Abstract
The structural basis of species specificity of transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy or “mad cow disease” and Creutzfeldt–Jakob disease in humans, has been investigated using the refined NMR structure of the C-terminal domain of the mouse prion protein with residues 121–231. A database search for mammalian prion proteins yielded 23 different sequences for the fragment 124–226, which display a high degree of sequence identity and show relevant amino acid substitutions in only 18 of the 103 positions. Except for a unique isolated negative surface charge in the bovine protein, the amino acid differences are clustered in three distinct regions of the three-dimensional structure of the cellular form of the prion protein. Two of these regions represent potential species-dependent surface recognition sites for protein–protein interactions, which have independently been implicated from in vitro and in vivo studies of prion protein transformation. The third region consists of a cluster of interior hydrophobic side chains that may affect prion protein transformation at later stages, after initial conformational changes in the cellular protein.
Transmissible spongiform encephalopathies (TSE) are neurodegenerative diseases for which there is evidence that they are related to a novel, so far unique, infectious agent, the prion (1, 2). TSEs have been reported to occur as infectious, inherited, and spontaneous diseases. Following the “protein-only hypothesis” (2, 3) the causative agent is a pathogenic conformation of the prion protein (PrP). PrP is ubiquitous in mammalian cells in a benign, cellular conformation (PrPC). In rare cases it may be transformed into the infectious scrapie conformation (PrPSc), which forms insoluble, protease-resistant aggregates in the brain of affected individuals (4, 5). The most widely discussed TSEs are Creutzfeldt–Jakob disease in humans, scrapie in sheep, and bovine spongiform encephalopathy. Other human prion diseases include kuru, the Gerstmann–Sträussler–Scheinker syndrome, and fatal familial insomnia.
With the background of the “mad cow crisis” in Europe, questions relating to the relative ease of infection between different individuals of the same species or between different species have attracted intensive interest, in particular regarding possible transmission of bovine spongiform encephalopathy from cows to humans through the food chain (6, 7). A species barrier for prion infection has indeed been convincingly documented (4, 5) and found to vary widely depending on the pair of species involved and the direction of transmission. Typical laboratory tests involve inoculation of mice or hamsters by injection of infectious material directly into the brain, and work with transgenic animals has led to the identification of polypeptide segments in PrP that appear to dominate the species barrier (surveyed in refs. 4 and 8). There is a general consensus that interspecies transfer, when compared with infection within one species, is inefficient and occurs, if at all, only after prolonged incubation times. However, uncertainty remains with regard to the stringency of the transmission barrier for certain pairs of species. In vitro studies of the cell-free conversion of PrPC to protease-resistant forms are overall in support of the aforementioned in vivo data (8).
The present paper correlates biological and biochemical data on the species barrier with the refined NMR structure of the recombinant C-terminal domain of the mouse PrP with residues 121–231 [PrP(121–231)] (9). The pairwise sequence identity for this domain of about 90% among mammalian species (10) implies that all these species should have identical three-dimensional PrP(121–231) folds (11); this is supported by the modeling of the three-dimensional structures of PrP(121–231) from different species in our laboratory (unpublished data). Within the framework of the protein-only hypothesis the contributions to the species barrier from the polypeptide segment 121–231 then must be related to part or all of the amino acid replacements between the PrP of a given pair of species. The aforementioned in vivo inoculation studies and in vitro conversion experiments support the hypothesis that a critical step of infectious transmission of transmissible spongiform encephalopathies involves a specific intermolecular contact between PrPC and PrPSc (4, 5, 8). From additional work with mouse/human chimeras, interactions of PrPC with a so far unknown additional protein X also have been postulated to contribute to efficient conversion (12). Here, we map the polypeptide segment 124–226 of all known mammalian PrP sequences (10) onto the three-dimensional structure of mouse PrP(121–231) and evaluate potential effects on protein–protein interactions with PrPC†.
METHODS
The structure of mouse PrP(121–231) used for the present analysis is a refinement of the previously reported NMR structure (9), which is based on an input of 1,586 nuclear Overhauser effect distance constraints. The structure determination will be described elsewhere (unpublished work). Key characteristics of the refined structure include that there are no residual violations of the aforementioned distance constraints in excess of 0.1 Å and that the well defined regions comprise residues 124–166 and 172–226, for which a root mean square distance for the backbone atoms N, Cα, and C′ of the 20 final conformers to their mean of 0.8 ± 0.1 Å was obtained. Figs. 2, 3, and 4 of this paper show an energy-refined mean structure of mouse PrP(121–231), of which the atom coordinates have been deposited in the Brookhaven Protein Data Bank (accession code 1AG2).
Figure 2.
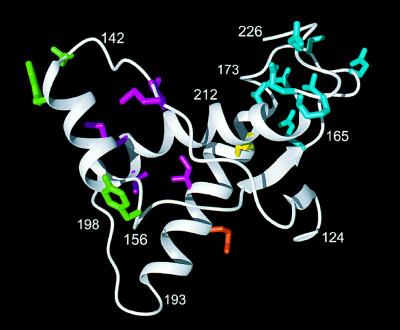
Mapping of the classes of A, B, C, and D of amino acid variability in mammalian PrP (Fig. 1) onto the refined NMR structure of the mouse PrP(121–231). The structure shown here and in the following figures is an energy-refined mean structure. The backbone is represented in white, and the single disulfide bridge Cys-179–Cys-214 is yellow. Side chains of the mouse PrP in the variable sites are blue for class A, magenta for B, green for C, and orange for D. This and all other color figures were prepared with the program molmol (15).
Figure 3.
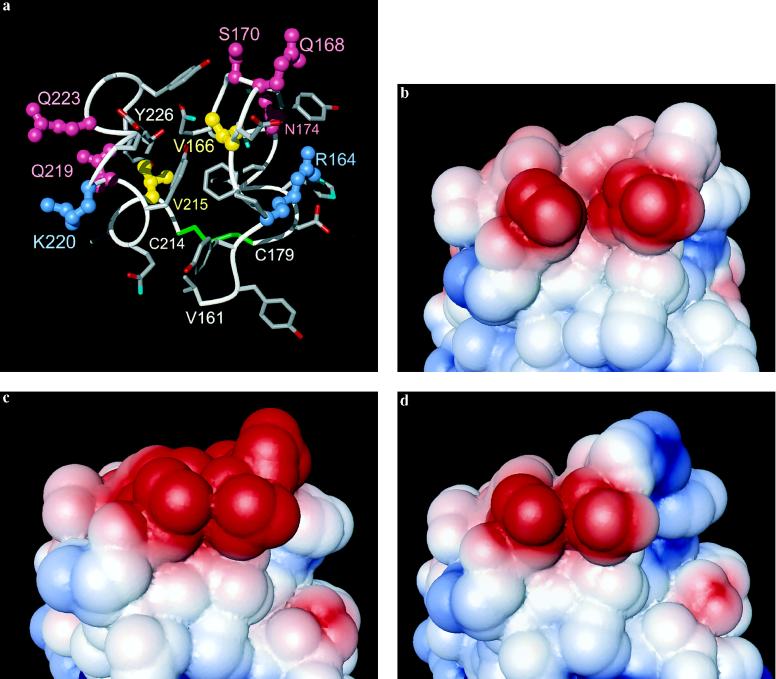
Molecular surface area of PrP(121–231) containing exchangeable residues of class A. The viewing angle is in the plane of Fig. 2 from the upper right corner. The polypeptide fragments 161–179 and 214–226 are shown. (a) The A-type residues in the mouse sequence are labeled and drawn as ball-and-stick models into the NMR structure, using yellow coloring for hydrophobic aliphatic residues, red for polar residues, and blue for positively charged residues. The backbone is white, the four chain ends are identified with white lettering, the cysteines 179 and 214 are green, and the other side chains are colored by atom type, with carbon gray, oxygen red, and nitrogen blue. (b–d) Solvent-accessible surface in three species containing different electrostatic charges. The surface is colored according to the electrostatic potential, with blue for positive charges and red for negative charges. The centrally located red area is common to PrPC from all three species and corresponds to the invariant residues Asp-167 and Glu-221 (see Fig. 1). (b) NMR structure of mouse PrPC, total charge of the molecular region shown −1 (same as a). This represents also bovine PrPC, which differs in this area only by the two conservative exchanges V215I and K220R (Fig. 1). (c) Model of human PrPC, total charge −3; with respect to b Val-166 has been changed to Met, Gln-168 to Glu, Val-215 to Ile, Gln-219 to Glu, and Lys-220 to Arg. (d) Model of sheep PrPC, total charge 0; with respect to b Gln-168 has been changed to Arg, Val-215 to Ile, and Lys-220 to Arg.
Figure 4.
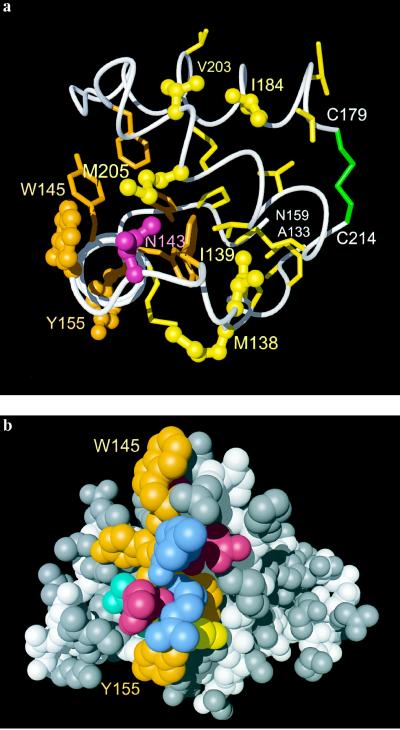
Location of the B- and C-class amino acid exchange sites (Fig. 1) in the NMR structure of mouse PrPC. (a) The view is along the axis of the first helix (on the left in Fig. 2) in the direction from Trp-145 in the front to Tyr-155 in the back. The display includes the residues 133–159 and 179–214, with helix 1 and its connections to the β-sheet, the loop connecting the helices 2 and 3, and these two helices up to the disulfide link 179–214. Variable residues are represented by ball-and-stick models and labeled, with yellow coloring for hydrophobic aliphatic side chains, red for polar residues, and orange for aromatic residues. The polypeptide backbone is white, the four chain ends are identified with white lettering, hydrophobic side chains are yellow, the aromatic side chains orange and the disulfide bridge green. (b) Space-filling model of the three-dimensional structure formed by residues 124–226. The viewing angle is in the plane of Fig. 2 from the left, so that the first helix of residues 145–155 runs from top to bottom in the foreground. The side chains of helix 1 are: orange, Tyr and Trp; red, Asp and Glu; blue, Arg; cyan, Asn-153; and yellow, Met-154. For the rest of the protein the polypeptide backbone is white and the side chains are gray.
Information on mammalian PrP sequences was collected from the protein databases swissprot (13) and the nucleic acid database genembl. The structure comparisons in the following section were based on the sequence alignment of Schätzl et al. (10), which is not a critical issue because the pairwise sequence identity is of the order of 90% for all combinations of mammalian species considered.
RESULTS
A group of 23 PrP sequences (Fig. 1) was identified in a database search for entries with over 60% identity and an overlap of at least 60 residues with the polypeptide fragment 124–226 of the mouse PrP. The following additional selection criteria were used to ensure that the focus was on statistically reliable data: (i) In the presence of the complete sequence, partial sequences of the same species were dropped. (ii) Where different sequences were reported for the same species, only the one with the smallest number of changes relative to the mouse sequence was retained. (iii) All sequences were omitted that differ from the sequence of the mouse protein only by conservative mutations in positions where all other species in the databases show identity with the mouse protein (see Fig. 1 legend). (iv) The sequence of the marsupial possum was not included because its evolutionary distance to all other mammals is outstandingly large (see Table II A, B. in ref. 14).
Figure 1.
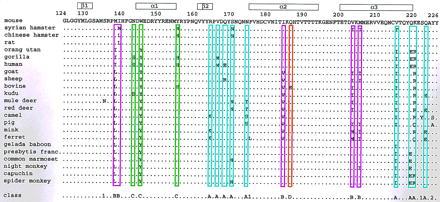
Sequence comparisons of the fragment 124–226 of mammalian PrP (see text for the selection criteria used), with the mouse sequence as a reference. At the top the locations of the regular secondary structures in mouse PrP(121–231) are indicated, and the sequence numbers refer to the residue under the third digit. The bottom line gives a classification of the observed variable sites into six classes A (blue), B (magenta), C (green), D (orange), 1, and 2 (see text). In addition to the 23 sequences listed here, swissprot and genembl provide three sequences (chimpanzee, gibbon, and siamang) that differ from gorilla only by the exchange N171S, two sequences that differ from that of Presbytis francoisi only by the exchanges N159S and I182V (squirrel monkey) and D144E (sooty mangabey), respectively; for all these sites no amino acid substitutions are observed in any other of the mammalian sequences. There are further two mouse sequences with the replacements M134V and T190V, respectively, relative to the mouse sequence listed here, two bovine sequences with the replacements G143S and G143S/M201K, respectively, a goat sequence with I139M, a pig sequence with N143S, and a squirrel monkey sequence with the exchange R164K.
Overall, as indicated in the bottom line of the figure, the 23 sequences in Fig. 1 contain 22 positions with amino acid replacements. Of these, 18 are grouped into the classes A, B, C, and D on the basis of their locations in the three-dimensional structure of PrP(121–231) (Fig. 2) and the chemical properties of the amino acid residues (Figs. 3 and 4). The three-dimensional fold of PrP(121–231) includes three helices with residues 144–154, 175–193, and 200–219, and an antiparallel β-sheet with residues 128–131 and 161–164 (Figs. 1 and 2). The helices 2 and 3 are linked by the disulfide bond Cys-179–Cys-214 (Fig. 2) and represent a stable scaffold for the global molecular structure (9). The class A sites form a dense cluster of nine residues in the loop between the second β-strand and the second helix (positions 164, 166, 168, 170, and 174), and near the C terminus (positions 215, 219, 220, and 223) (upper right in Fig. 2). Class B includes the five hydrophobic residues 138, 139, 184, 203, and 205, which are located in the interface between the first helix and the remainder of the protein. Class C sites include positions 145 and 155 at the two ends of helix 1, and the position 143 immediately preceding this helix. Class D includes only the exchange Q186E in the bovine protein, which introduces an isolated surface-exposed negative charge that might affect intermolecular interactions. The position 225 (“2”) near the C terminus and three positions with largely conservative single-species amino acid substitution (“1”) are not shown in Fig. 2 and are not further considered.
In class A the amino acid replacements in positions 168, 219, 220, and 223 (Fig. 3a) involve changes in the electrostatic charge of the side chains and may modify long-range forces effective in intermolecular recognition. Fig. 3 b-d shows the electrostatic surface potential for the mouse, human, and sheep proteins, where this molecular region has a total net charge of −1, −3, and 0, respectively. In total, seven of the nine sequence positions in class A contain polar or charged side chains in all species of Fig. 1. The class A residues thus form a surface region of PrPC that has the potential to function as a selective protein–protein interaction site. It is readily apparent from inspection of the NMR structure of mouse PrP(121–231) that many of the A-type amino acid exchanges will cause different surface hydrogen bonding patterns in the PrP from different species, and some of the amino acid replacements also might lead to different hydrogen bonding with a docked molecule. Overall, although the amino acid substitutions between different species can be expected to preserve the global shape of this putative binding site, they may modify its specificity for both long-range electrostatic interactions and short-range hydrogen bonding with other proteins.
Classes B and C contain residues from helix 1, part of helices 2 and 3, and helix-adjacent loops (Fig. 4a). The group C sites are related to a special feature of helix 1, which includes four largely or partially solvent-exposed aromatic side chains in the mouse sequence (Fig. 4b). With the sole exception of Met-154 all other side chains of this helix have polar character (Fig. 4b), and the positions 146–148, 151, and 152 actually contain charged side chains (Fig. 1). The combination of long-range electrostatic potentials, high hydrogen bonding propensity, and the presence of exposed aromatic residues on the solvent-accessible surface of helix 1 again represents structural features of a surface area that could well function as a specific recognition site for other proteins. Replacement of the C-type amino acid residues 145 and 155, in particular replacement of the unusual surface-exposed Trp-145, can be expected to modify the specificity of intermolecular interactions in this molecular region.
In all proteins of Fig. 1 the type B sites contain exclusively hydrophobic side chains of Val, Leu, Ile, or Met, which are in contact with other hydrophobic residues (Fig. 4a). Because of their limited surface accessibility, and considering the absence of electrostatic charges and hydrogen bonding propensity, the molecular region containing these sites is not reminiscent of a potential binding site that would mediate intermolecular interactions of the folded PrPC molecule. However, upon limited conformational rearrangement of PrPC, for example, by unfolding of the helix 1 (see ref. 9), several of these hydrophobic residues would become accessible and thus could support propagation of self-association of PrP. The observation in the spatially neighboring positions 184 and 203 (Fig. 4a) that replacement of Ile-184 by Val correlates in all but two species with the replacement of Val-203 by Ile or Met (Fig. 1), appears to further support the view that class B amino acid exchanges leave the PrPC surface properties largely unperturbed.
In summary, Fig. 2 shows that large regions of PrP(121–231) are not affected by species variations. These include the β-sheet, the central two turns of the first helix, the C-terminal two turns of the second helix, the loop linking the helices 2 and 3, and the third and fourth turn of the third helix. Part of this conservation can readily be explained by requirements to maintain local three-dimensional PrPC structure. However, some conservation also may be needed to secure further intramolecular or intermolecular interactions, for example, with the polypeptide segment 23–120 in intact PrPC, with the cell surface to which the PrP is attached, or with a natural ligand. The high frequency of amino acid exchanges between different species in the region of the three-dimensional structure that contains the class A residues makes this region a likely candidate for a specific binding site, where amino acid replacements would affect intermolecular recognition with PrPC (Fig. 3). The helix 1 with the C-type amino acid substitutions forms another potential binding site (Fig. 4b) on the opposite molecular surface (Fig. 2), where replacement of solvent-exposed aromatic side chains between different species would be expected to modify the specificity by which this region is recognized by other proteins.
DISCUSSION
Inoculation experiments with chimeric PrP suggested two protein binding sites in the PrP amino acid sequence. These experiments so far have been the only information available for mapping of species-related differences in PrPC. One of these binding sites would be located in the segment with residues 96–167, which would bind an infecting PrPSc particle. The second site would be composed of residues outside of this segment, which could possibly interact with the postulated protein X (4, 12). The variable C-type residues in helix 1 hence would contribute to a protein surface area that might be involved in interactions with PrPSc, and the cluster of type A residues would be a potential binding site for protein X. The in vitro PrPC conversion experiments of Kocisko et al. (8) suggested that the hamster/mouse species barrier is dependent on the three amino acid exchanges M139I, N155Y, and N170S (Fig. 1). It is striking that the segment 96–167 as well as the group of residues 139/155/170 includes at least one residue from each of the classes A, B, and C (Fig. 1). Other experimental data attribute an exclusive role for the species barrier between mouse and hamster to position 139 (16): The substitution of Met in hamster PrP by Ile in mouse PrP appears to be sufficient to protect hamster from infection by mouse PrPSc. This would place the critical structural feature for the species barrier in the interface between helix 1 and the protein core (Figs. 2 and 4a). The residue 139 presumably would become operational only after initial conformational transitions of PrPC induced by complexation with PrPSc and possibly protein X (9). The present structural observations can be expected to support future refinement of in vivo and in vitro experiments with chimeric PrP.
The class D exchange Q186 E in bovine PrP introduces a negative electrostatic charge in a surface location that is clearly separated in space from the two presumed binding sites characterized by A- and C-type residues, respectively (Fig. 2). Interestingly, recent results indicate the presence of Glu-186 also in the related species Watussi, Banteng, and Wisent (H. Schätzl, personal communication). The newly determined PrP sequences of the house dog, the Canadian wolf, and the dingo (H. Schätzl, personal communication) further extend the available information on D-type mutation sites, because the otherwise strictly conserved Asn-159 (Fig. 1) is replaced by Asp, and this negative charge is located in close spatial proximity to position 186 (Fig. 2). This leaves the intriguing option that a third variable surface area in PrPC might emerge by further extension of the sequence database, and that this site might be functional in the species barrier with cattle, including possibly also transmission of disease from bovine spongiform encephalopathy-infected cows to other species.
Finally, although the NMR structure of mouse PrP(121–231) represents only part of PrPC and the ensemble of all accumulated data indicates that N-terminal parts of the sequence also may influence the species barrier (for example, see refs. 4 and 5), there is evidence that the data presented in this paper are highly relevant with regard to the intact system, and that PrP(121–231) has a key part in PrPC physiology (8, 12, 16). Structure predictions and experimental observations (9, 17) indicate that PrP(121–231) is probably the only polypeptide segment with a global fold in intact PrPC, and initial NMR studies with intact PrPC performed in our laboratory indicate that the segment 121–231 in the intact protein has the same fold as in PrP(121–231) (unpublished data).
Acknowledgments
We thank Dr. H. M. Schätzl for the communication of unpublished data, Prof. C. Weissmann for helpful discussions, and R. Hug for the careful processing of the manuscript. Financial support was obtained from the Schweizerischer Nationalfonds (Grant 31.32035.91) and a fellowship of the Boehringer-Ingelheim-Fonds to S.H.
ABBREVIATIONS
- PrP
prion protein
- PrP(121–231)
recombinant C-terminal domain of the mouse Prp with residues 121–231
- PrPC
cellular form of PrP
- PrPSc
scrapie form of PrP
Footnotes
Data deposition: The atomic coordinates of mouse PrP(121–231) have been deposited in the Protein Data Bank, Brookhaven National Laboratory, Upton, NY 11973 (reference 1AG2).
Within the framework of the “protein-only” hypothesis, various different mechanisms are possible by which amino acid exchanges in the PrP sequence could cause a barrier for transmission between difference species. These include, for example, the presently discussed situation that the amino acid exchange affects a binding site in PrPC for PrPSc or protein X, that it belongs to a region in PrPSc, which interacts with PrPC, or that it is involved in subunit–subunit contacts in oligomeric PrPSc. The discussions in this paper are focused entirely on the influence of amino acid substitutions on structure and intermolecular interactions of PrPC.
References
- 1.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 3.Griffith J S. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S B. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 5.Weissmann C. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 6.Collinge J, Sidle K C L, Meads J, Ironside J, Hill A F. Nature (London) 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 7.Roberts G W, James S. Curr Biol. 1996;6:1247–1249. doi: 10.1016/s0960-9822(02)70708-2. [DOI] [PubMed] [Google Scholar]
- 8.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 10.Schätzl H M, Da Costa M, Taylor L, Cohen F E, Prusiner S B. J Mol Biol. 1995;245:362–374. doi: 10.1006/jmbi.1994.0030. [DOI] [PubMed] [Google Scholar]
- 11.Flores T P, Orengo C A, Moss D S, Thornton J M. Protein Sci. 1993;2:1811–1826. doi: 10.1002/pro.5560021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, De Armond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 13.Bairoch A, Apweiler R. Nucleic Acids Res. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czihak G, Langer H, Ziegler H, editors. Biologie. Berlin: Springer; 1981. [Google Scholar]
- 15.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 16.Priola S, Chesebro B. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornemann S, Glockshuber R. J Mol Biol. 1996;261:614–619. doi: 10.1006/jmbi.1996.0487. [DOI] [PubMed] [Google Scholar]