMed] [Google Scholar]
- 49.Choi EM, Chen JL, Wooldridge L, Salio M, Lissina A, Lissin N, Hermans IF, Silk JD, Mirza F, Palmowski MJ, Dunbar PR, Jakobsen BK, Sewell AK, Cerundolo V. J. Immunol. 2003;171:5116–5123. doi: 10.4049/jimmunol.171.10.5116. [DOI] [PubMed] [Google Scholar]
- 50.Pittet MJ, Rubio-Godoy V, Bioley G, Guillaume P, Batard P, Speiser D, Luescher I, Cerottini JC, Romero P, Zippelius A. J. Immunol. 2003;171:1844–1849. doi: 10.4049/jimmunol.171.4.1844. [DOI] [PubMed] [Google Scholar]
- 51.Holler PD, Kranz DM. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 52.Gao GF, Rao Z, Bell JI. Trends Immunol. 2002;23:408–413. doi: 10.1016/s1471-4906(02)02282-2. [DOI] [PubMed] [Google Scholar]
- 53.Gao GF, Jakobsen BK. Immunol. Today. 2000;21:630–636. doi: 10.1016/s0167-5699(00)01750-3. [DOI] [PubMed] [Google Scholar]
- 54.Tomonari K, Spencer S. Int. Immunol. 1990;2:1189–1194. doi: 10.1093/intimm/2.12.1189. [DOI] [PubMed] [Google Scholar]
- 55.Kwan-Lim GE, Ong T, Aosai F, Stauss H, Zamoyska R. Int. Immunol. 1993;5:1219–1228. doi: 10.1093/intimm/5.10.1219. [DOI] [PubMed] [Google Scholar]
- 56.Gabert J, Langlet C, Zamoyska R, Parnes JR, Schmitt-Verhulst AM, Malissen B. Cell. 1987;50:545–554. doi: 10.1016/0092-8674(87)90027-4. [DOI] [PubMed] [Google Scholar]
- 57.Buslepp J, Kerry SE, Loftus D, Frelinger JA, Appella E, Collins EJ. J. Immunol. 2003;170:373–383. doi: 10.4049/jimmunol.170.1.373. [DOI] [PubMed] [Google Scholar]
- 58.Kerry SE, Buslepp J, Cramer LA, Maile R, Hensley LL, Nielsen AI, Kavathas P, Vilen BJ, Collins EJ, Frelinger JA. J. Immunol. 2003;171:4493–4503. doi: 10.4049/jimmunol.171.9.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schott E, Bertho N, Ge Q, Maurice MM, Ploegh HL. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13735–13740. doi: 10.1073/pnas.212515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guillaume P, Legler DF, Boucheron N, Doucey MA, Cerottini JC, Luescher IF. J. Biol. Chem. 2003;278:4500–4509. doi: 10.1074/jbc.M208863200. [DOI] [PubMed] [Google Scholar]
- 61.Xu XN, Purbhoo MA, Chen N, Mongkolsapaya J, Cox JH, Meier UC, Tafuro S, Dunbar PR, Sewell AK, Hourigan CS, Appay V, Cerundolo V, Burrows SR, McMichael AJ, Screaton GR. Immunity. 2001;14:591–602. doi: 10.1016/s1074-7613(01)00133-9. [DOI] [PubMed] [Google Scholar]
- 62.Denkberg G, Cohen CJ, Reiter Y. J. Immunol. 2001;167:270–276. doi: 10.4049/jimmunol.167.1.270. [DOI] [PubMed] [Google Scholar]
- 63.Campanelli R, Palermo B, Garbelli S, Mantovani S, Lucchi P, Necker A, Lantelme E, Giachino C. Int. Immunol. 2002;14:39–44. doi: 10.1093/intimm/14.1.39. [DOI] [PubMed] [Google Scholar]
- 64.Daniels MA, Jameson SC. J. Exp. Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luescher IF, Vivier E, Layer A, Mahiou J, Godeau F, Malissen B, Romero P. Nature. 1995;373:353–356. doi: 10.1038/373353a0. [DOI] [PubMed] [Google Scholar]
- 66.Cho BK, Lian KC, Lee P, Brunmark A, McKinley C, Chen J, Kranz DM, Eisen HN. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1723–1727. doi: 10.1073/pnas.98.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Merwe PA, Davis SJ. Annu. Rev. Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 68.Xiong Y, Kern P, Chang H, Reinherz E. J. Biol. Chem. 2001;276:5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 69.Glaichenhaus N, Shastri N, Littman DR, Turner JM. Cell. 1991;64:511–520. doi: 10.1016/0092-8674(91)90235-q. [DOI] [PubMed] [Google Scholar]
- 70.Hamad AR, O'Herrin SM, Lebowitz MS, Srikrishnan A, Bieler J, Schneck J, Pardoll D. J. Exp. Med. 1998;188:1633–1640. doi: 10.1084/jem.188.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boniface JJ, Rabinowitz JD, Wulfing C, Hampl J, Reich Z, Altman JD, Kantor RM, Beeson C, McConnell HM, Davis MM. Immunity. 1998;9:459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 72.Crawford F, Kozono H, White J, Marrack P, Kappler J. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 73.Sewell AK, Price DA. Trends Immunol. 2001;22:595. doi: 10.1016/s1471-4906(01)01866-x. [DOI] [PubMed] [Google Scholar]
- 74.Laugel B, Boulter JM, Lissin N, Vuidepot A, Li Y, Gostick E, Crotty LE, Douek DC, Hemelaar J, Price DA, Jakobsen BK, Sewell AK. J. Biol. Chem. 2005;280:1882–1892. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- 75.Rosette C, Werlen G, Daniels MA, Holman PO, Alam SM, Travers PJ, Gascoigne NR, Palmer E, Jameson SC. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 76.Krogsgaard M, Prado N, Adams EJ, He XL, Chow DC, Wilson DB, Garcia KC, Davis MM. Mol. Cell. 2003;12:1367–1378. doi: 10.1016/s1097-2765(03)00474-x. [DOI] [PubMed] [Google Scholar]
- 77.Whelan JA, Dunbar PR, Price DA, Purbhoo MA, Lechner F, Ogg GS, Griffiths G, Phillips RE, Cerundolo V, Sewell AK. J. Immunol. 1999;163:4342–4348. [PubMed] [Google Scholar]
- 78.Savage PA, Boniface JJ, Davis MM. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 79.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. Nat. Immunol. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 80.van den Berg HA, Burroughs NJ, Rand DA. Bull. Math. Biol. 2002;64:781–808. doi: 10.1006/bulm.2002.0302. [DOI] [PubMed] [Google Scholar]
- 81.Mason D. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.