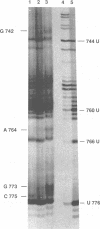
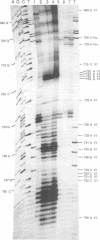
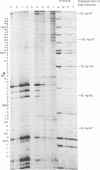
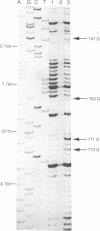
Abstract
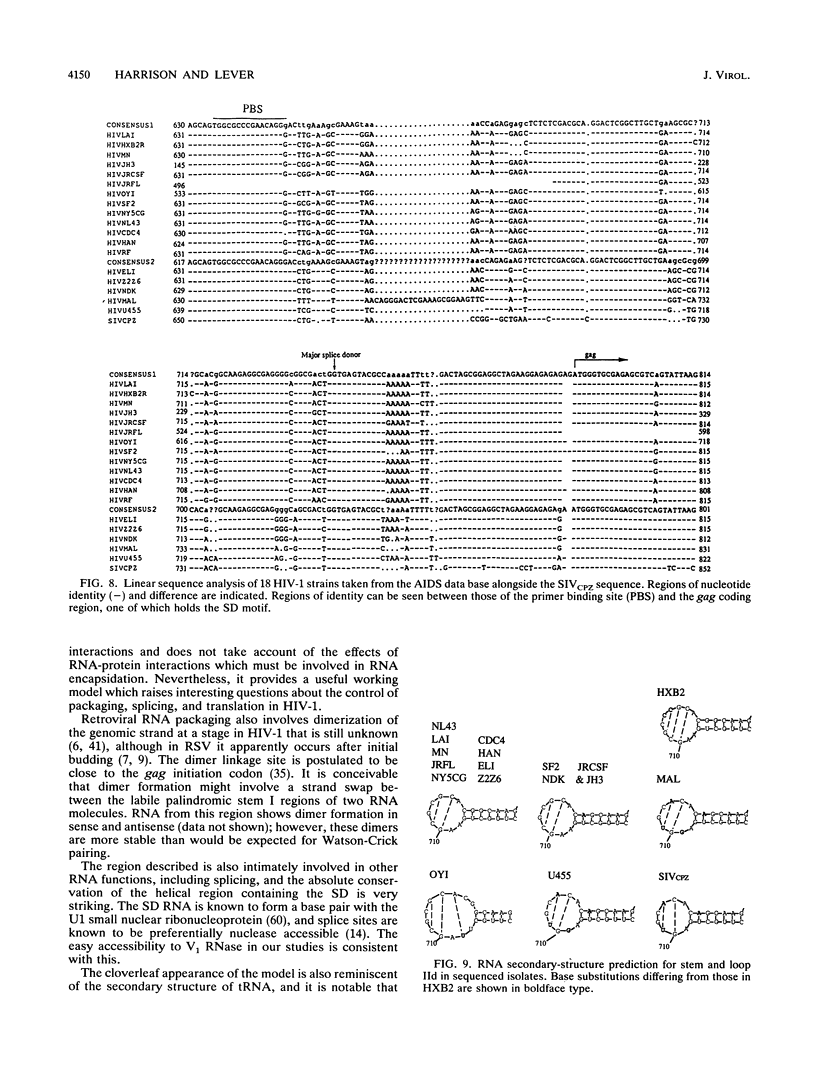
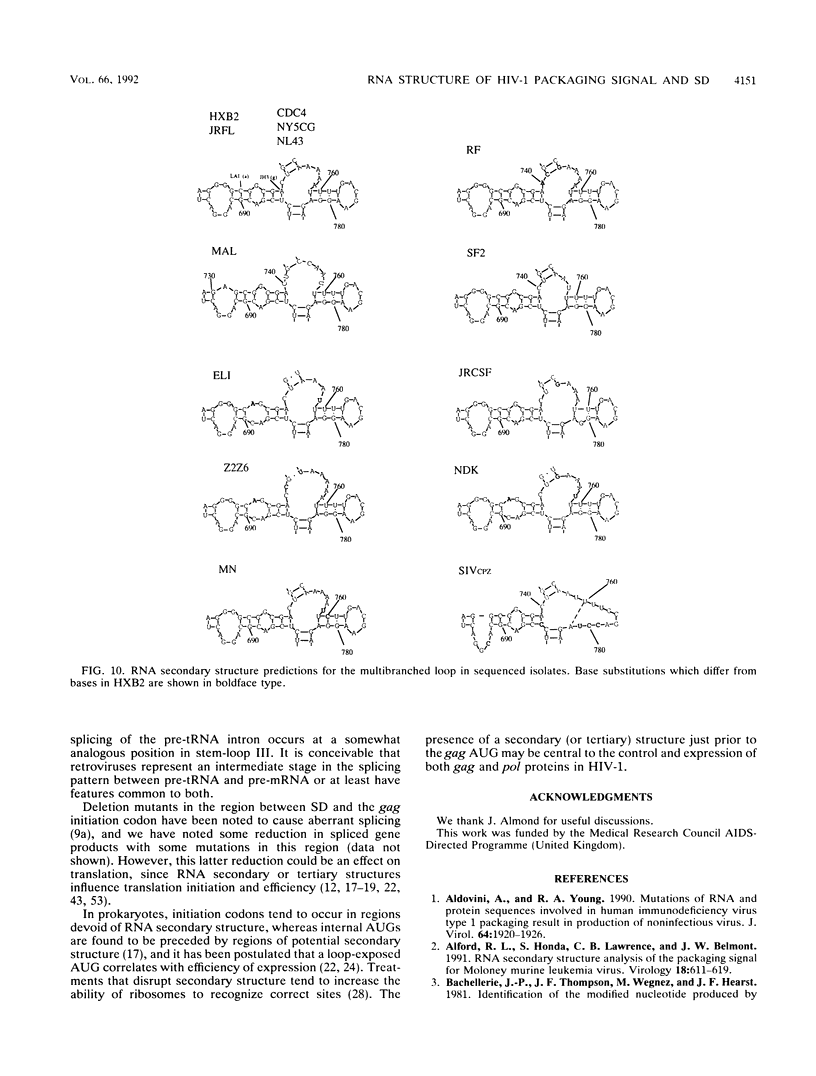
Interaction of cis-acting RNA sequences with nucleocapsid proteins is one of the critical events leading to retroviral genomic RNA packaging. We have derived a potentially stable secondary structure for the packaging signal region of human immunodeficiency virus strain IIIB, using a combination of biochemical analysis and computer modelling. This region encompasses the major splice donor (SD), which is found in a highly structured conserved stem-loop. Comparison with other published human immunodeficiency virus type 1 sequences shows almost absolute nucleotide conservation in base-paired regions required to maintain this structure. Presently and previously described packaging-defective mutants would disrupt the structure. The structure depends on base pairing between nucleotide sequences 5' of the major SD which are common to both genomic and subgenomic RNAs and sequences 3' of SD which are unique to the unspliced RNA. This may explain how in retroviruses such as Rous sarcoma virus, mutations in regions common to genomic and subgenomic RNA might prevent packaging of the unspliced mRNA by disrupting a signal structure which can exist only in the genomic RNA species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldovini A., Young R. A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990 May;64(5):1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. L., Honda S., Lawrence C. B., Belmont J. W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991 Aug;183(2):611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- Bachellerie J. P., Thompson J. F., Wegnez M. R., Hearst J. E. Identification of the modified nucleotides produced by covalent photoaddition of hydroxymethyltrimethylpsoralen to RNA. Nucleic Acids Res. 1981 May 11;9(9):2207–2222. doi: 10.1093/nar/9.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M. A., Palmer T. D., Gelinas R. E., Miller A. D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987 May;61(5):1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Bieth E., Gabus C., Darlix J. L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990 Jan 11;18(1):119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani E., Helm K. V., Duesberg P. Evidence for 30-40S RNA as precursor of the 60-70S RNA of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1973 Feb;70(2):401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K. S., Smith R. E., Stone M. P., Joklik W. K. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology. 1972 Dec;50(3):851–864. doi: 10.1016/0042-6822(72)90439-4. [DOI] [PubMed] [Google Scholar]
- Clavel F., Orenstein J. M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990 Oct;64(10):5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J. B., Engel J. D. The nucleotide sequence of the adult chicken alpha-globin genes. J Biol Chem. 1983 Apr 10;258(7):4623–4629. [PubMed] [Google Scholar]
- Favorova O. O., Fasiolo F., Keith G., Vassilenko S. K., Ebel J. P. Partial digestion of tRNA--aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry. 1981 Feb 17;20(4):1006–1011. doi: 10.1021/bi00507a055. [DOI] [PubMed] [Google Scholar]
- Fisher A. G., Collalti E., Ratner L., Gallo R. C., Wong-Staal F. A molecular clone of HTLV-III with biological activity. Nature. 1985 Jul 18;316(6025):262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Kofoid E. C., Marlière P., Louis B. G. Potential secondary structure at translation-initiation sites. Nucleic Acids Res. 1987 Jan 12;15(1):345–360. doi: 10.1093/nar/15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gualerzi C. O., Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990 Jun 26;29(25):5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Structure in solution of M1 RNA, the catalytic subunit of ribonuclease P from Escherichia coli. Biochemistry. 1984 Dec 18;23(26):6327–6334. doi: 10.1021/bi00321a006. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Fox G. E. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16 (Suppl):r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Pace N. R. Phylogenetic comparative analysis of RNA secondary structure. Methods Enzymol. 1989;180:227–239. doi: 10.1016/0076-6879(89)80104-1. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Terry R. W., Skalka A. M. A conserved cis-acting sequence in the 5' leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986 Jul;59(1):163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean J. M., Draper D. E. Secondary structure of a 345-base RNA fragment covering the S8/S15 protein binding domain of Escherichia coli 16S ribosomal RNA. Biochemistry. 1985 Sep 10;24(19):5052–5061. doi: 10.1021/bi00340a015. [DOI] [PubMed] [Google Scholar]
- Kozak M., Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol Rev. 1972 Mar;36(1):109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A. M., Richardson J. H., Harrison G. P. Retroviral RNA packaging. Biochem Soc Trans. 1991 Nov;19(4):963–966. doi: 10.1042/bst0190963. [DOI] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989 Sep;63(9):4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Medeiros E., Hayward W. S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell. 1978 Dec;15(4):1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- Lipson S. E., Hearst J. E. Psoralen cross-linking of ribosomal RNA. Methods Enzymol. 1988;164:330–341. doi: 10.1016/s0076-6879(88)64053-5. [DOI] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Marquet R., Baudin F., Gabus C., Darlix J. L., Mougel M., Ehresmann C., Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991 May 11;19(9):2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Pugatsch T., Stacey D. W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983 Jul 30;128(2):505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- Roy C., Tounekti N., Mougel M., Darlix J. L., Paoletti C., Ehresmann C., Ehresmann B., Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990 Dec 25;18(24):7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V. P., Reznikoff W. S. In vitro secondary structure analysis of mRNA from lacZ translation initiation mutants. J Mol Biol. 1990 Jan 20;211(2):427–445. doi: 10.1016/0022-2836(90)90363-Q. [DOI] [PubMed] [Google Scholar]
- Shelness G. S., Williams D. L. Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. Evaluation of primer extension for high resolution structure mapping of mRNA. J Biol Chem. 1985 Jul 15;260(14):8637–8646. [PubMed] [Google Scholar]
- Siliciano P. G., Jones M. H., Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987 Sep 18;237(4821):1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Skinner M. A., Racaniello V. R., Dunn G., Cooper J., Minor P. D., Almond J. W. New model for the secondary structure of the 5' non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989 May 20;207(2):379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- Sorge J., Ricci W., Hughes S. H. cis-Acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983 Dec;48(3):667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Meissner F., Moll J., Vorderwülbecke T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1987;15 (Suppl):r53–188. doi: 10.1093/nar/15.suppl.r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Teare J., Wollenzien P. The structure of a pre-mRNA molecule in solution determined with a site directed cross-linking reagent. Nucleic Acids Res. 1990 Feb 25;18(4):855–864. doi: 10.1093/nar/18.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure of E. coli 16S RNA elucidated by psoralen crosslinking. Cell. 1983 Apr;32(4):1355–1365. doi: 10.1016/0092-8674(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Vassilenko S. K., Carbon P., Ebel J. P., Ehresmann C. Topography of 16 S RNA in 30 S subunits and 70 S ribosomes accessibility to cobra venom ribonuclease. J Mol Biol. 1981 Nov 15;152(4):699–721. doi: 10.1016/0022-2836(81)90123-6. [DOI] [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5' long terminal repeat and the start of the gag gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. Sequencing psoralen photochemically reactive sites in Escherichia coli 16 S rRNA. Anal Biochem. 1982 Jan 1;119(1):86–89. doi: 10.1016/0003-2697(82)90669-8. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zuker M., Jaeger J. A., Turner D. H. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991 May 25;19(10):2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- van de Guchte M., van der Lende T., Kok J., Venema G. A possible contribution of mRNA secondary structure to translation initiation efficiency in Lactococcus lactis. FEMS Microbiol Lett. 1991 Jun 15;65(2):201–208. doi: 10.1111/j.1574-6968.1991.tb04746.x. [DOI] [PubMed] [Google Scholar]