Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that are involved in the regulation of thousands of gene targets. Recent studies indicate that miRNAs are likely to be master regulators of many important biological processes. Due to their functional importance, miRNAs are under intense study at present, and many studies have been published in recent years on miRNA functional characterization. The rapid accumulation of miRNA knowledge makes it challenging to properly organize and present miRNA function data. Although several miRNA functional databases have been developed recently, this remains a major bioinformatics challenge to miRNA research community. Here, we describe a new online database system, miRDB, on miRNA target prediction and functional annotation. Flexible web search interface was developed for the retrieval of target prediction results, which were generated with a new bioinformatics algorithm we developed recently. Unlike most other miRNA databases, miRNA functional annotations in miRDB are presented with a primary focus on mature miRNAs, which are the functional carriers of miRNA-mediated gene expression regulation. In addition, a wiki editing interface was established to allow anyone with Internet access to make contributions on miRNA functional annotation. This is a new attempt to develop an interactive community-annotated miRNA functional catalog. All data stored in miRDB are freely accessible at http://mirdb.org.
Keywords: microRNA, database, target prediction, functional annotation, wiki
INTRODUCTION
MicroRNAs (miRNAs) are short noncoding RNAs that regulate a variety of biological processes such as cell growth, tissue differentiation, apoptosis, and viral infection (Ambros 2004; Miska 2005). Although only ∼22 nucleotides long, one miRNA may potentially regulate the expression of many gene targets, and thus, they are likely to be master switches in many biological pathways (Lewis et al. 2005; Lim et al. 2005; Miranda et al. 2006).
miRNA functional characterization is currently a very active research field in biology, and there have been a rapid accumulation of miRNA knowledge in the past few years. While the large quantity of existing data is certainly very helpful to guide future studies, at the same time it also makes it challenging for miRNA researchers to quickly retrieve information relevant to their studies. Several miRNA databases have been established to systematically organize miRNA data. The most prominent one is miRBase, which was set up to provide official nomenclatures to miRNA research community (Griffiths-Jones et al. 2006). Authoritative names, sequences, and genomic locations were systematically assigned to each miRNA according to community-adopted standards (Ambros et al. 2003). The standardization on miRNA nomenclatures is important to cross-reference results from different studies as well as to automatically integrate miRNA data from heterogeneous sources.
Although miRBase is a valuable source for providing standard miRNA nomenclatures, it contains limited information on miRNA functional annotation. Besides miRBase, a few other databases have been developed to focus more on miRNA function. For example, TargetScan and PicTar host miRNA targets predicted by different computational algorithms (Krek et al. 2005; Grimson et al. 2007). TarBase has a main focus of collecting and presenting experimentally validated miRNA targets (Sethupathy et al. 2006). More comprehensive databases have also been developed recently, attempting to integrate target prediction results with other functional annotations (Hsu et al. 2006; Shahi et al. 2006; Chiromatzo et al. 2007; Megraw et al. 2007; Nam et al. 2008). These databases usually include data from miRBase for miRNA sequences and standard nomenclatures, as well as target prediction results from other prediction databases.
Here, we describe a new database, which we called miRDB, for miRNA target prediction and functional annotation. miRDB is different from existing miRNA functional databases mainly as the following: (1) Originality of the target prediction results; (2) a new database design strategy centered on mature miRNAs; and (3) a wiki editing interface for community-provided miRNA annotations. miRDB is freely accessible at http://mirdb.org.
With the rapid progress on miRNA functional studies, it is challenging for any single team to keep track of all the latest development in the field. To address this issue, miRDB provides an open platform using the wiki model, which has been proved to be widely successful for many other Internet-based projects. One well-known example is Wikipedia (http://wikipedia.org), which is a free encyclopedia anyone can edit. As of November 2007, there were close to 10 million entries in Wikipedia, making it the largest reference work on the Internet. Wikipedia's articles are written collaboratively by volunteers around the world and most of them can be edited by anyone with Internet access.
Although the wiki model could potentially be applied to any Internet-based projects, it has not been widely used in biological research (Salzberg 2007). The gene annotation database is similar to the encyclopedia in many ways. The information required to construct the database is broad-based, and the expert knowledge is scattered around the scientific community. miRDB represents a new attempt to build a collaborative miRNA knowledge base by providing a wiki web interface for community editing. All miRNA researchers are invited to continuously provide miRNA functional annotations and actively interact with each other. A comprehensive and dynamic catalog of miRNA functions will be a valuable asset to the whole miRNA community.
RESULTS
miRDB consists of two child databases and the related web interfaces for (1) the retrieval of computationally predicted miRNA targets, and (2) miRNA functional annotations with a wiki editing interface. Perl and PHP were used to construct miRDB website, which links to a backend MySQL database server.
Presentation of predicted miRNA targets
We have recently developed a new computational algorithm for miRNA target prediction (Wang and El Naqa 2008). Relevant features associated with miRNA target binding were identified by analyzing thousands of miRNA down-regulated genes and these features were used to train a bioinformatics target prediction model with machine learning. This prediction model has been validated with independent experimental data. Among the miRNA down-regulated genes from a miR-124a data set and curated human genes from TarBase (Sethupathy et al. 2006; Wang and Wang 2006), 43% and 40% were identified by our prediction algorithm, respectively. A large number of down-regulated genes from these validation data sets could be suppressed by miRNA indirectly. Thus, the actual prediction sensitivity is likely to be higher if only direct miRNA targets are considered. Overall, our target prediction algorithm has been demonstrated to have superior performance in prediction sensitivity and specificity over a few other popular algorithms included in our comparative analysis (Wang and El Naqa 2008). Despite the improvement in performance, a significant number of bona fide miRNA targets could still be missed by our prediction algorithm, indicating the need for further algorithmic improvement in future. Readers who are interested in learning more details about this new prediction algorithm and its performance are referred to our recently published article (Wang and El Naqa 2008). Current challenges for miRNA target prediction in general have also been extensively discussed in that article.
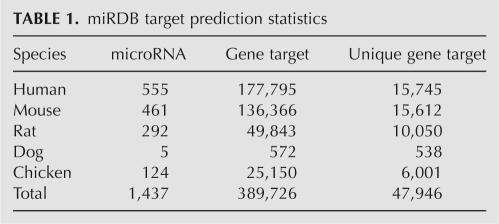
To help other miRNA researchers to take advantage of this new algorithm, genome-wide target prediction was performed, and the predicted targets were imported into miRDB. miRDB hosts predicted miRNA targets in five species: human, mouse, rat, dog, and chicken. The detailed statistics are listed in Table 1. As of version 2.0, miRDB contains 1437 miRNAs targeting 47,946 unique genes. All predicted targets are freely accessible via web search. Alternatively, the prediction data can be batch downloaded for current and all previous miRDB versions.
TABLE 1.
miRDB target prediction statistics
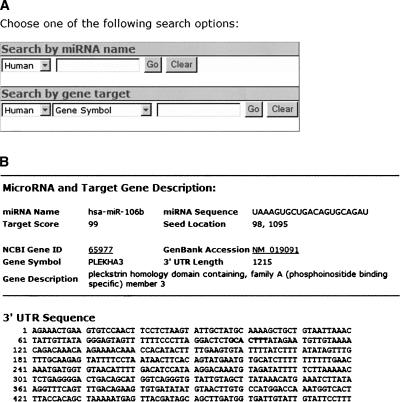
A web query interface was developed to access target prediction results by miRNA name, target GenBank accession, NCBI Gene ID, or gene symbol (Fig. 1A). The search result is sorted by target score, which represents the confidence level for target prediction. A screen shot of target search result is presented in Figure 1B. The detailed result page contains information about the miRNA and its gene target. In addition, the target sites in the 3′-untranslated region (UTR) are also highlighted.
FIGURE 1.
miRNA target search with the standard query interface in miRDB. (A) A screen shot of the web search page. There are two search options for either miRNA or the gene target. (B) A screen shot of one detailed result page.
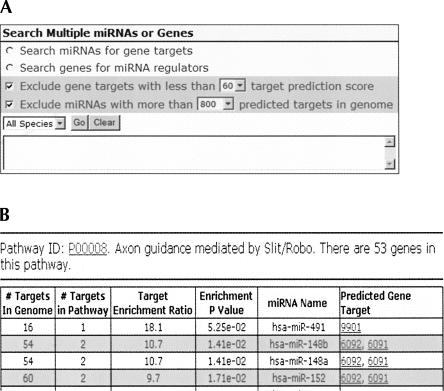
The simple search interface described above is for the analysis of one miRNA or gene target. In addition, miRDB also provides a more advanced query interface for analyzing multiple miRNAs or gene targets together (Fig. 2A). For example, one may want to check whether a group of genes in a biological pathway are targeted by any miRNA. To provide more flexibility in target retrieval, search filter options are also provided to exclude less interesting miRNAs or gene targets. Besides the dynamic query interface for user-provided genes or miRNAs, miRDB also presents miRNA target prediction results for precompiled pathways (Fig. 2B). The biological pathways were imported from PANTHER (Mi et al. 2005), and potential miRNA target enrichment in the pathways was evaluated. The enrichment ratio was defined as (the fraction of miRNA targets among all genes in a pathway)/(the fraction of targets among all genes in the genome). In this way, interesting links between miRNAs and biological pathways may be discovered.
FIGURE 2.
Advanced target search for multiple miRNAs or genes. (A) A screen shot of the advance web search page. Target search may be performed using either miRNA names or target gene IDs. Search filters were also available to further exclude less relevant miRNAs or gene targets. (B) A screen shot of one pathway target search result. The pathway information was imported from PANTHER database.
A miRNA functional catalog
Most existing miRNA databases organize miRNA annotations by precursor name. While useful to represent miRNA gene structures in the genome, this database design strategy also creates major challenges for the annotation of functional miRNA molecules. For example, mature miRNA hsa-let-7f has three precursors in the genome, and thus, there are three separate pages describing hsa-let-7f. As a result, there is no centralized place to present the functional annotations of hsa-let-7f and database redundancy is inevitable. To address this issue, miRDB is designed to focus primarily on mature miRNAs, which are the primary carriers of miRNA function. Functional annotations associated with one mature miRNA (including multiple precursors in the genome) are organized together in one web page. In this way, miRDB provides a centralized view for the annotations of functional miRNA molecules. This strategy is analogous to that used in protein annotation databases, where each page is focused on one protein even though there may be multiple gene copies in the genome.
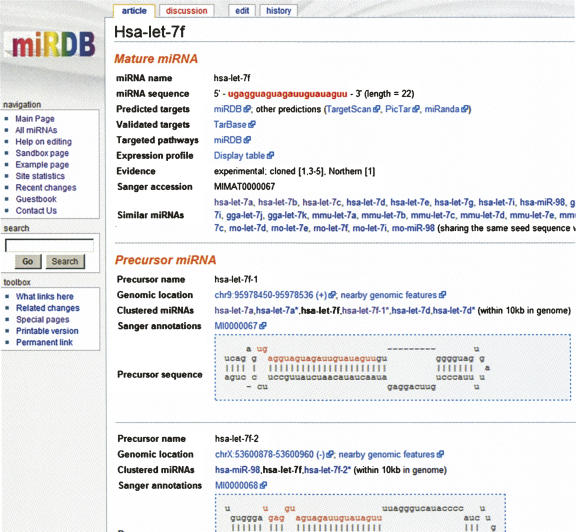
A screen shot of one miRNA functional page is presented in Figure 3. The official miRNA names and sequences were imported from Sanger miRBase. Most other annotations from miRBase are also presented in miRDB. In addition, each miRNA page also contains dynamic links to predicted targets stored in miRDB and other target prediction databases. A dynamic link is also included in each page pointing to validated miRNA targets hosted in TarBase (Sethupathy et al. 2006). Targeted PANTHER pathways (Mi et al. 2005) are presented if there is any miRNA target enrichment in those pathways. The tissue expression profile of a miRNA is also presented based on a recent miRNA profiling study on 40 normal human tissues (Liang et al. 2007). As demonstrated by many previous studies, miRNAs sharing the same seed sequence target similar sets of genes (Lewis et al. 2005; Linsley et al. 2007). Therefore, these miRNAs are considered to be “functionally similar” and are presented as part of the functional annotations. Associated precursor information, including official name, sequence, and genomic location, was imported from Sanger miRBase. The secondary structure of a precursor was calculated with RNAfold (Hofacker 2003) and then presented in web format by parsing the RNAfold output. All the annotations described here were stored in standard web pages, which were later converted to wiki format and used as template files for the construction of the wiki miRNA annotation catalog.
FIGURE 3.
A screen shot of one miRNA functional annotation page. hsa-let-7f is presented here as an example.
Wiki editing interface for user-provided annotations
The wiki server of miRDB was developed with the MediaWiki package (http://www.mediawiki.org), which is widely used to build wiki applications including Wikipedia. All miRNA annotation pages in miRDB can be edited by anyone with Internet access. There are two annotation sections on each page. The first section was prepared by transcluding the miRNA template files. Transclusion is a computer science technique to include one document into another document by reference. The template files were generated with an automated annotation pipeline, and they can only be updated by miRDB administrator. The second section of the page is open for editing by anyone who is interested in providing more miRNA functional descriptions. A history tab is associated with each miRNA page for version control. Undesired changes can be easily rolled back by the author or other miRDB users. There is also a discussion tab on each miRNA page for a group of users to discuss how to best develop the associated miRNA page. In this way, each annotation page can be developed by a group of users in a collaborative way.
By separating miRNA annotations into two sections, miRDB provides a platform that allows both regular batch updates (based on Sanger miRBase and high-throughput data analyses) and manual annotations (contributions from individual researchers) at the same time. These two sections were seamlessly integrated together into one miRNA page with the transclusion technique. The wiki annotation pages and target prediction pages for the same miRNAs are also cross-referenced to each other by dynamic web links to provide an integrated annotation system for miRNA functional studies.
DISCUSSION
miRNA database design
At present, information about miRNA functions is scattered around at many different places. It is a major challenge to properly organize the functional annotations for easy retrieval. One database design strategy is to present the annotations by associating them with miRNA precursors. This design strategy has been adopted by miRBase and most other miRNA databases. This is an effective way to study miRNA gene structures in the genome. However, the same strategy also brings challenges to miRNA functional annotations. Most miRNA functional studies are focused on mature miRNAs, since they are the functional carriers of miRNA-mediated gene expression regulation. For examples, most miRNA microarrays and real-time RT-PCR assays were designed to detect the expression of mature miRNAs; mature miRNAs, but not their precursors, are directly involved in target down-regulation. For these reasons, it is better to focus primarily on mature miRNAs when functional annotations are presented. However, one mature miRNA can be generated from multiple precursors, and there is no central place for functional annotations if miRNA precursors are the main focus. To address this issue, miRDB presents a functional catalog whose pages are organized by mature miRNAs. Annotations on precursors are also presented by associating to the corresponding mature miRNAs (Fig. 3).
A wiki strategy for miRNA annotations
New functional data are constantly generated from high-throughput experiments, such as sequencing and microarrays. These data, typically, are processed with automated bioinformatics pipelines. Besides high-throughput experiments, there are many other “traditional” biological experiments focusing on the functions of one or few genes. At present, computational data processing algorithms are inefficient at retrieving relevant information from these studies. Thus, human input is required to interpret the results and associate new functional annotations to the corresponding genes. With the large number of genes in the genome, it is very challenging for a single team or even an institute to carry out the annotation task alone.
The wiki model could potentially be very helpful to collect gene annotations contributed by researchers around the world (Salzberg 2007). The open environment provided by wiki allows anyone with Internet access to make contributions. Although widely successful in many other projects, the wiki model has not been fully embraced by the biological research community. One major concern is that most existing gene annotations were generated by automated bioinformatics pipelines. As new high-throughput data emerge, it is important to continue to use these pipelines to systematically update the gene annotations. In contrast, wiki pages are primarily developed with human input and are not compatible with the automated update process. To address this apparent discrepancy, each miRNA annotation page in miRDB is composed of two sections. The first section was “transcluded” from a noneditable template file generated by bioinformatics processing. The second section resembles a typical wiki page that provides an open platform to collect manual annotations from miRNA researchers. This is a new attempt to introduce the wiki concept to miRNA data management. If successful, the same concept may also be applied to other biological data management projects, such as functional annotations for all known human genes.
MATERIALS AND METHODS
Target prediction
miRNA sequences and nomenclatures were downloaded from miRBase (Griffiths-Jones et al. 2006). All database tables from miRBase were imported and linked to other tables in miRDB. mRNA sequences and annotation files were downloaded from the NCBI databases (Benson et al. 2007; Maglott et al. 2007). Transcript 3′-UTR sequences from human, mouse, rat, dog, and chicken were parsed from the GenBank files with BioPerl (http://www.bioperl.org), and genome-wide miRNA target prediction was performed with a newly developed bioinformatics tool, MirTarget2 (Wang and El Naqa 2008). Predicted transcript targets were then imported into miRDB. Multiple transcripts from the same genes were mapped using NCBI gene index files, and the transcript with the highest target prediction score was presented on the website. Target prediction results for all the transcripts were also available for batch download from the Data Download page on the miRDB website.
Pathway data were downloaded from the PANTHER database (Mi et al. 2005). For each pathway, target prediction was performed to identify miRNAs that were significantly associated with this pathway. Statistical significance for pathway-specific target enrichment was calculated with hypergeometric test using all genes in the genome as background.
miRNA functional annotations
The functional page for each miRNA consists of annotations from multiple sources. Most annotations stored in Sanger miRBase, such as sequences, nomenclatures, experimental evidence, and references, were also included in the miRDB functional pages. In a recent study, miRNA expression profiles were determined in 40 human tissues with real-time RT-PCR (Liang et al. 2007). The profiling result was downloaded from the journal website and imported into miRDB. A link was created on the miRNA page pointing to an expression profile table, which was dynamically generated from the backend database.
The MediaWiki application package was downloaded from http://mediawiki.org. This package was deployed on a Linux operating system running Apache and PHP5 web server. All wiki miRNA pages and server administration pages were stored in a MySQL database. miRNA annotation template pages were initially prepared in html format with an automated scripting pipeline and later converted to wiki format. There are different levels of access control for this wiki server. miRDB system administrators have the privilege for batch processing and updating of miRNA annotation template files, as well as managing all user accounts. Standard users have the privilege of editing all miRNA functional pages, but not the transcluded template files.
ACKNOWLEDGMENTS
This research was supported by a start-up fund from Washington University School of Medicine in St. Louis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.965408.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiromatzo A.O., Oliveira T.Y., Pereira G., Costa A.Y., Montesco C.A., Gras D.E., Yosetake F., Vilar J.B., Cervato M., Prado P.R., et al. miRNApath: A database of miRNAs, target genes and metabolic pathways. Genet. Mol. Res. 2007;6:859–865. [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: Genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L., Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Linsley P.S., Schelter J., Burchard J., Kibukawa M., Martin M.M., Bartz S.R., Johnson J.M., Cummins J.M., Raymond C.K., Dai H., et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Sethupathy P., Corda B., Hatzigeorgiou A.G. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Miska E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Nam S., Kim B., Shin S., Lee S. miRGator: An integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S.L. Genome re-annotation: A wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P., Loukianiouk S., Bohne-Lang A., Kenzelmann M., Kuffer S., Maertens S., Eils R., Grone H.J., Gretz N., Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., El Naqa I.M. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;34:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]