Abstract
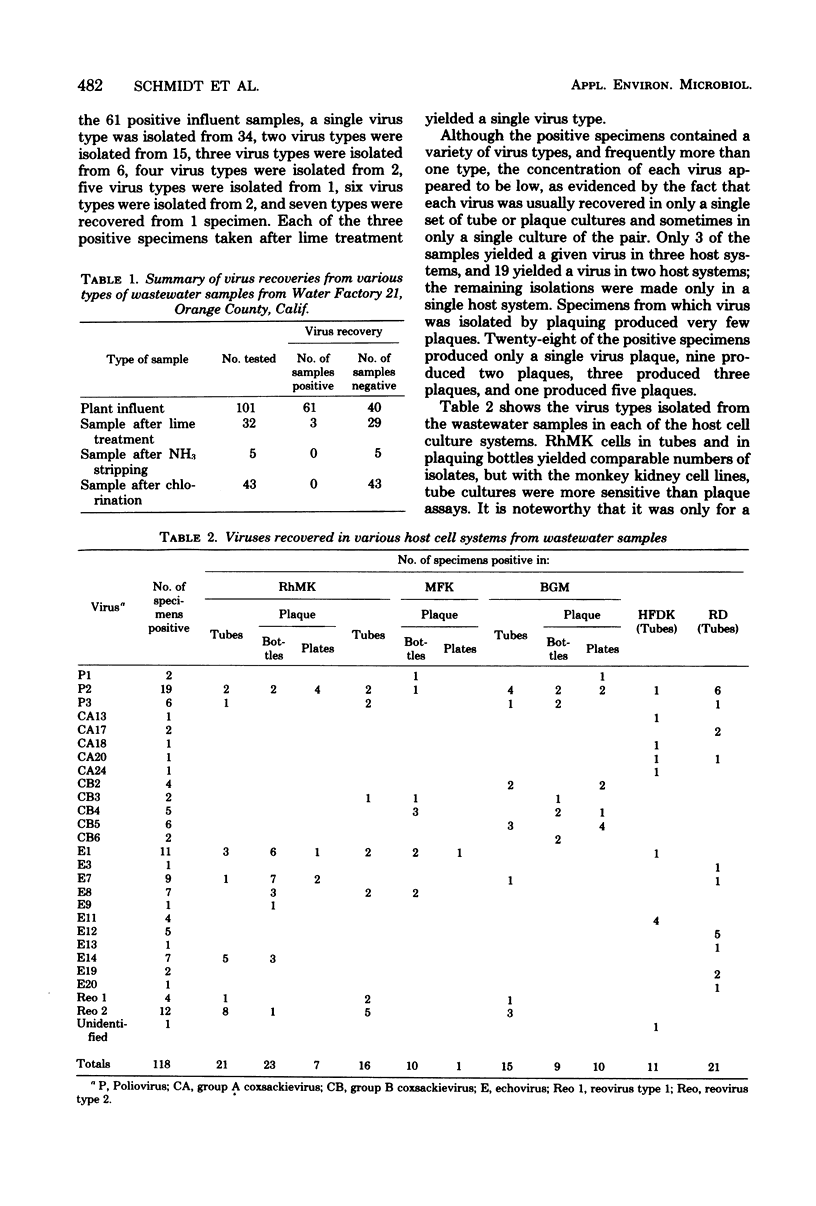
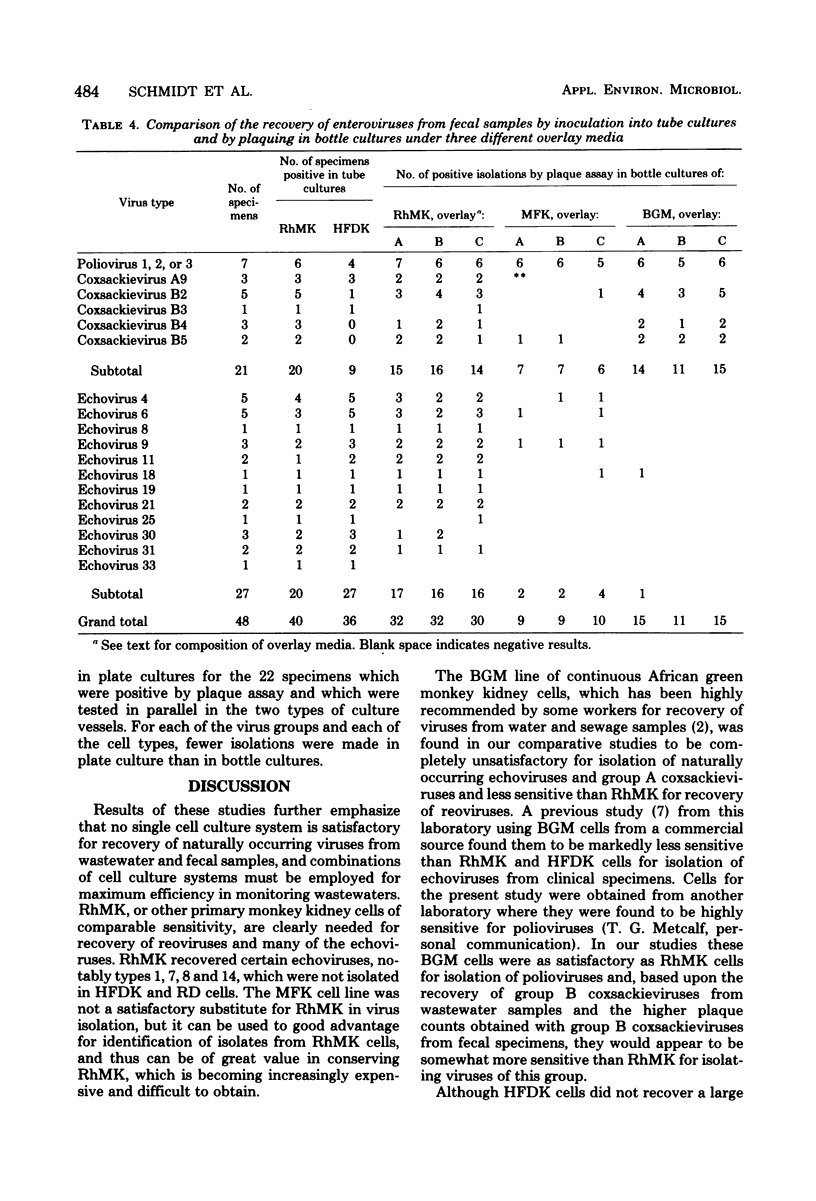
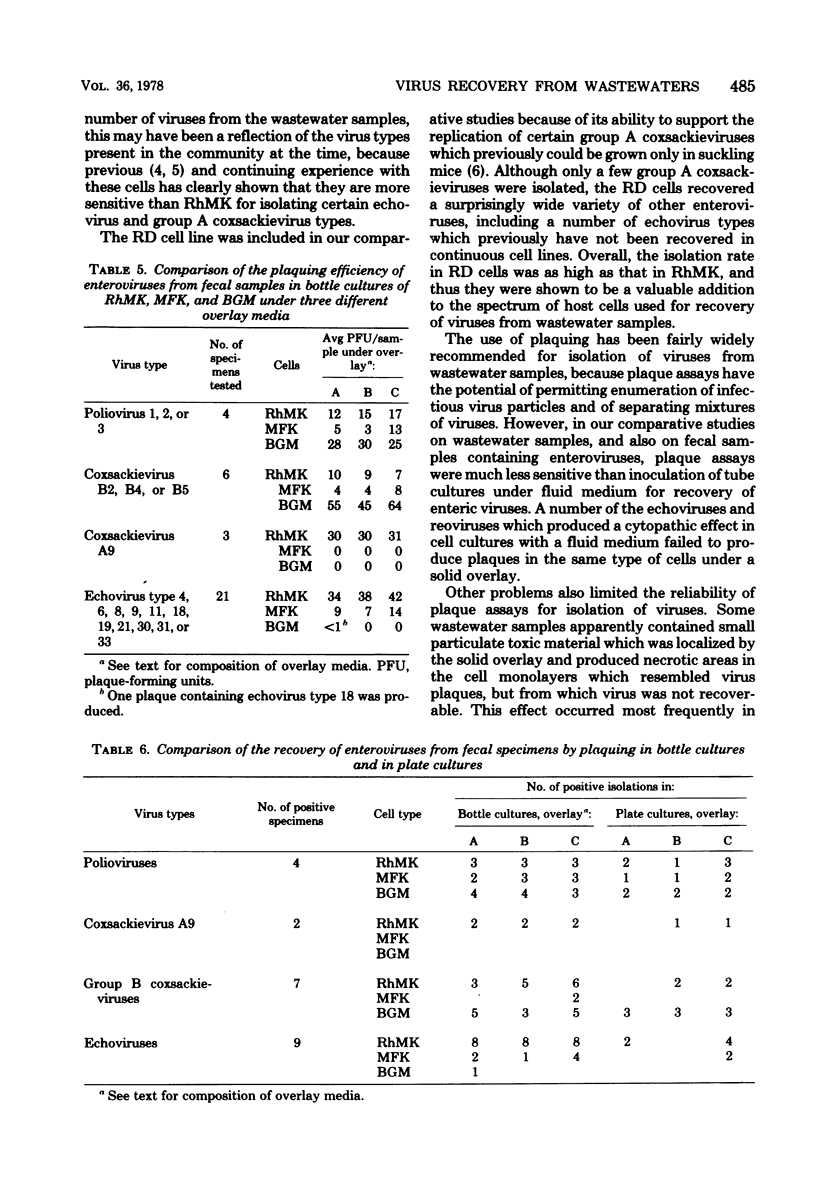
In efforts to define the most sensitive cell culture systems for recovery of viruses from wastewaters, 181 samples were inoculated in parallel into tube cultures of various cell types and were plaqued in bottle and petri dish cultures of three types of monkey kidney cells. Polioviruses were recovered most frequently in the RD line of human rhabdomyosarcoma cells, group A coxsackieviruses in RD and human fetal diploid kidney (HFDK) cells, group B coxsackieviruses in the BGM line of African green monkey kidney cells, echoviruses in RD and primary rhesus monkey kidney (RhMK) cells, and reoviruses in RhMK cells. BGM cells were unsatisfactory for recovery of viruses other than polioviruses and group B coxsackieviruses, and a line of fetal rhesus monkey kidney (MFK) was not a satisfactory substitute for primary RhMK. With RhMK cells, comparable numbers of virus isolations were made in tube cultures and in plaque assays conducted in bottle cultures, but with BGM and MFK cells, fewer isolations were made by plaquing than by inoculation of tube cultures. In comparative plaque assays on fecal samples under three different overlays in bottle and plate cultures of RhMK, BGM, and MFK cells, it was found that plaquing in the most sensitive system, RhMK, was less efficient for virus recovery than was inoculation of tube cultures of RhMK or HFDK cells. Overall, plaque assays performed in petri dishes in a CO2 incubator yielded fewer virus isolates than did parallel plaque assays performed in closed bottle cultures. Other limitations of plaque assays for recovery of human enteric viruses are discussed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barron A. L., Olshevsky C., Cohen M. M. Characteristics of the BGM line of cells from African green monkey kidney. Brief report. Arch Gesamte Virusforsch. 1970;32(4):389–392. doi: 10.1007/BF01250067. [DOI] [PubMed] [Google Scholar]
- Dahling D. R., Berg G., Berman D. BGM, a continuous cell line more sensitive than primary rhesus and African green kidney cells for the recovery of viruses from water. Health Lab Sci. 1974 Oct;11(4):275–282. [PubMed] [Google Scholar]
- McAllister R. M., Melnyk J., Finkelstein J. Z., Adams E. C., Jr, Gardner M. B. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer. 1969 Sep;24(3):520–526. doi: 10.1002/1097-0142(196909)24:3<520::aid-cncr2820240313>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- SCHMIDT N. J., LENNETTE E. H., HO H. H. COMPARATIVE SENSITIVITY OF HUMAN FETAL DIPLOID KIDNEY CELL STRAINS AND MONKEY KIDNEY CELL CULTURES FOR ISOLATION OF CERTAIN HUMAN VIRUSES. Am J Clin Pathol. 1965 Apr;43:297–301. doi: 10.1093/ajcp/43.4.297. [DOI] [PubMed] [Google Scholar]
- Schmidt N. J., Ho H. H., Lennette E. H. Comparative sensitivity of the BGM cell line for isolation of enteric viruses. Health Lab Sci. 1976 Apr;13(2):115–117. [PubMed] [Google Scholar]
- Schmidt N. J., Ho H. H., Lennette E. H. Propagation and isolation of group A coxsackieviruses in RD cells. J Clin Microbiol. 1975 Sep;2(3):183–185. doi: 10.1128/jcm.2.3.183-185.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N. J. Tissue culture in the laboratory diagnosis of viral infections. Am J Clin Pathol. 1972 Jun;57(6):820–828. doi: 10.1093/ajcp/57.6.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis C., Melnick J. L. Mechanism of enhancement of virus plaques by cationic polymers. J Virol. 1968 Apr;2(4):267–274. doi: 10.1128/jvi.2.4.267-274.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]


