Abstract
Osteosarcoma is the most common primary malignant tumor of bone. Analysis of familial cancer syndromes and sporadic cases has strongly implicated both p53 and pRb in its pathogenesis; however, the relative contribution of these mutations to the initiation of osteosarcoma is unclear. We describe here the generation and characterization of a genetically engineered mouse model in which all animals develop short latency malignant osteosarcoma. The genetically engineered mouse model is based on osteoblast-restricted deletion of p53 and pRb. Osteosarcoma development is dependent on loss of p53 and potentiated by loss of pRb, revealing a dominance of p53 mutation in the development of osteosarcoma. The model reproduces many of the defining features of human osteosarcoma including cytogenetic complexity and comparable gene expression signatures, histology, and metastatic behavior. Using a novel in silico methodology termed cytogenetic region enrichment analysis, we demonstrate high conservation of gene expression changes between murine osteosarcoma and known cytogentically rearranged loci from human osteosarcoma. Due to the strong similarity between murine osteosarcoma and human osteosarcoma in this model, this should provide a valuable platform for addressing the molecular genetics of osteosarcoma and for developing novel therapeutic strategies.
Keywords: Cancer, mouse model, osteocarcinoma
Osteosarcoma (OS) is the most common primary malignant nonhematological tumor of bone. OS has a peak incidence in adolescence (Unni et al. 2005), and cure rates for patients with metastatic or relapsed disease remain poor (<20% survival) (Meyers et al. 1992, 2005; Sandberg and Bridge 2003). Progress on the genetics and pathogenesis of OS has been limited, in part due to the low incidence of the disease.
Most OSs occur sporadically. However, important insights have been gained from familial cancer predisposition syndromes. The incidence of OS is appreciably increased in patients with hereditary retinoblastoma (Gurney et al. 1995). Among this patient population, ∼60% of second tumors are sarcomas, of which about one half are OSs (Wadayama et al. 1994). Moreover, sporadic OSs frequently harbor alterations in the RB pathway, and specifically the Rb gene. Germline mutation of the p53 gene in Li-Fraumeni syndrome also predisposes patients to OS (Porter et al. 1992). In addition, somatic mutation of p53 is frequently observed in sporadic OS (Miller et al. 1990; Nishio et al. 2006). Alterations of p53 correlate with markedly reduced event-free survival (Tsuchiya et al. 2000; Pakos et al. 2004; Wunder et al. 2005). The relative importance of mutations in the p53 or pRb pathways and the degree to which they cooperate in the initiation of OS are unclear. Patients with RECQ helicase-associated disorders (Rothmund-Thomson, Werner, and Bloom syndromes) are predisposed to OS, but to a lesser extent than those associated with either hereditary retinoblastoma or Li-Fraumeni syndrome (Kansara and Thomas 2007). Other genetic contributions to OS pathogenesis are largely unknown. Numerous cytogenetic abnormalities have been described, including chromosomal segment loss, rearrangement, and amplification with karyotypic complexity in the absence of recurrent clonal translocations (Helman and Meltzer 2003).
The development of a tractable animal model of OS that mimics the genetics and pathology of the human malignancy would provide new opportunities for probing the genetics of OS, identifying candidate genes for contributing somatic events in the generation or progression of the disease, and devising new therapies based either on inactivation of specific targets or promotion of differentiation. Initial OS models were generated using radiation-induced or chemically induced lesions in mice. The unpredictability of tumor formation rendered these models impractical (for review, see Ek et al. 2006). Orthotopic transplantation of mouse OS cell lines has been used to examine metastasis and screen drugs for antitumor effects. Several human OS cell lines have been characterized and studied in an orthotopic setting using immunocompromised mice. Such approaches are limited as they depend on cells that can survive in both cell culture conditions and in a xenograft, which may not provide an appropriate microenvironment for support of all human cells (Kelly et al. 2007). Furthermore, the tumor microenvironment can contribute significantly to tumor behavior, and such interactions are lost when established disease is introduced directly into the recipient animal (Becher and Holland 2006; Sharpless and Depinho 2006; Frese and Tuveson 2007).
Several genetically modified mouse strains have been reported to develop OS. Notably, p53-deficient germline mutants or animals carrying a pathogenic mutant p53 allele develop OS among other malignancies (Jacks et al. 1994; Lang et al. 2004; Olive et al. 2004). Osteoblast-restricted deletion of p53 has been reported recently to result in the development of OS with 60% penetrance, the remainder of animals developing lymphoma or fibrosarcoma (Lengner et al. 2006). Interestingly, mice germline heterozygous for pRb develop neither OS nor retinoblastoma (Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992), unlike that observed for germline heterozygosity of p53. Transgenic mice overexpressing c-Fos develop OS and chondrosarcoma, and Fos overexpression has also frequently been observed in human OS (Ruther et al. 1989; Wu et al. 1990; Wang et al. 1995). Heterozygous mutation of Nf2 results in the development of OS in mouse (>60% penetrance), yet human neurofibromatosis 2 patients do not normally develop OS nor are mutations in Nf2 found in human OS samples (McClatchey et al. 1998; Stemmer-Rachamimov et al. 1998). While these models provide important information regarding the genetics of OS, the long latency combined with low penetrance makes utilization of these models impractical.
Improvements in the generation of genetically engineered mice have led to the establishment of several mouse models that recapitulate critical features of human cancers (Jonkers and Berns 2002; Tuveson and Jacks 2002; Isakoff et al. 2005; Kim et al. 2005a; Sweet-Cordero et al. 2005; Haldar et al. 2007; Li et al. 2007; Liu et al. 2007). For the most part, these models are based on the generation of conditionally altered alleles that permit either expression of a somatically rearranged oncogene, such as a fusion gene, or deletion of a tumor suppressor gene in a cell-type-specific manner. The extent to which mouse models recapitulate the underlying human biology on which they are based will determine their usefulness. A valid murine model should faithfully reproduce both the genetics and behavior of the corresponding human disease.
We describe here the generation and characterization of a genetically engineered mouse model that exhibits highly penetrant and short latency malignant OS. This model recapitulates many of the defining features of human OS, including cytogenetic complexity, gene expression signatures, pathology, age of onset, and metastatic behavior. We demonstrate a striking correlation between known cytogenetic rearrangements in human OS and gene expression changes in mouse OS. Tumor development is strictly dependent on p53 mutation, whereas pRb mutation potentiates development of a p53-dependent disease but is insufficient in isolation to initiate OS. This high penetrance, short latency mouse model closely resembles human OS and will provide new opportunities for dissecting the molecular genetics of OS and developing novel therapeutic strategies.
Results
Osterix-Cre-mediated deletion of p53 and pRb results in OS
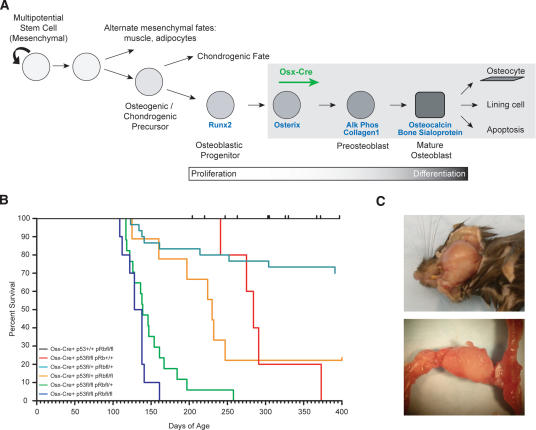
Based on the near ubiquitous loss of function of p53 and pRb pathways in human OS, we hypothesized that osteoblast-specific loss of these genes may be sufficient to initiate OS development (murineOS, mOS). We made use of conditional (floxed) alleles of both p53 (p53fl/fl) and pRb (pRbfl/fl) to allow for the tissue-restricted inactivation of these genes following Cre expression (Jonkers et al. 2001; MacPherson et al. 2003; Sage et al. 2003). To achieve osteoblast-restricted deletion, we utilized the Osterix-Cre transgenic mouse (Osx-Cre) to direct Cre expression to committed osteoblast progenitors (Rodda and McMahon 2006). Osterix is a bone-specific transcription factor required for osteoblast development, and the Osterix-Cre transgene expresses Cre recombinase in a manner that follows that of endogenous Osterix (Nakashima et al. 2002). Animals were intercrossed to develop Osx-Cre+p53fl/flpRbfl/fl animals with all intermediate genotypes also generated in addition to Osx-Cre+ animals bearing only a single conditional allele. All genotypes were born at the expected ratios and mutant animals are viable, fertile, and developmentally normal with the exception of a slight growth delay in Osx-Cre+ animals compared with Osx-Cre− animals, although this did not affect fertility and largely resolved with age.
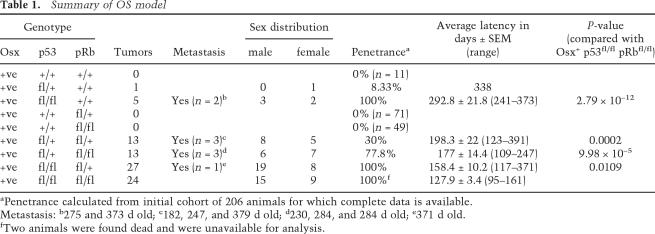
We have not observed the development of mOS in Osx-Cre+pRbfl/fl and Osx-Cre+pRbfl/+ mice with an observation time of ∼18 mo of age (Fig. 1B; Table 1). Unlike osteoblast-restricted deletion of pRb, both Osx-Cre+p53fl/fl and Osx-Cre+p53fl/+ animals develop mOS. Osx-Cre+p53fl/+ mice develop disease with low penetrance and long latency. However, deletion of both alleles of p53 resulted in mOS development with complete penetrance and an average survival to 292 d of age (Fig. 1B; Table 1; Supplemental Table 1). Heterozygosity for both p53 and pRb shortened the latency and increased the penetrance of mOS compared with either heterozygous mutant alone (Fig. 1B; Table 1; Supplemental Table 1).
Figure 1.
Complete penetrance short Latency OS development. (A) Model of osteoblast differentiation and the putative stage of Osx-Cre expression. A and Figure 5D are composite diagrams based on the work of Aubin (2001), Franz-Odendaal et al. (2006), and Rodda and McMahon (2006). (B) Kaplan-Meier survival plots for the indicated genotypes: Osx-Cre+p53+/+pRbfl/fl, n = 49; Osx-Cre+ p53fl/flpRb+/+, n = 5; Osx-Cre+p53fl/+pRbfl/+, n = 30; Osx-Cre+p53fl/+pRbfl/fl, n = 9; Osx-Cre+p53fl/flpRbfl/+, n = 17; Osx-Cre+p53fl/fl pRbfl/fl, n = 12. (C) Representative presentation of OS of the lower jaw and femur, respectively.
Table 1.
Summary of OS model
aPenetrance calculated from initial cohort of 206 animals for which complete data is available.
Metastasis: b275 and 373 d old; c182, 247, and 379 d old; d230, 284, and 284 d old; e371 d old.
fTwo animals were found dead and were unavailable for analysis.
Osx-Cre+p53fl/flpRbfl/+ and Osx-Cre+p53fl/flpRbfl/fl animals develop mOS at ∼4 mo of age, a time point within the first quarter of the expected lifespan of a laboratory mouse. Completely penetrant mOS development was observed in both genotypes (Fig. 1B,C; Table 1; Supplemental Table 1). Interestingly, Osx-Cre+p53fl/+pRbfl/fl animals are much more susceptible to OS development than double heterozygous animals with 77% penetrance, whereas complete loss of p53 resulted in 100% development of mOS (Fig. 1B; Table 1). We also observed the development of adipogenic tumors, restricted to the Osx-Cre+p53fl/flpRbfl/fl animals (five of 24 animals to date, ∼20%), which occurred on the outer chest or abdomen (data not shown). All mice presenting with these tumors also had OS. Primary mOS tumors and mOS cell lines derived from them were serially transplantable when injected subcutaneously into Rag2−/− immunodeficient animals (data not shown). Thus, Osx-Cre-dependent deletion of p53 and pRb results in the development of completely penetrant malignant OS, and this disease is strictly dependent on disruption of p53.
Multifocal nature of disease and metastatic potential
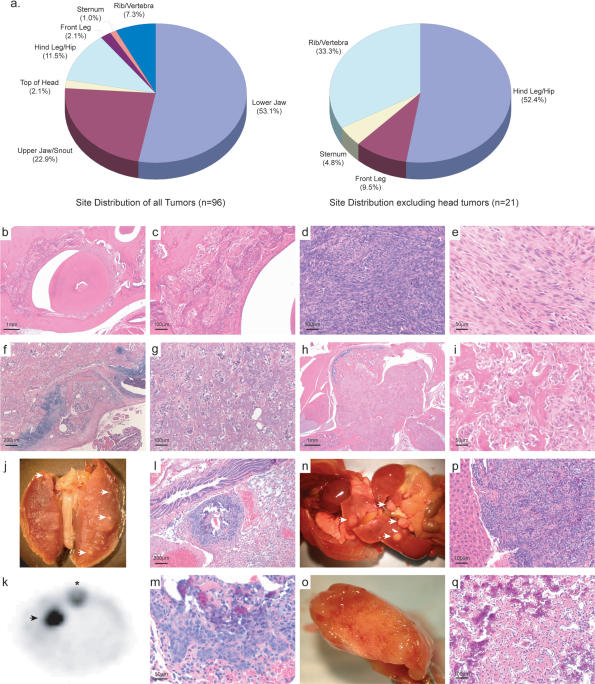
The most frequent site of mOS was the jaw and head, followed by the hind leg/hip and ribs and vertebra (Fig. 2a; Supplemental Table 2). This contrasts with human OS, in which jaw tumors account for only 9% of all tumors (Unni et al. 2005). When the tumor site data are analyzed exclusive of those tumors arising on the head, the site distribution more closely approximates that observed in human OS (Tan et al. 2006). The tumors presented with histology most consistent with that of human medullary OS. Tumor histology ranged from almost purely fibroblastic tumors with minimal mineralization/osteoid to tumors with marked mineralization of the osteoid (Fig. 2b–i). The distribution of the mOS was typically metaphyseal, with growth into the central medullary cavity and with extraosseous extension into the soft tissues. As in human OS, the tumor bone was woven bone formed by the tumor cells. Areas of cartilage formation were also observed within the mOS, a finding also seen in human tumors. The tumor cells were predominately spindle shaped with moderate to marked atypia (Fig. 2d,e). Mitotic figures were variable in number, and abnormal mitoses are seen among the more atypical cells. Occasional multinucleated giant cells were seen among the tumor cells. In nearly all respects, the mOS bears a close histopathological resemblance to human OS (Unni et al. 2005).
Figure 2.
Tumor histology, site distribution, and metastatic disease. (a) Distribution of tumors in all animals and in a subset of animals excluding those that arise on the head respectively. (b,c) Osteoid-rich, partially mineralized tumor surrounding a tooth. (d) Osteoid-poor high-grade tumor. (e) Osteoid-poor fibroblastic tumor. (f) Osteoid-rich, partially mineralized tumor. (g) Osteoid-rich tumor. (h,i) Extraosseous extension of an osteoid-rich tumor. (j) Lung metastasis as indicated by the arrows. (k) Lung metastasis as revealed by F18 sodium fluoride microPET imaging; asterisk indicates the spine. (l,m) Histology of lung metastasis with focal areas of osteoid. (n) Disseminated metastasis through the liver and intestines as indicated by arrows. (o) Liver metastasis. (p,q) Histology of liver metastasis with osteoid. All sections stained with hematoxylin and eosin. Scale is as indicated on each image.
A major cause of mortality in patients with OS is metastasis, which is often present at initial diagnosis. Human OS shows a preference for metastasis to the lung and brain. We observed metastasis in 9.4% of all mOS-bearing mice. The metastatic frequency was >39% among animals with primary tumors at sites other than the head. The most frequent site of metastasis was the lung (seven of nine) followed by the liver (six of nine), and one animal presented with disseminated metastasis throughout the intestinal tissues (Fig. 2j–q). Several animals had metastatic disease in both the lung and the liver, which were apparent by microPET (Fig. 2k; Supplemental Fig. 1). Metastatic disease was observed across genotypes, however it most often presented with a longer latency than the genotype median and rarely in animals that developed a tumor on the jaw or snout, which significantly decreased lifespan (Table 1). We did not observe metastasis in Osx-Cre+p53fl/flpRbfl/fl animals, perhaps due to extremely rapid tumor development in these animals, which may not provide sufficient time for metastatic disease to become evident. The expression of Cre from the Osx-Cre transgene is able to be temporally repressed by the administration of doxycycline (dox). When Osx-Cre+p53fl/flpRbfl/fl animals were maintained on dox from conception until 4 wk of age, preventing gene inactivation until the removal of dox, the time to development of OS was delayed by ∼100 d and all animals presented with metastatic OS but no animals had tumors on the head (Supplemental Fig. 2). Cell lines derived from the metastatic lesions were transferred subcutaneously to Rag2−/− immunocompromised recipients, and mOS readily ensued (data not shown).
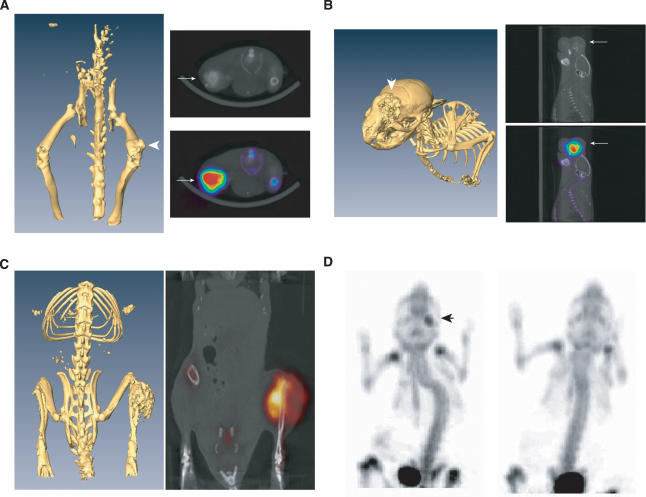
Multifocal lesions were observed in a significant number of animals (22.3%). Their appearance did not correlate with genotype. Tumors most frequently presented as multiple lesions on the jaw and snout (Supplemental Fig. 3; Supplemental Movie 1). To enable more accurate assessment of tumor burden, we utilized micro-PET/CT imaging to determine noninvasively the numbers of tumors. For microPET imaging, fluorine-18 sodium fluoride (Na18F) was used as the imaging agent (Brenner et al. 2004). Na18F binds to the hypoxanthine matrix of bone with highest deposition at sites of turnover (Even-Sapir et al. 2004; Lim et al. 2007). mOS showed highly avid uptake of Na18F, which directly corresponded to lesions as visualized by microCT (Fig. 3A–C). Histology was performed verifying the detected lesions as mOS. Interestingly, Na18F uptake was most pronounced in regions of the tumor that contained less areas of mineralized osteoid as determined by microCT (Fig. 3C). The capacity of microPET to detect small lesions, such as that on the calvaria in Supplemental Figure 3, which would not otherwise be readily detectable on conventional autopsy, suggested that the estimate of tumor burden may underrepresent the true burden due to the microscopic, multifocal nature of some lesions.
Figure 3.
In vivo imaging with microPET/CT demonstrates rapid tumor formation. (A) microCT reconstruction of an animal with a femoral tumor with the overlay of microPET and microCT images. (B) microPET/CT of a calvarial tumor. (C) microPET/CT of a tibial tumor. The most F18 avid area of tumor corresponds with an area of lesser osteoid formation as revealed by microCT. (D) Eight-week-old animals were assessed for tumor burden by N18F revealing a positive lesion on the lower jaw; animal on the left is Osx-Cre+p53fl/flpRbfl/fl, animal on the right is Osx-Cre−p53fl/flpRbfl/+. Arrows indicate OS lesion.
Given the sensitivity of Na18F microPET, we imaged several 2-mo-old animals to determine when lesions developed. The Osx-Cre+p53fl/flpRbfl/fl animal we assessed had a detectable jaw lesion at 2 mo of age, which was not externally visible, while two Osx-Cre+p53fl/flpRbfl/+ animals did not have detectable lesions at this time point (Fig. 3D; Supplemental Movies 2, 3). By 3 mo of age, one of the Osx-Cre+p53fl/flpRbfl/+ animals had also developed a PET positive lesion in the tibia (Supplemental Movie 4). Collectively, these studies demonstrate that the tumor burden may be underrepresented using conventional autopsy coupled with histology and that the mOS can develop by at least 2 mo of age, consistent with the pediatric or adolescent nature of human OS.
Cytogenetic complexity of mOS
A feature of human OS is cytogenetic complexity with the absence of recurrent clonal chromosomal translocations (Bridge et al. 1997; Helman and Meltzer 2003). To determine whether the mOS also displayed cytogenetic complexity, we performed spectral karyotyping (SKY) on early passage cell lines derived from primary tumors. mOS cells were isolated by enzymatic digestion of primary tumor tissue, placed into culture, and then following metaphase arrest, cells were assessed for cytogenetic integrity using SKY. We assessed ploidy and the presence of translocations in cell lines derived from six independent tumors. mOS cells consistently exhibited marked aneuploidy, a characteristic of human OS (Supplemental Fig. 4; Supplemental Table 3). Two of the tumors assessed showed aberrations of chromosomes 8 and 14, with multiple small chromosomes and fragments. Translocations were present, although these were nonclonal and, in general, nonreciprocal (Supplemental Fig. 4).
OS cell of origin
The mOS we generated is based on the deletion of p53 and pRb in cells that express Osterix. Osterix expression identifies a stage of osteoblast commitment after Runx2 expression that defines the first commitment step to the osteoblast lineage from a multi/bipotential precursor (Aubin 2001; Nakashima et al. 2002). Thus, OS arises from a cell at, or after, the Osterix stage of osteoblast differentiation. To determine the most likely stage of OS development in this model, we undertook expression analysis of a panel of genes representative of different stages of osteoblast differentiation on the cell lines derived from the primary tumors. The mOS cell lines were utilized as they are relatively homogenous, while they retain the capacity for efficient in vivo tumor formation and display an expression profile consistent with human OS (Supplemental Fig. 4; data not shown).
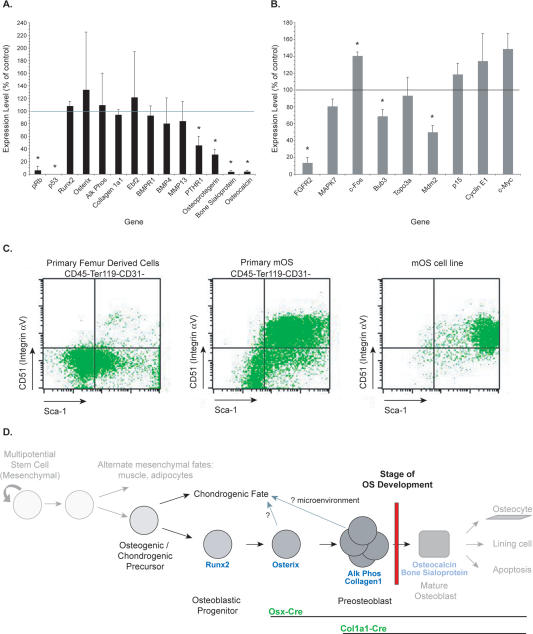
Markers of osteoblast progenitors (Runx2, Osterix), preosteoblasts (alkaline phosphatase, Collagen1a1, Ebf2, BMP4), and mature osteoblasts (osteoprotegerin, bone sialoprotein, osteocalcin) were assessed by quantitative real-time PCR, and the expression in the OS cell lines was compared with that of in vitro differentiated primary osteoblasts (Aubin 2001; Franz-Odendaal et al. 2006). mOS cell lines express markers of osteoblast progenitors and preosteoblasts but fail to express markers of mature osteoblasts. These results indicate that the mOS cell lines are arrested at the preosteoblast stage of differentiation and that the candidate target cell for transformation in this model is a lineage-committed immature osteoblast (Fig. 4A).
Figure 4.
The preosteoblast as the candidate cell or origin of OS. (A) Analysis of differentiation status of mOS cell lines by quantitative real-time PCR. Expression levels of the indicated genes are compared with primary in vitro differentiated osteoblasts (normalized to 100%). Data are expressed as mean ± SEM; n = 4 OS cell lines, 3 wild-type primary Ob. (*) P < 0.05 (Student’s t-test). (B) Analysis of expression of genes implicated in human OS in the mOS cell lines by quantitative real-time PCR. Expression levels of the indicated genes are compared with primary in vitro differentiated osteoblasts (normalized to 100%). Data are expressed as mean ± SEM; n = 4 OS cell lines, 3 wild-type primary Ob. (*) P < 0.05 (Student’s t-test). (C) Representative flow cytometry assessment of expression of Sca-1 and CD51 on primary femur compact bone, primary mOS, and mOS cell lines, respectively. (D) Proposed model for OS development.
We also analyzed the expression of other genes associated with human OS. We found significantly increased expression of the proto-oncogene c-Fos in the mOS cell lines compared with primary osteoblasts. Constitutive transgenic overexpression of Fos results in the development of osteogenic tumors in mice and is frequently observed in human OS and in human OS cell lines (Ruther et al. 1989; Wu et al. 1990; Wang et al. 1995; Thomas et al. 2004). The expression levels of both the fibroblast growth factor receptor 2 (FGFR2) and Bub3 were reduced. Their expression is reduced frequently in human OS, likely due to allelic loss at 10q26 (Mendoza et al. 2005). Reduced expression of the p53 target gene Mdm2 was also detected (Fig. 4B). Lowered Mdm2 expression in osteoblasts in vivo was associated with reduced differentiation and Runx2 expression (Lengner et al. 2006). Changes in the expression from the INK4 locus were noted with both p16Ink4a and p19Arf expression increased nearly 10-fold compared with control osteoblasts (P < 0.04); however, in the absence of pRb and p53, both p16Ink4a and p19Arf lack their respective molecular targets.
We also performed an assessment by flow cytometry to determine the cell surface phenotype of both primary mOS and mOS cell lines. Limited information is available regarding the cell surface phenotype of murine osteoblasts. However, previous studies demonstrate that osteoblasts have a surface phenotype negative for hematopoietic (CD45 and Ter119) and vascular (CD31) markers and positive for Sca-1 and CD51 (Integrin αv) (Semerad et al. 2005). Both primary mOS and mOS cell lines contain a prominent population of Sca-1+CD51+ double positive cells that are negative for hematopoietic and vascular markers (Fig. 4C). Other markers associated with mesenchymal cells and osteoblasts, such as SSEA-4, CD34, and VCAM, showed variable, if any, expression, on both primary mOS and cell lines (Gang et al. 2007). Taken together, these data suggest that OS development in this model arises from a cell stage at or near the preosteoblast and that many of the gene expression changes observed in human OS are mirrored in mOS (Fig. 4C).
Recapitulation of the transcriptional profile of human OS
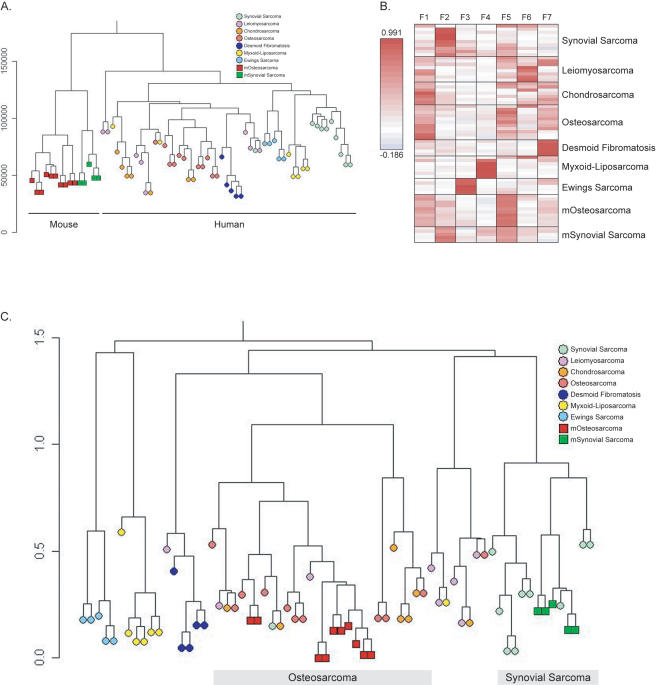
The mOS we describe recapitulates many of the characteristic biologic features of human OS. To determine whether mOS reflects the molecular features of human OS, we undertook expression profiling using Affymetrix A430 2.0 gene chips. To enable a direct comparison between murine and human samples, we performed metagene projection analysis (Isakoff et al. 2005; Tamayo et al. 2007). Metagene projection analysis permits a robust cross-species and cross-platform comparison enabling an assessment of the degree to which mOS displays a transcriptional profile comparable to human OS, as distinguished from other human sarcomas. In the absence of metagene analysis, unsupervised hierarchical clustering of the mouse and human tumor data sets is uninformative, as it results in the separation of these data based on both species and microarray platform, consistent with that previously described (Fig. 5A).
Figure 5.
Mouse OS shares a transcriptional profile with human OS. (A) Hierarchical clustering of mouse and human tumor data sets separates data based on species and platform. Using this analysis technique mouse and human data sets separate because of species and platform differences. (B) Heat maps of the metagene projections that were defined for each of the seven human sarcoma types. The metagene expression level for each of the individual sarcoma types is shown. Following definition of the human metagene for each tumor type, the murine tumor sets were added, and the metagene projection as it applies to these data sets is shown. (C) Hierarchical clustering of the data set after metagene projection. mOS (red square) and human OS (salmon circle) associate, as do mouse synovial sarcoma (green square) and human synovial sarcoma (green circle).
To define the metagene for human OS, we utilized a previously published set of human sarcoma data (Henderson et al. 2005). This data set contained 11 OS samples, as well as a variety of other defined human sarcoma types of mesenchymal origin. All samples, both human and mouse, were normalized together and then the metagenes were defined from the human data set alone. A metagene was defined for each of the human sarcoma types (Fig. 5B; Supplemental Fig. 5). Once the metagene that defined human OS was identified, the transcriptional information from our primary mOS (n = 10 primary tumors, two independent data sets) and also five previously published primary murine synovial sarcoma samples (Haldar et al. 2007) were added, and the metagene information was used to cluster the mouse samples within the defined human tumor types (Fig. 5C; Supplemental Fig. 6). Mouse synovial sarcomas (green squares) associated with their human counterparts (light green circle), providing independent validation of this approach as this model was previously defined using an alternative comparison technique to be a faithful model of human synovial sarcoma (Haldar et al. 2007). Strikingly, the mOS expression signature (Fig. 5C, red squares) closely resembled that of the human OS samples (Fig. 5C, salmon circles). The mOS samples cluster within the human OS samples, which also associate with the human chondrosarcoma samples, reflecting the common origin of osteoblasts and chondrocytes. Also, as for human OS, we observed areas of cartilage formation within the mOS. These analyses demonstrate that mOS arising in our engineered mice exhibit a transcriptional profile very comparable with human OS.
Conservation of cytogenetic rearrangements between mOS and human OS
Numerous alterations and rearrangements in human OS, targeting many regions throughout the genome, have been observed by conventional cytogenetics (Bridge et al. 1993; Batanian et al. 2002). To query the mOS model independent of metagene analysis, we sought to determine whether the mouse tumors display altered expression of genes contained within regions defined cytogenetically in human OS.
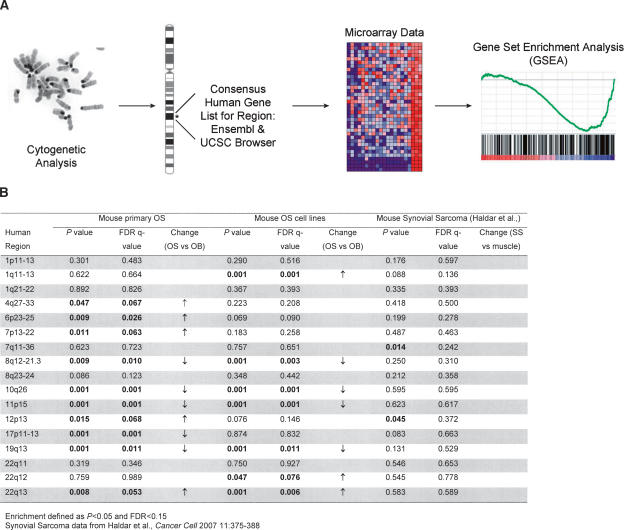
To achieve this, we devised a novel in silico approach, based on the gene set enrichment analysis (GSEA) methodology, which we term cytogenetic region enrichment analysis (CREA) (Mootha et al. 2003; Subramanian et al. 2005). Cytogenetic regions of interest were chosen based on those frequently reported in the literature as rearranged in human OS (Bridge et al. 1993; Batanian et al. 2002). The human genes contained within these chromosomal bands were compiled from the consensus human genome sequence (UCSC Genome Browser, release March 2006, and Ensembl human genome sequence, release 46.36h), and GSEA gene lists were constructed for each region containing all known genes (outlined in Fig. 6A). GSEA was then performed using the primary mOS expression profiles, the expression profile of mOS cell lines, and also the murine synovial sarcoma data set that we previously utilized for metagene analysis. The mOS samples (both primary tumor and cell line) were compared with the gene expression signatures of in vitro differentiated primary osteoblasts, while the synovial sarcoma samples were compared with wild-type muscle. Enrichment of a given region was defined as a P-value of <0.05 and a false discovery rate (FDR) Q-value of <0.15.
Figure 6.
Cytogenetic region enrichment analysis (CREA). (A) Schematic of the application of CREA to the analysis of OS. (B) Summary of CREA results for each of the indicated cytogenetic regions for primary mOS samples, mOS cell lines, and mouse synovial sarcoma.
Strikingly, primary mOS showed significant enrichment at 10 of 17 regions tested by CREA. Moreover, mOS cell lines showed enrichment at 7 of 17 regions when compared with primary in vitro-differentiated osteoblasts (Fig. 6B; Supplemental Figs. 7 and 8). In contrast, we did not observe enrichment for any of the 17 regions for the murine synovial sarcoma samples compared with control muscle samples (Fig. 6B; Supplemental Fig. 9). We further analyzed the gene expression changes at the 17 regions in two additional data sets derived from either breast or lung cancer data sets (Sweet-Cordero et al. 2005; Li et al. 2007), observing 3 and 4 significantly altered regions, respectively (Supplemental Table 4). Thus, changes in these regions are not a common occurrence in tumors per se but appear specific for OS. These results underscore how closely mOS resembles human OS. Furthermore, our findings suggest that combining cytogenetic changes with gene expression signatures via CREA may be a useful strategy for validation of other mouse tumor models as faithful representations of the corresponding human disease.
Discussion
Here we describe the generation of a genetically engineered mouse model of OS that bears striking resemblance to human OS. This model is based on the osteoblast-restricted loss of both p53 and pRb. mOS exhibited many of the features characteristic of human OS, including comparable histology, metastatic site preference, karyotypic complexity, and transcriptional profiles. These results demonstrate that disease in these animals represents a faithful and biologically meaningful recapitulation of human OS.
p53-dependent disease
Inherited mutation of RB in humans predisposes patients to OS, the second most frequent tumor in this population with an incidence increased 500-fold compared with the general population (Gurney et al. 1995). Moreover, sporadic OS also frequently harbors alterations in the Rb gene (Wadayama et al. 1994). These observations have led to the hypothesis that the retinoblastoma gene plays a central role in osteoblast biology, and in OS genesis in particular (Thomas et al. 2003). Loss of Rb was reported to impair terminal osteoblast differentiation due to a loss of coactivator activity from CBFA1, which was not observed in the absence of either of the related pocket proteins p107 or p130 (Thomas et al. 2001). However, mice bearing germline heterozygous Rb mutation have not been reported to develop OS but do develop other malignancies (Jacks et al. 1992). We did not observe OS development in Osx-CrepRbfl/fl animals up to 18 mo of age, nor did we observe any gross skeletal abnormalities in these animals. These in vivo results contrast with those derived from in vitro analysis of cell lines in which pRb expression was shown to be required for osteoblast differentiation. We propose that Rb acts as a potentiator in the context of OS, but its mutation alone is insufficient to act as an initiating event.
The results of Rb deficiency contrast with the clearly established link between p53 mutation and OS development both in human and murine malignancies (Jacks et al. 1994; Lang et al. 2004; Olive et al. 2004; Lengner et al. 2006). Consistent with the importance of p53 loss or mutation to the genesis of OS, we observed loss of heterozygosity of p53 but did not observe this for pRb (Supplemental Fig. 10). The effects of missense mutation of p53, as distinct from the loss of function allele utilized here, would be interesting to determine if this alters any characteristics of the OS generated in these animals. Other cancer-associated mutations in the “Rb pathway,” such as p16INK4a and p15INK4b, have been reported in human OS at low frequency, and animals mutant for these genes can develop OS at low penetrance (Miller et al. 1996, 1997; Sharpless et al. 2001, 2004; Krimpenfort et al. 2007). In the model we report that Rb mutation is unable to initiate OS in the absence of mutation of p53, consistent with observations from radiation-induced OS in retinoblastoma patients (Gonin-Laurent et al. 2006, 2007). These results argue strongly that loss of p53 is the rate-limiting step in initiation of OS and that pRb loss cooperates in the steps leading to frank malignancy.
Cell of origin of OS
Lengner and colleagues previously reported OS following conditional p53 deletion with Collagen1a1-Cre (Col3.6-Cre transgenic) (Lengner et al. 2006). The Col3.6-Cre transgenic resulted in 60% penetrance of OS with a survival time similar to that observed in the Osx-Cre model. Within the osteoblast lineage, Osterix expression precedes the expression of Collagen1a1 expression, which is thought to occur at the preosteoblast stage of differentiation (Aubin 2001; Nakashima et al. 2002). Despite the difference in stages of Cre expression, both models develop OS with a very similar latency for p53fl/fl animals. Analysis of OS arising in c-Fos transgenic animals also suggested that the cell of origin was contained within the committed osteoblast population (Grigoriadis et al. 1993). Additionally, we observed tumors in the Osx-Cre animals from intramembranous bone, such as the calvaria and periosteal surface of the femur, demonstrating that OS can arise from osteo-restricted precursors independently of chondrogenic and adipogenic cells. Collectively, these independent lines of evidence support a model in which OS arises as a result of transformation events that occur at or near the preosteoblast stage of osteoblast development (Fig. 5D).
Identification of the cell of origin of the majority of human tumors has remained elusive. The tumor initiating cell, or cancer stem cell as it also termed, has been postulated to arise from within the normal tissue stem cell pool given the similarities that exist between these cells and cells capable of sustaining the tumor (Reya et al. 2001; Kim et al. 2005b; Polyak and Hahn 2006). The mesenchymal stem cell (MSC) is the stem cell from which osteoblasts arise. Our data are inconsistent with the MSC as the cell of origin of OS and are most consistent with disease arising from an osteoblast lineage-committed progenitor, unlike that recently postulated for Ewing sarcoma (Tirode et al. 2007). Interestingly, despite the deletion occurring in an Osterix-dependent manner, we have observed areas of chondrogenic differentiation within some of the primary OS and have observed adipogenic tumors on several double mutant animals. These results suggest either that the expression of the Osx-Cre transgene is not absolutely restricted to the osteoblast population or that, once immortalized, the OS cells are able to undergo multilineage differentiation in response to appropriate environmental cues. We also cannot exclude that more differentiated cells than preosteoblasts are also capable of initiating OS, and this will need to be experimentally determined. Therefore, sarcomas, like hematopoietic tumors and also recently proposed for breast cancer, may arise from the lineage-committed fraction of cells and do not require transformation of the multipotential tissue-restricted stem cell (Huntly et al. 2004; Ince et al. 2007).
Molecular profiling demonstrates similarity to human OS
In addition to the similar transcriptional profiles of human and mOS as observed using metagene projection analysis, we documented transcriptional changes in murine tumors that were reflective of the common cytogenetic alterations observed in human OS using a novel in silico approach, CREA. CREA is based on the gene set enrichment analysis methodology and utilizes cytogenetic information to define genomic regions of interest for which gene expression information can be assessed. Conserved cytogenetic regions that display expression alterations are of particular interest. These may represent chromosomal regions of importance in the genesis, maintenance, or progression of OS. Regions, such as 10q26, that show loss of heterozygosity in human OS and contain both FGFR2 and Bub3 (which have been implicated as important in OS biology) show significantly reduced expression in both primary mOS and mOS cell lines by CREA (Mendoza et al. 2005). Reduced expression of FGFR2 and Bub3 was also confirmed by quantitative real-time PCR. Analysis of the genes within the other significantly altered cytogenetic regions, and those that show alterations in both the primary mOS and cell lines, may yield important insights into the molecular genetics of OS and highlight conserved genomic regions of importance in OS. One intriguing observation from analysis of the gene expression data is that independent of the initial genotype of the animals the OS that arises appears to be the same disease based on the correlation coefficient of gene expression of the tumors as measured by microarray (Supplemental Fig. 11).
The genetically engineered mouse OS we describe recapitulates multiple defining features of human OS, a devastating malignancy for which new therapies have not been developed in more than two decades. Metastasis is a major cause of mortality in OS and with temporal control of Cre expression it is possible to generate a model in which all animals develop metastatic disease. Murine models of human cancer serve as valuable platforms for probing cancer genetics and preclinical therapeutics. The model we report provides new opportunities for exploration of the molecular pathogenesis of OS and should constitute a convenient platform in which to test or screen for novel therapeutic agents to treat the disease.
Materials and methods
A detailed version of the Materials and Methods can be found in the Supplemental Material.
Animals
pRbfl/fl animals were generously provided by Dr. T. Jacks (Massachusetts Institute of Technology) and were on a C57Bl6/J 129 hybrid background (MacPherson et al. 2003; Sage et al. 2003); p53fl/fl animals were obtained from the National Cancer Institute Mouse Models of Human Cancer Consortium Mouse Repository, have been described previously, and were on a C57Bl6/J 129 FVB/n hybrid background (Jonkers et al. 2001). Osx-Cre animals have been described previously and were on a C57Bl6/J background (Rodda and McMahon 2006). All animals were genotyped using published protocols. Rag2−/− immunodeficient mice were used as recipients for transplant of primary tumor samples and mOS cell lines (subcutaneous transplant). All experiments were approved by the Animal Ethics Committee (Children’s Hospital Boston).
Histology
Tissue was fixed in 10% neutral buffered formalin and then paraffin embedded, sectioned, and stained with hematoxylin and eosin. All pathology was performed blinded to the sample genotype.
SKY
SKY was performed as previously described (Franco et al. 2006). Briefly, metaphase spreads of OS cell lines were hybridized using a SkyPaint DNA Kit (Applied Spectral Imaging), following manufacturer’s instructions. Spectral images were captured and analyzed using an interferometer and software from Applied Spectral Imaging.
microPET/CT imaging
PET imaging studies were performed using a Siemens Focus 120 high-resolution, small-animal PET scanner. Each animal was injected via tail vein with a bolus injection, <0.2 mL per animal, of the radiopharmaceutical (18F sodium fluoride). Between 3.7 and 37 MBq (0.1 to 1.0 mCi) of the radiopharmaceutical was administered. Animals were anesthetized to facilitate imaging by inhalation of isoflurane (2%–4%) through a nose cone for the duration of the imaging study. Image acquisition was performed after an uptake period of 30 min. Each animal was positioned on the imaging table (head first, prone) and imaged for 15 min. MicroCT imaging was performed on a Siemens MicroCAT II scanner at 27-μm resolution. Data were acquired with the system with X-ray tube voltage of 60–80 kVp, tube currents of 200–500 uA, and exposures per view of 300 msec for 200–400 views.
Microarray sample preparation and analysis
Primary tumor samples and cell lines were homogenized in Trizol (Invitrogen) to extract RNA purified using an RNeasy kit (Qiagen) using standard procedures (a more detailed method is provided in the Supplemental Data). Samples were hybridized to Affymetrix A430 2.0 mouse expression arrays, according to the manufacturer’s instructions, at the Dana-Farber Cancer Institute’s Microarray Core Facility. Data from microarray was normalized using dChip software (Li and Wong 2001). The microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE9460.
Metagene analysis
Metagene analysis was performed essentially as described by Tamayo et al. (2007) using the available program from http://www.broad.mit.edu/cgi-bin/cancer/publications/pub_paper.cgi?mode=view&paper_id=161. Human sarcoma data were obtained from a previously published data set (Henderson et al. 2005).
CREA
Common recurrent cytogenetic rearrangements reported in the literature were complied and then the consensus human genes contained within these bands were collated from both the Ensembl and UCSC human genome browsers. Lists of consensus genes were compiled, and GSEA was performed using gene expression data obtained from the comparisons of primary mOS versus in vitro differentiated primary osteoblasts, mOS cell lines versus in vitro differentiated primary osteoblasts, and mouse synovial sarcoma versus muscle (Haldar et al. 2007). GSEA was performed as described (http://www.broad.mit.edu/gsea). Each data set underwent 1000 permutations. A complete list of human genes for each chromosomal region can be found in the Supplemental Material (Supplemental Table 5).
Quantitative real-time PCR
Genomic DNA and cDNA were prepared and quantitative real-time PCR performed as described previously (Walkley et al. 2007). Oligonucleotide sequences are listed in the Supplemental Material.
Acknowledgments
We thank Drs. L Purton, J. Wu, T.J. Martin, K. Janeway, S.T. Treves, C. Roberts, and P. Tamayo for helpful discussion; F. Godinho and J. Shea for technical assistance; and the Dana-Farber Cancer Institute/Harvard Cancer Centre Rodent Histopathology Core for specimen processing. Work in A.P.M’s laboratory was supported by a grant from the National Institutes of Health (P01 056246). S.J.R. was supported by post-doctoral fellowships from the NHMRC of Australia (#301299) and the Arthritis Foundation (#401683). This work was supported in part by an NCI U01 Mouse Models of Human Cancer Consortium award to S.H.O. V.G.S. was supported by MSTP and NRSA awards from the NIH. C.R.W. is a Special Fellow of the Leukemia and Lymphoma Society, and S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1656808.
References
- Aubin J.E. Regulation of osteoblast formation and function. Rev. Endocr. Metab. Disord. 2001;2:81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- Batanian J.R., Cavalli L.R., Aldosari N.M., Ma E., Sotelo-Avila C., Ramos M.B., Rone J.D., Thorpe C.M., Haddad B.R. Evaluation of paediatric osteosarcomas by classic cytogenetic and CGH analyses. Mol. Pathol. 2002;55:389–393. doi: 10.1136/mp.55.6.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher O.J., Holland E.C. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66:3355–3358. doi: 10.1158/0008-5472.CAN-05-3827. [DOI] [PubMed] [Google Scholar]
- Brenner W., Vernon C., Muzi M., Mankoff D.A., Link J.M., Conrad E.U., Eary J.F. Comparison of different quantitative approaches to 18F-fluoride PET scans. J. Nucl. Med. 2004;45:1493–1500. [PubMed] [Google Scholar]
- Bridge J.A., Bhatia P.S., Anderson J.R., Neff J.R. Biologic and clinical significance of cytogenetic and molecular cytogenetic abnormalities in benign and malignant cartilaginous lesions. Cancer Genet. Cytogenet. 1993;69:79–90. doi: 10.1016/0165-4608(93)90080-6. [DOI] [PubMed] [Google Scholar]
- Bridge J.A., Nelson M., McComb E., McGuire M.H., Rosenthal H., Vergara G., Maale G.E., Spanier S., Neff J.R. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet. Cytogenet. 1997;95:74–87. doi: 10.1016/s0165-4608(96)00306-8. [DOI] [PubMed] [Google Scholar]
- Clarke A.R., Maandag E.R., van Roon M., van der Lugt N.M., van der Valk M., Hooper M.L., Berns A., te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Ek E.T., Dass C.R., Choong P.F. Commonly used mouse models of osteosarcoma. Crit. Rev. Oncol. Hematol. 2006;60:1–8. doi: 10.1016/j.critrevonc.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Even-Sapir E., Metser U., Flusser G., Zuriel L., Kollender Y., Lerman H., Lievshitz G., Ron I., Mishani E. Assessment of malignant skeletal disease: Initial experience with 18F-fluoride PET/CT and comparison between 18F-fluoride PET and 18F-fluoride PET/CT. J. Nucl. Med. 2004;45:272–278. [PubMed] [Google Scholar]
- Franco S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M., et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Franz-Odendaal T.A., Hall B.K., Witten P.E. Buried alive: How osteoblasts become osteocytes. Dev. Dyn. 2006;235:176–190. doi: 10.1002/dvdy.20603. [DOI] [PubMed] [Google Scholar]
- Frese K.K., Tuveson D.A. Maximizing mouse cancer models. Nat. Rev. Cancer. 2007;7:654–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- Gang E.J., Bosnakovski D., Figueiredo C.A., Visser J.W., Perlingeiro R.C. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- Gonin-Laurent N., Gibaud A., Huygue M., Lefevre S.H., Le Bras M., Chauveinc L., Sastre-Garau X., Doz F., Lumbroso L., Chevillard S., et al. Specific TP53 mutation pattern in radiation-induced sarcomas. Carcinogenesis. 2006;27:1266– 1272. doi: 10.1093/carcin/bgi356. [DOI] [PubMed] [Google Scholar]
- Gonin-Laurent N., Hadj-Hamou N.S., Vogt N., Houdayer C., Gauthiers-Villars M., Dehainault C., Sastre-Garau X., Chevillard S., Malfoy B. RB1 and TP53 pathways in radiation-induced sarcomas. Oncogene. 2007;26:6106–6112. doi: 10.1038/sj.onc.1210404. [DOI] [PubMed] [Google Scholar]
- Grigoriadis A.E., Schellander K., Wang Z.Q., Wagner E.F. Osteoblasts are target cells for transformation in c-fos transgenic mice. J. Cell Biol. 1993;122:685–701. doi: 10.1083/jcb.122.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney J.G., Severson R.K., Davis S., Robison L.L. Incidence of cancer in children in the United States. Sex-, race-, and 1-year age-specific rates by histologic type. Cancer. 1995;75:2186–2195. doi: 10.1002/1097-0142(19950415)75:8<2186::aid-cncr2820750825>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Haldar M., Hancock J.D., Coffin C.M., Lessnick S.L., Capecchi M.R. A conditional mouse model of synovial sarcoma: Insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Helman L.J., Meltzer P. Mechanisms of sarcoma development. Nat. Rev. Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- Henderson S.R., Guiliano D., Presneau N., McLean S., Frow R., Vujovic S., Anderson J., Sebire N., Whelan J., Athanasou N., et al. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly B.J., Shigematsu H., Deguchi K., Lee B.H., Mizuno S., Duclos N., Rowan R., Amaral S., Curley D., Williams I.R., et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Ince T.A., Richardson A.L., Bell G.W., Saitoh M., Godar S., Karnoub A.E., Iglehart J.D., Weinberg R.A. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Isakoff M.S., Sansam C.G., Tamayo P., Subramanian A., Evans J.A., Fillmore C.M., Wang X., Biegel J.A., Pomeroy S.L., Mesirov J.P., et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jonkers J., Berns A. Conditional mouse models of sporadic cancer. Nat. Rev. Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kansara M., Thomas D.M. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- Kelly P.N., Dakic A., Adams J.M., Nutt S.L., Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Kirsch D.G., Grimm J., Shaw A.T., Lane K., Kissil J., Olive K.P., Sweet-Cordero A., Weissleder R., et al. Mouse models of human non-small-cell lung cancer: Raising the bar. Cold Spring Harb. Symp. Quant. Biol. 2005a;70:241–250. doi: 10.1101/sqb.2005.70.037. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005b;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., IJpenberg A., Sing J.Y., van der Valk M., Nawijn M., Zevenhoven J., Berns A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–947. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lee E.Y., Chang C.Y., Hu N., Wang Y.C., Lai C.C., Herrup K., Lee W.H., Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lengner C.J., Steinman H.A., Gagnon J., Smith T.W., Henderson J.E., Kream B.E., Stein G.S., Lian J.B., Jones S.N. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J. Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tognon C.E., Godinho F.J., Yasaitis L., Hock H., Herschkowitz J.I., Lannon C.L., Cho E., Kim S.J., Bronson R.T., et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–558. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R., Fahey F.H., Drubach L.A., Connolly L.P., Treves S.T. Early experience with fluorine-18 sodium fluoride bone PET in young patients with back pain. J. Pediatr. Orthop. 2007;27:277–282. doi: 10.1097/BPO.0b013e31803409ba. [DOI] [PubMed] [Google Scholar]
- Liu X., Holstege H., van der Gulden H., Treur-Mulder M., Zevenhoven J., Velds A., Kerkhoven R.M., van Vliet M.H., Wessels L.F., Peterse J.L., et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D., Sage J., Crowley D., Trumpp A., Bronson R.T., Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey A.I., Saotome I., Mercer K., Crowley D., Gusella J.F., Bronson R.T., Jacks T. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes & Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza S., David H., Gaylord G.M., Miller C.W. Allelic loss at 10q26 in osteosarcoma in the region of the BUB3 and FGFR2 genes. Cancer Genet. Cytogenet. 2005;158:142–147. doi: 10.1016/j.cancergencyto.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Meyers P.A., Heller G., Healey J. Retrospective review of neoadjuvant chemotherapy for osteogenic sarcoma. J. Natl. Cancer Inst. 1992;84:202–204. doi: 10.1093/jnci/84.3.202-b. [DOI] [PubMed] [Google Scholar]
- Meyers P.A., Schwartz C.L., Krailo M., Kleinerman E.S., Betcher D., Bernstein M.L., Conrad E., Ferguson W., Gebhardt M., Goorin A.M., et al. Osteosarcoma: A randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J. Clin. Oncol. 2005;23:2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- Miller C.W., Aslo A., Tsay C., Slamon D., Ishizaki K., Toguchida J., Yamamuro T., Lampkin B., Koeffler H.P. Frequency and structure of p53 rearrangements in human osteosarcoma. Cancer Res. 1990;50:7950–7954. [PubMed] [Google Scholar]
- Miller C.W., Aslo A., Campbell M.J., Kawamata N., Lampkin B.C., Koeffler H.P. Alterations of the p15, p16, and p18 genes in osteosarcoma. Cancer Genet. Cytogenet. 1996;86:136–142. doi: 10.1016/0165-4608(95)00216-2. [DOI] [PubMed] [Google Scholar]
- Miller C.W., Yeon C., Aslo A., Mendoza S., Aytac U., Koeffler H.P. The p19INK4D cyclin dependent kinase inhibitor gene is altered in osteosarcoma. Oncogene. 1997;15:231–235. doi: 10.1038/sj.onc.1201185. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nishio J., Gentry J.D., Neff J.R., Nelson M., Daniels W., Perry D., Gatalica Z., Bridge J.A. Monoallelic deletion of the p53 gene through chromosomal translocation in a small cell osteosarcoma. Virchows Arch. 2006;448:852–856. doi: 10.1007/s00428-006-0181-x. [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pakos E.E., Kyzas P.A., Ioannidis J.P. Prognostic significance of TP53 tumor suppressor gene expression and mutations in human osteosarcoma: A meta-analysis. Clin. Cancer Res. 2004;10:6208–6214. doi: 10.1158/1078-0432.CCR-04-0246. [DOI] [PubMed] [Google Scholar]
- Polyak K., Hahn W.C. Roots and stems: Stem cells in cancer. Nat. Med. 2006;12:296–300. doi: 10.1038/nm1379. [DOI] [PubMed] [Google Scholar]
- Porter D.E., Holden S.T., Steel C.M., Cohen B.B., Wallace M.R., Reid R. A significant proportion of patients with osteosarcoma may belong to Li-Fraumeni cancer families. J. Bone Joint Surg. Br. 1992;74:883–886. doi: 10.1302/0301-620X.74B6.1447251. [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rodda S.J., McMahon A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Ruther U., Komitowski D., Schubert F.R., Wagner E.F. c-fos expression induces bone tumors in transgenic mice. Oncogene. 1989;4:861–865. [PubMed] [Google Scholar]
- Sage J., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T. Acute mutation of retinoblastoma is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sandberg A.A., Bridge J.A. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet. Cytogenet. 2003;145:1–30. [PubMed] [Google Scholar]
- Semerad C.L., Christopher M.J., Liu F., Short B., Simmons P.J., Winkler I., Levesque J.P., Chappel J., Ross F.P., Link D.C. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood. 2005;106:3020–3027. doi: 10.1182/blood-2004-01-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E., Depinho R.A. The mighty mouse: Genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy N., Lee K.H., Carrasco D., Castrillon D.H., Aguirre A.J., Wu E.A., Horner J.W., DePinho R.A. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Ramsey M.R., Balasubramanian P., Castrillon D.H., DePinho R.A. The differential impact of p16INK4a or p19ARF deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- Stemmer-Rachamimov A.O., Nielsen G.P., Rosenberg A.E., Louis D.N., Jones D., Ramesh V., Gusella J.F., Jacoby L.B. The NF2 gene and merlin protein in human osteosarcomas. Neurogenetics. 1998;2:73–74. doi: 10.1007/s100480050054. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero A., Mukherjee S., Subramanian A., You H., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Tamayo P., Scanfeld D., Ebert B.L., Gillette M.A., Roberts C.W., Mesirov J.P. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc. Natl. Acad. Sci. 2007;104:5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.Z., Schlicht S.M., Powell G.J., Thomas D., Slavin J.L., Smith P.J., Choong P.F. Multidisciplinary approach to diagnosis and management of osteosarcoma—A review of the St Vincent’s Hospital experience. Int. Semin. Surg. Oncol. 2006;3:38. doi: 10.1186/1477-7800-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.M., Carty S.A., Piscopo D.M., Lee J.S., Wang W.F., Forrester W.C., Hinds P.W. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Thomas D.M., Yang H.S., Alexander K., Hinds P.W. Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol. Ther. 2003;2:124–130. [PubMed] [Google Scholar]
- Thomas D.M., Johnson S.A., Sims N.A., Trivett M.K., Slavin J.L., Rubin B.P., Waring P., McArthur G.A., Walkley C.R., Holloway A.J., et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F., Laud-Duval K., Prieur A., Delorme B., Charbord P., Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Sekine K., Hinohara S., Namiki T., Nobori T., Kaneko Y. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet. Cytogenet. 2000;120:91–98. doi: 10.1016/s0165-4608(99)00255-1. [DOI] [PubMed] [Google Scholar]
- Tuveson D.A., Jacks T. Technologically advanced cancer modeling in mice. Curr. Opin. Genet. Dev. 2002;12:105–110. doi: 10.1016/s0959-437x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- Unni K.K., Bridge J.A., Kindblom L.-G., Wold L.E. Tumors of the bones and joints. American Registry of Pathology; Washington, D.C: 2005. [Google Scholar]
- Wadayama B., Toguchida J., Shimizu T., Ishizaki K., Sasaki M.S., Kotoura Y., Yamamuro T. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994;54:3042–3048. [PubMed] [Google Scholar]
- Walkley C.R., Shea J.M., Sims N.A., Purton L.E., Orkin S.H. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Liang J., Schellander K., Wagner E.F., Grigoriadis A.E. c-fos-induced osteosarcoma formation in transgenic mice: Cooperativity with c-jun and the role of endogenous c-fos. Cancer Res. 1995;55:6244–6251. [PubMed] [Google Scholar]
- Wu J.X., Carpenter P.M., Gressens C., Keh R., Niman H., Norris J.W., Mercola D. The proto-oncogene c-fos is over-expressed in the majority of human osteosarcoma. Oncogene. 1990;5:989–1000. [PubMed] [Google Scholar]
- Wunder J.S., Gokgoz N., Parkes R., Bull S.B., Eskandarian S., Davis A.M., Beauchamp C.P., Conrad E.U., Grimer R.J., Healey J.H., et al. TP53 mutations and outcome in osteosarcoma: A prospective, multicenter study. J. Clin. Oncol. 2005;23:1483–1490. doi: 10.1200/JCO.2005.04.074. [DOI] [PubMed] [Google Scholar]