Abstract
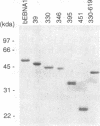
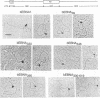
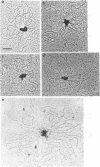
Epstein-Barr nuclear antigen 1 (EBNA1) activates DNA replication from the Epstein-Barr virus latent origin, oriP. This activation involves the direct interaction of EBNA1 dimers with multiple sites within the two noncontiguous functional elements of the origin, the family of repeats (FR) element and the dyad symmetry (DS) element. The efficient interaction of EBNA1 dimers bound to these two elements in oriP results in the formation of DNA loops in which the FR and DS elements are bound together through EBNA1. In order to elucidate the mechanism by which EBNA1 induces oriP DNA looping, we have investigated the DNA sequences and EBNA1 amino acids required for EBNA1-mediated DNA looping. Using a series of truncation mutants of EBNA1 produced in baculovirus and purified to apparent homogeneity, we have demonstrated that the EBNA1 DNA binding and dimerization domain is not sufficient to mediate oriP DNA looping and that an additional region(s) located between amino acids 346 and 450 is required. Single EBNA1-binding sites, separated by 930 bp of plasmid DNA, were also shown to support EBNA1-mediated looping, indicating that the formation of large EBNA1 complexes, such as those observed on oriP FR and DS elements, is not a requirement for looping.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987 May;61(5):1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambinder R. F., Mullen M. A., Chang Y. N., Hayward G. S., Hayward S. D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991 Mar;65(3):1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Mason R. J., Wickner S. H. Mini-P1 plasmid replication: the autoregulation-sequestration paradox. Cell. 1988 Feb 26;52(4):551–557. doi: 10.1016/0092-8674(88)90468-0. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J Virol. 1992 Mar;66(3):1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991 Apr 25;266(12):7819–7826. [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Dailey L., Held P., Heintz N. Eukaryotic replication origins as promoters of bidirectional DNA synthesis. Trends Genet. 1992 Nov;8(11):376–381. doi: 10.1016/0168-9525(92)90298-i. [DOI] [PubMed] [Google Scholar]
- Koff A., Schwedes J. F., Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J Virol. 1991 Jun;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Haase S. B., Calos M. P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989 Mar;9(3):1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. S. DNA looping. Microbiol Rev. 1992 Mar;56(1):123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton T., Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992 Jan;66(1):489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G., Hwang E. S. Epstein-Barr virus nuclear antigen forms a complex that binds with high concentration dependence to a single DNA-binding site. J Virol. 1987 Feb;61(2):465–471. doi: 10.1128/jvi.61.2.465-471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A., Mukherjee S., Bastia D. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J. 1992 Mar;11(3):1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6287–6291. doi: 10.1073/pnas.85.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988 Feb 12;52(3):375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Polvino-Bodnar M., Kiso J., Schaffer P. A. Mutational analysis of Epstein-Barr virus nuclear antigen 1 (EBNA 1). Nucleic Acids Res. 1988 Apr 25;16(8):3415–3435. doi: 10.1093/nar/16.8.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvino-Bodnar M., Schaffer P. A. DNA binding activity is required for EBNA 1-dependent transcriptional activation and DNA replication. Virology. 1992 Apr;187(2):591–603. doi: 10.1016/0042-6822(92)90461-w. [DOI] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah W. A., Ambinder R. F., Hayward G. S., Hayward S. D. Binding of EBNA-1 to DNA creates a protease-resistant domain that encompasses the DNA recognition and dimerization functions. J Virol. 1992 Jun;66(6):3355–3362. doi: 10.1128/jvi.66.6.3355-3362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Middleton T., Sugden B., Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989 Jun;63(6):2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher R., Griffith J. Electron microscopic visualization of DNA and DNA-protein complexes as adjunct to biochemical studies. Methods Enzymol. 1992;211:481–490. doi: 10.1016/0076-6879(92)11026-f. [DOI] [PubMed] [Google Scholar]
- Wysokenski D. A., Yates J. L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989 Jun;63(6):2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991 Jan;65(1):483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]