Abstract
Gene expression profiling of diffuse large B-cell lymphoma (DLBCL) has revealed biologically and prognostically distinct subgroups: germinal center B-cell-like (GCB), activated B-cell-like (ABC) and primary mediastinal (PM) DLBCL. The BCL6 gene is often translocated and/or mutated in DLBCL. Therefore, we examined the BCL6 molecular alterations in these DLBCL subgroups, and their impact on BCL6 expression and BCL6 target gene repression. BCL6 translocations at the major breakpoint region (MBR) were detected in 25 (18.8%) of 133 DLBCL cases, with a higher frequency in the PM (33%) and ABC (24%) subgroups than in the GCB (10%) subgroup. Translocations at the alternative breakpoint region (ABR) were detected in five (6.4%) of 78 DLBCL cases, with three cases in ABC and one case each in the GCB and the unclassifiable subgroups. The translocated cases involved IgH and non-IgH partners in about equal frequency and were not associated with different levels of BCL6 mRNA and protein expression. BCL6 mutations were detected in 61% of DLBCL cases, with a significantly higher frequency in the GCB and PM subgroups (> 70%) than in the ABC subgroup (44%). Exon-1 mutations were mostly observed in the GCB subgroup. The repression of known BCL6 target genes correlated with the level of BCL6 mRNA and protein expression in GCB and ABC subgroups but not with BCL6 translocation and intronic mutations. No clear inverse correlation between BCL6 expression and p53 expression was observed. Patients with higher BCL6 mRNA or protein expression had a significantly better overall survival. The biological role of BCL6 in translocated cases where repression of known target genes is not demonstrated is intriguing and warrants further investigation.
Keywords: BCL6, mutation, translocation, protein expression, gene expression profiling, diffuse large B-cell lymphoma
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin's lymphoma (NHL), comprising approximately 30–40% of newly diagnosed NHL cases in the United States.1 Patients with DLBCL exhibit significant diversity in their clinical presentation, cytomorphology, immunophenotype and genotype.2,3 Gene expression profiling (GEP) of DLBCL has revealed biologically and prognostically distinct subgroups: germinal center B-cell-like (GCB), activated B-cell-like (ABC) and primary mediastinal (PM) DLBCL.4–7 The GCB subgroup of DLBCL overexpresses gene characteristics of normal GC B cells, whereas the ABC subgroup overexpresses a group of genes that are highly transcribed by in vitro-activated peripheral blood B cells. The PM subgroup is characterized by the deregulated expression of genes involved in B-cell receptor signaling and cytokine pathways. There are also a small number of cases that cannot be assigned to any of these three subgroups, that is unclassifiable DLBCL. The overall survival (OS) of patients with GCB- and PM-DLBCL is significantly better than that of patients with ABC-DLBCL. These molecularly defined subgroups have also been shown to carry distinct profiles of cytogenetic abnormalities.2,6,8
The human protooncogene BCL6 was identified from chromosomal breaks at 3q279,10 and encodes a 92–98 kDa nuclear zinc finger phosphoprotein. BCL6 is expressed predominantly in GC B cells11 and functions as a sequence-specific transcriptional repressor.12 BCL6 is essential for GC formation,13 and the BCL6 transgene promotes the development of B-cell lymphomas in the mice, indicating that deregulation of BCL6 can lead to B-cell lymphoma.14
Translocation and somatic hypermutation are the two major types of molecular alterations involving the BCL6 gene in DLBCL. Chromosomal translocations lead to the substitution of the BCL6 promoter by heterologous sequences derived from the different translocation partners. Clinically, BCL6 translocation occurs primarily in de novo DLBCL.9 There is no consensus on the effect of BCL6 translocation on prognosis, with studies showing either favorable15,16 or unfavorable outcomes,17 or no effect.18 As in the IgVH genes, the BCL6 gene undergoes somatic hypermutations in the first intron, and at a lower frequency in the first non-coding exon,19,20 and BCL6 expression can be deregulated as a consequence of some somatic mutations.21 However, the accumulation of BCL6 mutations has not been shown to have any association with disease progression.22 GCB-DLBCL, with generally high expression of BCL6 has a better OS than the ABC subgroup.4,5,23 However, the influence of BCL6 translocations and mutations on mRNA and protein expression, and their functional consequences have not been studied in the context of DLBCL subgroups. Therefore, we analyzed a series of DLBCL to determine: (1) the frequency of BCL6 mutations and translocations among the subgroups of DLBCL as identified by GEP; (2) the correlation of BCL6 alterations with BCL6 mRNA and protein expression, and the repression of BCL6 target genes; and (3) the predictive value of BCL6 gene alterations, and mRNA and protein expression, on survival in the different subgroups of DLBCL.
Materials and methods
Patient population
We studied previously 240 cases of DLBCL profiled with gene expression by complementary DNA (cDNA) microarray technology.4 A panel of hematopathologists confirmed the diagnosis of DLBCL in all patients. The cases were classified into different molecularly defined subgroups using the Bayesian method.24 Of these 240 cases, 92 cases were classified as GCB-DLBCL, 82 cases ABC-DLBCL, 20 cases PM-DLBCL and 46 cases unclassifiable DLBCL. All patients included in this series had received an anthracycline-containing chemotherapy regimen. The Institutional Review Board of the University of Nebraska Medical center approved this study.
Immunohistochemical staining of tissue sections
Immunohistochemical (IHC) staining for CD20 and BCL6 was performed on 138 cases with available archival paraffin blocks as described previously.25 A prior comparative study of IHC staining in three different laboratories validated the reproducibility of staining and evaluation.26 Briefly, tissue microarrays (TMAs) were prepared from cases with adequate archival paraffin-embedded tissue. Sections of 5 µm were cut from each TMA and stained with an antibody to BCL6 (Polyclonal, Santa Cruz Biotechnology, Santa Cruz, CA, USA), as described previously.25 CD20 stains were performed to evaluate each core for involvement by tumor. The percentage of tumor cells stained with each antibody was recorded in 10% increments and for each case, the core with the highest percentage of tumor cells stained was used for analysis. Cases were considered positive if 30% or more of the tumor cells stained for BCL6.25
Detection of the 3q27 translocation by fluorescence in situ hybridization
Interphase fluorescence in situ hybridization (FISH) analysis for chromosome 3q27 (BCL6) translocations was performed on 133 cases using formalin-fixed, paraffin-embedded tissue sections, as described previously with minor modification.8 Briefly, a BCL6 break-apart probe (BAP; Abbott-Vysis, Downers Grove, IL, USA) was used to detect BCL6 translocations at major breakpoint region (MBR) and a BCL6 home-brew BAP (Clone RP11-1144D2 and RP11-67E18) at the alternative breakpoint region (ABR) (78/133 cases).27,28 Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) in Antifade solution and the slides were visualized using an Olympus BX51 fluorescence microscope. Images were captured and archived using Cyto Vision software (Applied Imaging, Santa Clara, CA, USA). To analyze the hybridization, a total of 50–100 nuclei per case were scored for the presence of the BCL6 translocation. The normal cutoff for this FISH assay in paraffin tissue sections has been established to be 20% by prior studies. The gain/amplification data on the 3q27 region of these 133 cases was obtained from a previous study,2 using comparative genomic hybridization (CGH) and compared with BCL6 mRNA and protein expression. The FISH data for t(14;18) were obtained from our previous study.8 To examine the correlation between BCL6 translocation and p53 genomic abnormalities, 66 cases of the cohort with p53 gene mutation as well as deletion data were obtained from a previous study for analysis.29
Mutational analysis of BCL6 intron-1 and exon-1
Genomic DNA extracted from 128 frozen tissue samples used for GEP was available for this analysis. The major mutation cluster (MMC) in BCL6 intron-1 (~ 700 bp) was divided into three overlapping fragments for PCR amplification (accession no.: AC072022.19; 37735–38012 (F1); 37975–38220(F2); 38200–38391(F3)) using the following PCR primer sets (F1′/F1″: 5′-(TTTTCCGCTCTTGCCAAATGCTTT/5′-GGAGGGGAATTAGGGG; F2′/F2″: 5′-GGTTTTTGGAAAGGAGGT/5′-CTGCTTTCCTTGCTCCGTC and F3′/F3″: 5′-TGCGGCTGTGTTTTTT/5′-ATTCCCTGCCTTCGAGCCG, respectively), see Figure 3 also. Mutational analysis of exon-1 (E1; 37375–37636) was performed using a previously described primer set (E1/E1″: 5′-ACGCTCTGCTTATGAGGA/5′-CGGCAGCAACAGCAATAA, respectively).21 The PCR mixtures contained 500 ng of genomic DNA, 200 µm dNTP, 1.5 mm MgCl2, 50 µm primer and 1.5 U Amplitaq polymerase in a reaction volume of 100 µl. Forty-five cycles of PCR were performed after an initial denaturation (94°C, 9 min) followed by denaturation at 94°C for 75 s., annealing at 60–68°C for 75 s (60°C for E1, 68°C for F1, 60°C for F2 and 64°C for F3), extension at 72°C for 40 s, and final extension at 72°C for 7 min. Mutants were then identified using denaturing high-performance liquid chromatography in a WAVE DNA Fragment Analysis System (Transgenomic, Omaha, NE, USA).30 Mutant fractions were collected on the Wave system on DNA samples that exhibited shifted peaks compared with wild-type DNA. These fractions were reamplified and then sequenced. The detailed analysis of the mutations was performed with the Vector NTI software program (Invitrogen Inc., CA, USA), and compared with germ line BCL6.
Figure 3.
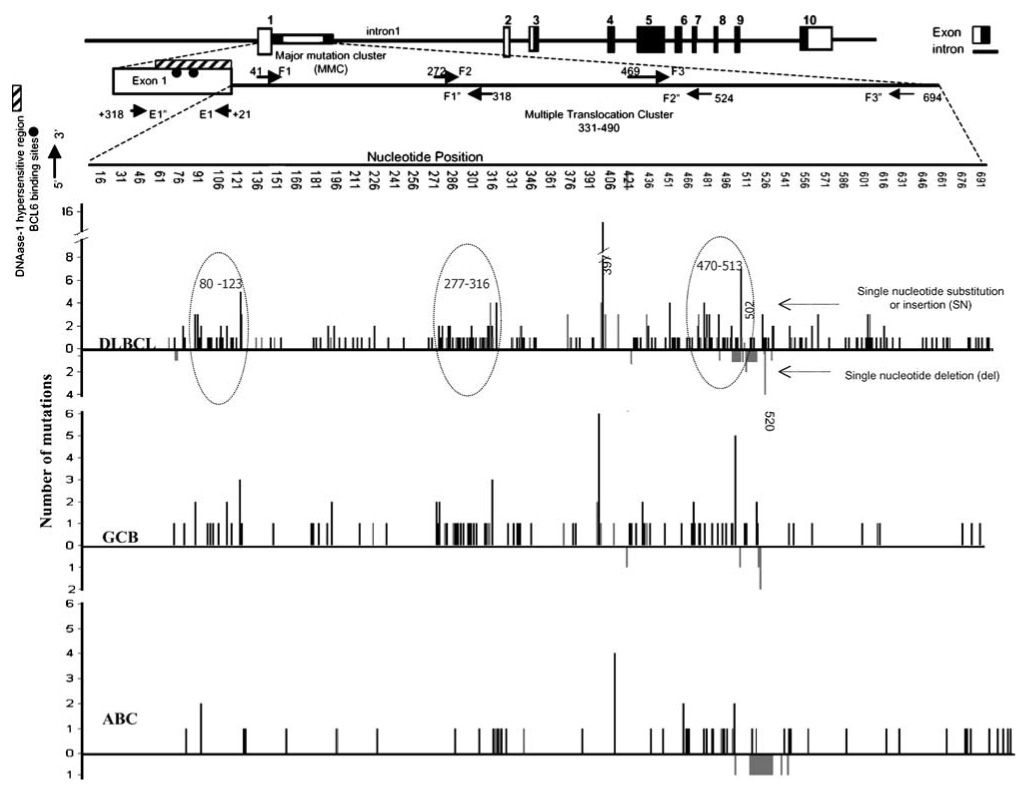
Spectrum of BCL6 mutations along the major mutation cluster (MMC) in intron 1. BCL6 mutations were clustered within the MMC, in which three regions were observed to have an increased frequency of mutations. This phenomenon was observed in the GCB subgroup (B), but not readily discernable in the ABC subgroup, except for the third cluster (470–513). Insertions and substitutions are indicated by lines above the baseline, whereas deletions are represented below the baseline. Grey lines indicate single-base deletion or insertion and black lines indicate a single-base substitution. GCB, germinal center B-cell-like.
Analysis of BCL6 and target gene expression
We evaluated BCL6 mRNA levels measured by the Lymphochip microarray in all DLBCL cases from our previous study.4 Expression levels were log-transformed and mean-centered across samples. The value was expressed as the number of fold changes over the reference pool mRNA.5 The median of hybridization values from four BCL6 cDNA clones, immobilized on the Lymphochip, was used as BCL6 mRNA expression values. We also used BCL6 transcript level, BCL6 protein expression, and BCL6 translocation and mutation data to supervise the analysis of the expression of known BCL6 targets31–34 (see Supplementary Reference list for known target genes) present on the Lymphochip for each subgroup of DLBCL. When the GCB cases were divided into quartiles according to their BCL6 mRNA expression levels, we observed repression of most of these targets and there was a general trend toward increased suppression with higher transcript levels. We selected a subgroup of these genes that showed a consistent pattern of repression and considered this subgroup as the most reliable BCL6 targets in the GCB environment. This set of target genes was then used to assess BCL6 activity in other subgroups. For target genes represented by three or more cDNA clones, the median hybridization values were used for analysis. If two clones were present, the mean value was used. The three clones of nuclear factor-κB1 (NF-κB1) on the Lymphochip showed inconsistent expression levels and, therefore we examined the cases that were also profiled with Affymetrix chips. The clone on the Lymphochip (UniqID: 31267), which showed maximum correlation (r=0.75) with expression levels measured with Affymetrix chips was selected for analysis.
Statistical analysis
The Kaplan–Meier method was used to estimate OS of the patients, and the log-rank test was used to compare the survival distributions between subgroups with and without BCL6 mutation/translocation. OS was defined as the time from diagnosis to death resulting from any cause or, for patients remaining alive, the time from diagnosis to last contact. Event-free survival was defined as the time from diagnosis to the first occurrence of relapse or death from any cause or, for patients remaining alive and relapse-free, the time from diagnosis to last contact. Fisher's exact test was used to analyze categorical data and the Wilcoxon's rank-sum test was used to analyze continuous data between groups. P-values for the Kaplan–Meier curves were not adjusted for subgroup analysis. For other comparisons, if the overall P-value comparing subgroups and BCL6 variable was significant, then pair-wise comparisons between the subgroups were performed. These post hoc tests were adjusted for multiple comparisons using the Bonferroni method. SAS software was used for the data analysis (SAS Institute Inc., Cary, NC, USA).
Results
Case characteristics
The 240 DLBCL cases profiled with the Lymphochip4 were divided into GCB (92 cases; 38.7%), ABC (82 cases; 34.2%), PM (20 cases; 7.9%) and unclassifiable (46 cases; 19.2%) subgroups. BCL6 mRNA expression data were available in all of the above cases. Of the 240 cases, BCL6 IHC data were available in 138 cases (57 GCB, 44 ABC, 10 PM and 27 unclassifiable). In the same series of patients, FISH data on BCL6 MBR translocation and gain/amplification were obtained in 133 cases (51 GCB, 46 ABC, 12 PM and 24 unclassifiable) and ABR translocation on 78 cases. BCL6 mutational data was obtained in 128 cases (50 GCB, 43 ABC, 12 PM and 23 unclassifiable). All datasets were available in 106 cases. Clinical data were available on 236 cases (91 GCB, 82 ABC, 19 PM and 44 unclassifiable). No significant differences were observed for any of the clinical parameters in the subgroups with IHC data or FISH data when compared with the entire group of DLBCL.
Prevalence of BCL6 translocations in DLBCL subgroups
The BCL6 gene translocation at the MBR was analyzed in 133 cases by FISH and observed in 18.8% (25/133) of DLBCL cases. The incidence of translocation was higher in the ABC (24%) and PM (33%) subgroups, as compared with the GCB subgroup (10%) (Table 1). We also compared the t(14;18) data obtained previously on these cases8 and observed that only two of 23 BCL6 translocated cases were also positive for the t(14;18), one a GCB- and the other a PM-DLBCL. There does not seem to be any association between these two translocations (Supplementary Table ST1).
Table 1.
BCL6 translocation at MBR in the DLBCL subgroups
| Subgroups (n) | Positive | Negative |
|---|---|---|
| ABC (46) | 11 (24%) | 35 (76%) |
| GCB (51) | 5 (10%) | 46 (90%) |
| PM (12) | 4 (33%) | 8 (67%) |
| UNCL (24) | 5 (21%) | 19 (79%) |
| Total (133) | 25 (19%) | 108 (81%) |
Abbreviations: ABC, activated B-cell-like; GCB, germinal center B-cell-like; MBR, major breakpoint region; PM, primary mediastinal; UNCL, unclassifiable.
We also evaluated BCL6 rearrangement at ABR in 78 informative DLBCL cases. Of these, five cases (6.4%) were positive using a BAP for this region. Of the five positive cases, three cases were in the ABC and one case each in the GCB and unclassifiable subgroup. As we expected that BCL6 translocation at MBR would be associated with deregulated BCL6 expression, we compared BCL6 mRNA and protein expression in cases with and without BCL6 translocation. When all the DLBCL cases were analyzed as a group, we observed no significant difference in either BCL6 mRNA expression (P=0.58) or protein (P=0.56) in cases with or without translocation. When the subgroups were analyzed separately, the only significant difference was observed in the ABC subgroup (Figure 1), where BCL6 mRNA expression was increased by 1.5-fold in the translocated cases (P=0.045). Similar results were obtained when the cases with translocation at the ABR were added to the analysis. Actually, cases (n=5) with translocation at the ABR were associated with low BCL6 protein (P<0.025, χ² test) and mRNA expression (threefold difference, P=0.05) compared with cases without translocation.
Figure 1.
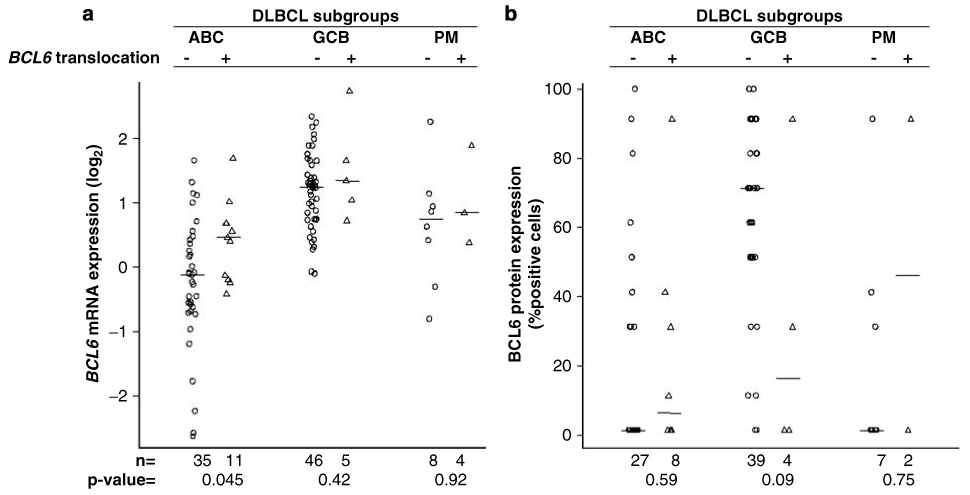
(a) Correlation of BCL6 (MBR) translocation with BCL6 transcript level. The activated B-cell-like (ABC) subgroup had a significant increase in mRNA expression in the translocated cases as compared with non-translocated cases (P=0.045), but no increase was observed in the other subgroups. (b) Correlation of BCL6 translocation with BCL6 protein expression. There is a higher percentage of cases with detectable protein expression in the translocated cases in the ABC subgroup but it does not reach statistical significance. The other subgroups contain too few translocated cases for evaluation.
Of the 133 cases studied for BCL6 translocation, 66 cases were analyzed for p53 mutation and deletion29 (Supplementary Table ST2). There was no significant difference (P=0.80 by the χ² test) in the frequency of p53 abnormalities in cases with and without BCL6 translocation. Similar findings were obtained when cases with translocation at ABR were added (P=0.2 by χ² test).
Among the 25 BCL6 translocated cases, data on IgH BAP were available in 16 cases, of which six were negative and therefore had a non-IgH translocation partner. Of the remaining 10 cases positive for IgH BAP, FISH using IgH/BCL6 fusion probes (Cancer Genetics Inc., River Vale, NJ, USA) were performed and seven cases had IgH/BCL6 translocation, two cases had variant BCL6 translocation and one was inconclusive. Of the seven cases with IgH/BCL6 translocation, four cases were in the ABC, two in the GCB and one in the unclassifiable subgroups and all the cases with variant translocations were in the ABC subgroup. Therefore, the incidence of IgH/BCL6 vs non-IgH/BCL6 translocation in DLBCL was almost same the (8:7). No differences in BCL6 mRNA or protein expression were observed between cases with or without t(3;14).
BCL6 gene mutations in DLBCL subgroups and correlation with mRNA and protein expression
We defined the mutational spectrum in our cases, including the frequently mutated 5′ region in intron-1 (MMC),22 and a newly defined region containing the negative regulatory elements for BCL6 in exon-1.21 Of the 128 analyzed cases, intron-1 mutations were detected in 78 cases (61%), with higher frequencies in the GCB (74%) and PM (75%) subgroups than in the ABC subgroup (44%) (Table 2). There were 302 different mutations consisting mostly of single-base substitutions (n=251), followed by single-base insertions (n=38) and single-base deletions (n=13). The number of mutations per case ranged from 1 to 24 with an average of 3.9 mutations per case (Table 2). G was the most frequently mutated nucleotide, and transversions were more frequent than transitions (140 vs 110) (see Supplementary Figure 1). The average number of mutations was highest in PM-DLBCL subgroup (Table 2). Cases of DLBCL with mutations had higher BCL6 mRNA expression than cases with wild-type BCL6 (P=0.0013); but this was not statistically significant in individual DLBCL subgroups (Figure 2). The GCB subgroup had high BCL6 protein and there was a trend toward higher BCL6 protein level in mutated cases (Figure 2).
Table 2.
BCL6 mutations in the DLBCL subgroups
| Subgroups (n) | Mutant cases | Mean mutations/case (range) | Mutational incidence in MMC |
|---|---|---|---|
| ABC (43) | 19 (44%) | 3.6 (1–8) | 5.4 × 10−3/bp |
| GCB (50) | 37 (74%) | 3.4 (1–9) | 4.8 × 10−3/bp |
| PM (12) | 9 (75%) | 6.2 (1–24) | 8.9 × 10−3/bp |
| UNCL (23) | 12 (50%) | 4.3 (1–12) | 6.4 × 10−3/bp |
| Total (128) | 77 (60%) | 3.9 (1–24) | 5.5 × 10−3/bp |
Abbreviations: ABC, activated B-cell-like; GCB, germinal center B-cell-like; MMC, major mutation cluster; PM, primary mediastinal; UNCL, unclassifiable.
The polymorphisms at positions 397(G-C), 502(A-T) and 520(Del T) have not been included.
n: total number of cases evaluated using the WAVE system.
Figure 2.
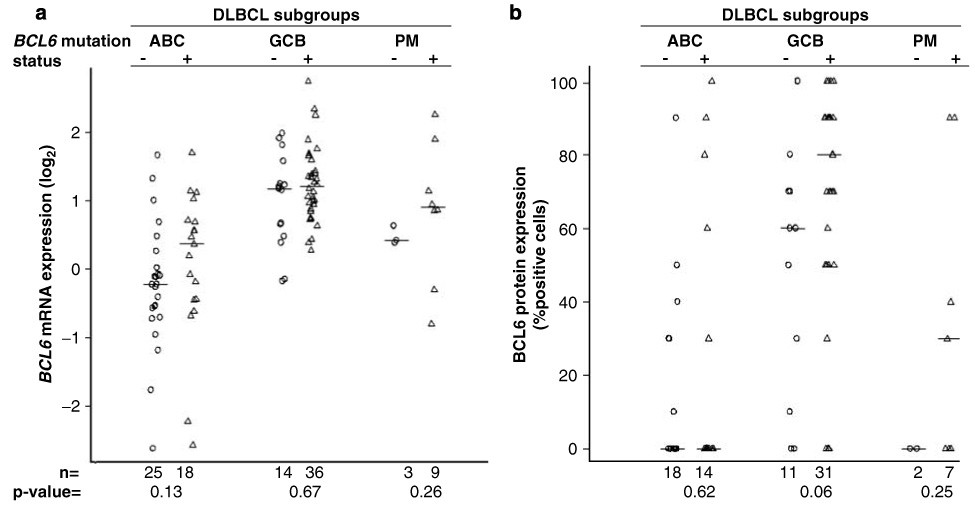
Correlation of BCL6 mutation status with (a) BCL6 transcript level, and (b) BCL6 protein expression. No significant association of BCL6 mRNA or protein expression was observed with mutation status in any of the DLBCL subgroups. DLBCL, diffuse large B-cell lymphoma.
Previous studies have reported clustering of mutations in the MMC35,36 and postulated the presence of two37 or three38 major clusters. We also observed a similar clustering pattern and defined three MMCs that had a mutational frequency ≥3-fold higher than the entire MMC region (1–1.5 × 10−2/bp vs 5.4 × 10−3/bp). These clusters were located at positions 80–123, 277–316 and 470–513 in the MMC region (Figure 3). The first mutation cluster (80–123) has been reported by others.37,38 In subgroup analysis, the presence of two major clusters was observed in the GCB subgroup (277–316 and 470–513), but clustering was not discernable in the other subgroups probably because of the lower number of cases with mutations. In agreement with previous studies, three single-nucleotide polymorphisms (SNPs) in the MMC were observed at positions 397 (C → G; 20%), 502 (A → G; 10%) and 520 (del T; 10%). The SNPs at 397 and 520 were observed with similar frequencies in the GCB (6 of 14) and ABC (4 of 14) subgroups; however, the SNP at 502 was observed more frequently in the GCB subgroup (6 of 8) than in the other subgroups (PM and ABC with one case each). Exon-1 mutations were detected in 10% (13 of 128) of the cases and the majority of these cases were in the GCB subgroup (9 of 13). These mutations were generally clustered in the DNAse-I hypersensitive region, particularly within the BCL6, STAT1 and predicted CEBP binding sites. In general, mutations in the BCL6-binding sites were associated with increased BCL6 mRNA and protein expression (see Supplementary Figure 2).
Relationship between BCL6 translocation and 3q27gains with BCL6 mRNA and protein expression
Using a 30% cutoff for positive cellular staining, the expression of BCL6 protein was detected in 78 of 138 DLBCL cases (55%) with the highest expression in the GCB subgroup (Table 3). In the ABC subgroup, there was a statistically significant association between BCL6 mRNA and protein expression (P=0.0025), but not in other subgroups (Figure 4). BCL6 mRNA and protein expression was not significantly associated with BCL6 translocation status at the MBR, but significantly lower expression was observed in cases with ABR (mRNA: P=0.05 and protein P=0.025).
Table 3.
BCL6 protein expression in the DLBCL subgroups
| Subgroups (n) | Positive | Negative |
|---|---|---|
| ABC (44) | 15 (34%) | 29 (66%) |
| GCB (57) | 49 (86%) | 8 (14%) |
| PM (10) | 4 (40%) | 6 (60%) |
| UNCL (27) | 10 (37%) | 17 (63%) |
| Total (138) | 78 (56%) | 60 (44%) |
Abbreviations: ABC, activated B-cell-like; GCB, germinal center B-celllike; MMC, major mutation cluster; PM, primary mediastinal; UNCL, unclassifiable.
Figure 4.
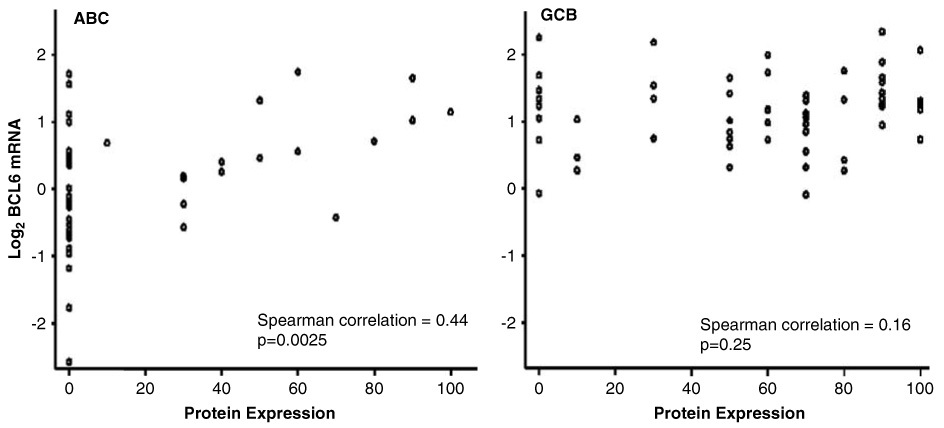
Correlation between BCL6 mRNA and protein expression. The ABC subgroup showed a significant association of BCL6 mRNA with protein expression, but not the GCB or the PM subgroup- (data not shown). *The immunoperoxidase assay indicated the percentage positive cells for BCL6. ABC, activated B-cell-like; GCB, germinal center B-cell-like.
We also correlated BCL6 protein and mRNA expression with 3q27 gains obtained from CGH performed separately.2 Of the 123 DLBCL cases evaluated, 3q27 gain was observed in 22 cases (17.8 %) cases and the majority of 3q27 gains were observed in the ABC subgroup (17/42; 40% vs 3/45; 6% in GCB; P<0.001); however, these gains were not associated with increased BCL6 mRNA or protein expression.
Association of BCL6 gene abnormalities and BCL6 target gene repression
We used BCL6 mRNA and protein expression, and mutation and translocation data, to perform supervised analyses of the expression of BCL6 target genes31,39,40 in the different subgroups of DLBCL. Target gene selection was described in the Materials and methods section and repression of the majority of the target genes was observed in all three subsets when the level of BCL6 expression was sufficiently high at both protein or mRNA level. For example, repression of the target genes was observed in the GCB subgroup from the second to fourth quartile of BCL6 transcript levels, but in the ABC and PM subgroup significant repression was observed only in the fourth quartile (Figure 5a). However, the pattern of repression differed among the subsets and some of the target genes, for example, CD80, STAT3, NFκB1 and BCL-xl, were not consistently repressed in the ABC or PM subgroups. P53 expression levels did not show any significant inverse correlation with BCL6 expression in any subgroup (Figures 5a and b).
Figure 5.
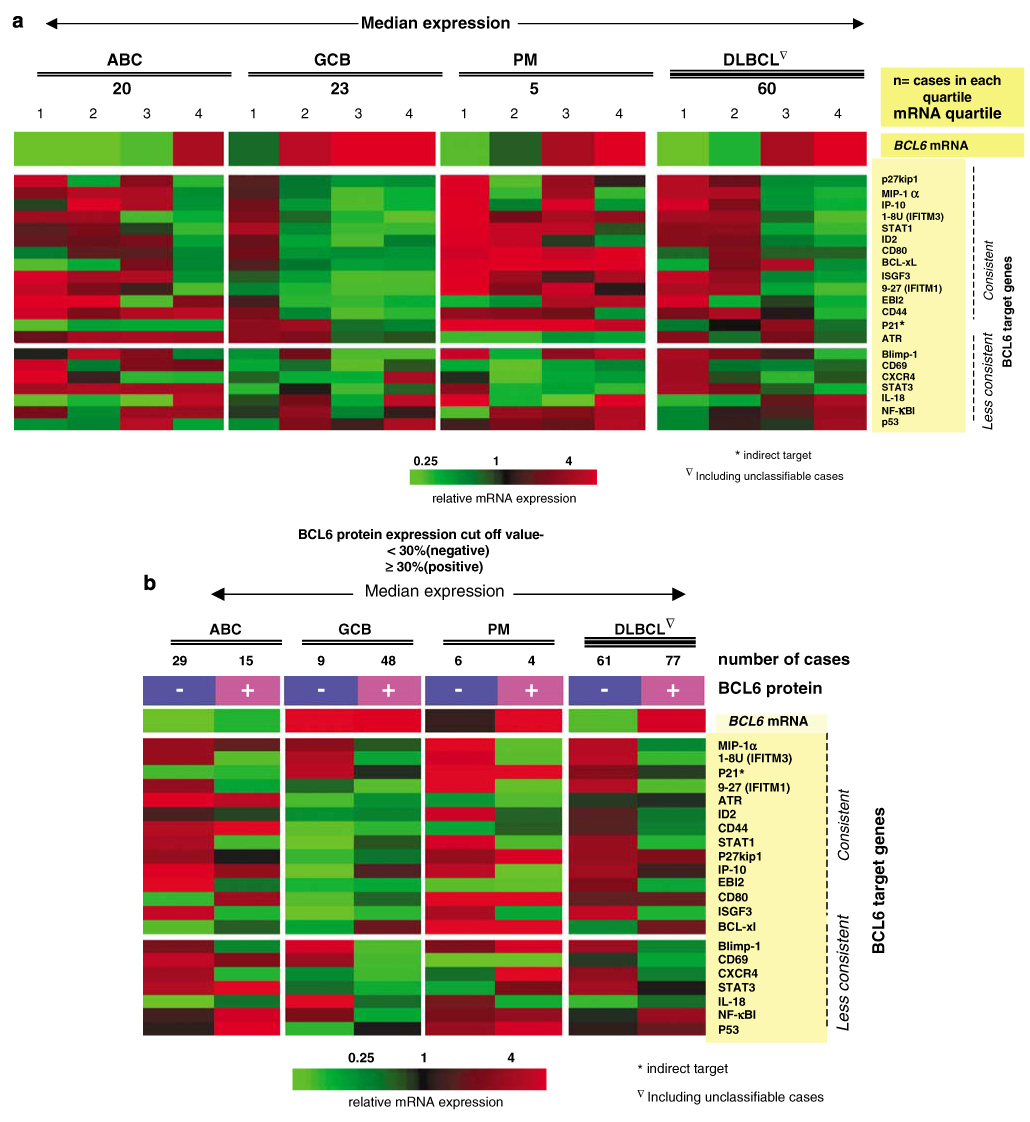
Association of BCL6 expression and BCL6 target repression. (a) BCL6 mRNA expression is associated with BCL6 target gene repression but the pattern is variable among the different subgroups. An inverse relationship is observed between BCL6 mRNA level and BCL6 target gene expression. (b) BCL6 protein expression is associated with repression of BCL6 target genes. Again, the pattern of repression varies with the subtypes of DLBCL. 30% and 50% (not shown) cutoff for protein expression showed same pattern. DLBCL, diffuse large B-cell lymphoma.
We did not observe any significant correlation between the mutational status in intron-1 and the transcriptional levels of target genes; however, in the GCB subgroup, the majority of target genes showed lower expression in cases carrying exon-1 mutations (see Supplementary Figure 3).
There was no evidence of repression of known target genes in cases with BCL6 translocation in any of the subgroups of DLBCL. We also examined the expression of eight known translocation partner genes (IL-21R, MBNL, Pim-1 kinase, JAW1, CD71, eIFF4G1, CIITA-8 and MBNL)41,42 in BCL6 translocated cases. Cases with non-IgH partners showed higher (≥1.5-fold) expression of five (out of eight) known translocation partners genes (IL-21R, MBNL, Pim-1 kinase, JAW1, CD71 and eIFF4G1) compared with cases with IgH/BCL6 translocation (Figure 6).
Figure 6.
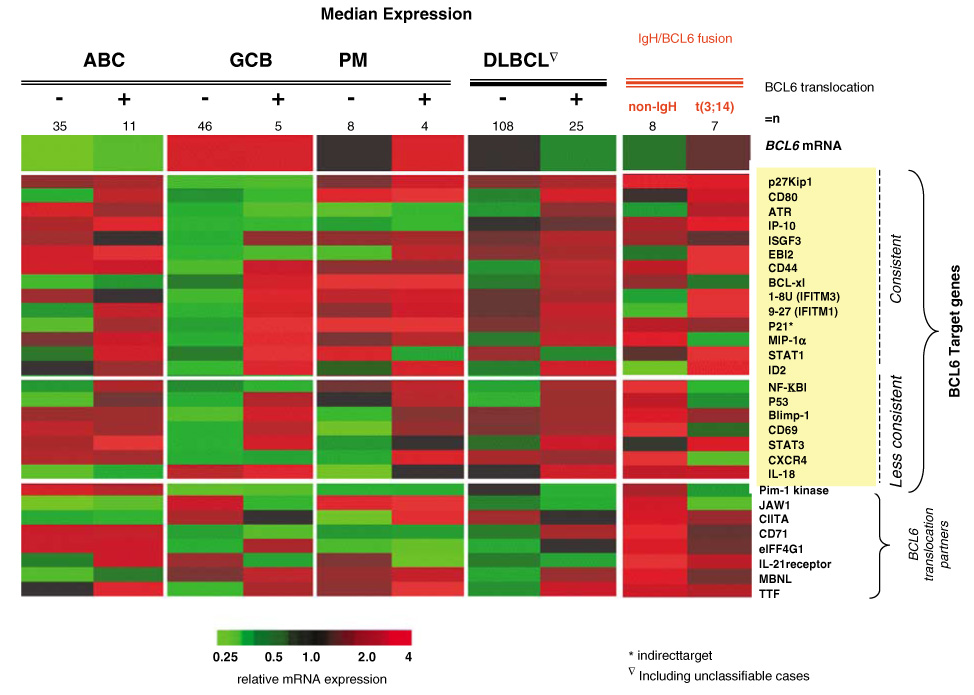
Relationship between BCL6 translocation and BCL6 target gene or translocation partner gene expression. Most of the BCL6 target genes showed increased rather than decreased expression in both the ABC and GCB subgroups (for example, Id2, Blimp-1, CD80, MIP-αIP-10 and P53). Some of the known BCL6 translocation partner genes (IL-21R, MBNL, Pim-1 kinase, JAW1, CD71 and eIFF4G1) were highly expressed in non-IgH/BCL6 translocated cases. GCB, germinal center B-cell-like.
Clinical characteristics and OS
The major clinical characteristics of patients with or without BCL6 mutations or translocations and with different levels of BCL6 mRNA and protein expression are given in the supplementary tables (see Supplementary Tables ST2–5). Patients with BCL6 mutations were more likely to be less than 60 years old than the wild-type cases (P=0.01), but were similar in the other clinical characteristics examined. GCB-DLBCL cases with lower BCL6 protein expression were more likely to be males (P=0.005). There were no other major clinical differences.
BCL6 protein expression (≥30%) was predictive of favorable OS (P=0.001) in the entire group of DLBCL (Figure 7a), and this prediction was also observed with other cutoffs (≥10%, P=0.004; ≥50%, P<0.001). Higher mRNA levels were also associated with a more favorable outcome (P<0.001) when the cases were divided into quartiles or halves according to their mRNA expression levels (Figure 7b). In subgroup analysis, high levels of BCL6 protein expression were associated with better OS in the ABC subgroup (BCL6 ≥50%; P=0.026). BCL6 protein expression was also a significant predictor of survival in the low International Prognostic Index (IPI) group of patients (P=0.001). This association was also observed for mRNA expression (P=0.009 for quartiles and 0.003 for halves) (Figure 7 c–d). In the high IPI risk group, high BCL6 protein expression was significantly associated with a favorable outcome (P=0.005); however, this association was marginally significant in BCL6 mRNA expression (P=0.06) (see Supplementary Figure 4). BCL6 translocation showed no association with OS in DLBCL as a single entity or in subgroup analysis.
Figure 7.
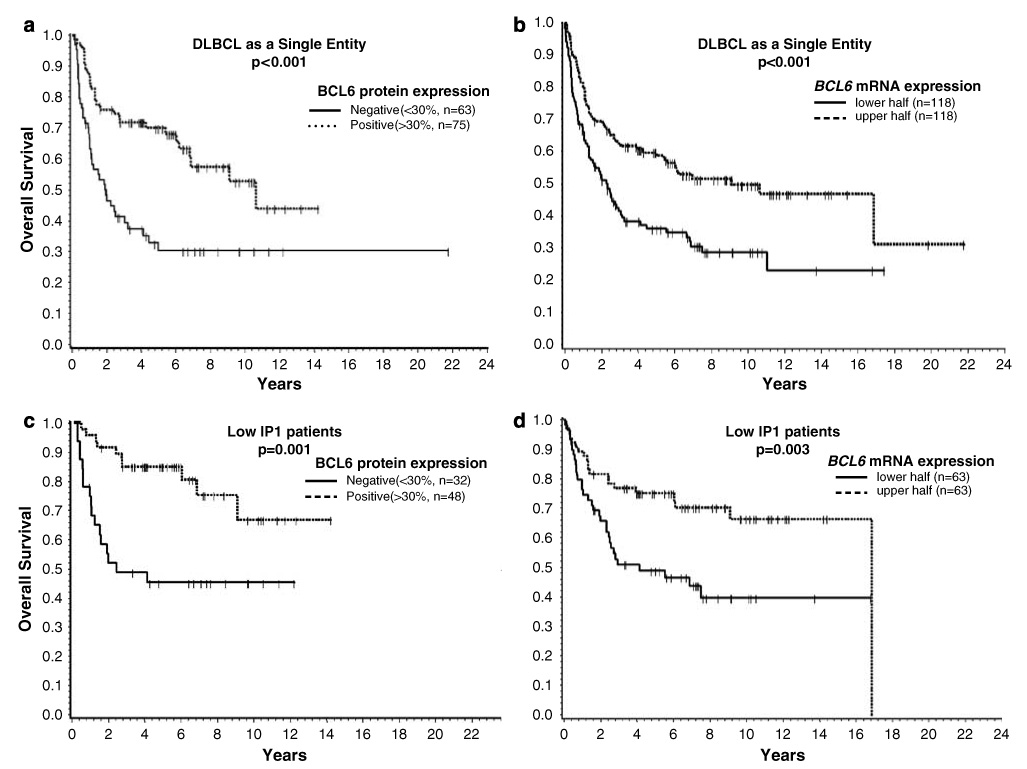
Association of (a) BCL6 protein, and (b) mRNA expression, with overall survival (OS) in the entire group of DLBCL. BCL6 protein expression or high mRNA expression is associated with a favorable outcome. (c) BCL6 protein expression or (d) high mRNA expression is also associated with a favorable outcome in patients with low IPI scores (0–2). DLBCL, diffuse large B-cell lymphoma.
The presence of BCL6 mutations was a significant predictor of favorable OS (P<0.001) in DLBCL and in the ABC subgroup (P=0.04), but was of marginal significance in the GCB subgroup (P=0.08). The association of mutational status with favorable outcome was also observed in both the low IPI (P=0.02) and high IPI risk groups (P=0.008) (see Supplementary Figure 5).
Discussion
DLBCL is an aggressive lymphoma that frequently harbors BCL6 genetic alterations, including mutations in the 5′ non-coding region and translocations involving Ig genes and several non-Ig partners. BCL6 mutations show the characteristic features of hypermutation involving the Ig gene loci, but at a lower frequency. BCL6 is a transcriptional repressor regulating important genes in B-cell differentiation, survival, cell-cycle control and inflammatory reactions, and is essential for GC formation. The two common genomic alterations (translocation and mutation) affecting BCL6 may lead to dysregulation of B-cell maturation and contribute to lymphomagenesis in a significant proportion of DLBCL.13,43,44 This study aimed at defining the incidence of BCL6 gene alterations in the different subgroups of DLBCL as defined by GEP. In addition, we have explored the biological and clinical consequences of these genomic alterations in DLBCL and its subgroups.
BCL6 mutations arise in normal GC B cells due to somatic hypermutation and the mutations are, therefore, present in tumors that are derived from GC or post-GC B-cells.22,35,37 In our series, BCL6 mutations in the intronic region (MMC) were detected in 61% of all DLBCL cases, which is similar to previous findings.20,35 Both the GCB and PM subgroups of DLBCL had a high frequency of mutated cases (Table 2), suggesting that these lymphomas maintain an active hypermutation program for a prolonged period during their development.45 The lower incidence of mutation in the ABC subgroup probably indicates that the GCB program is inactivated earlier in this subgroup. A preferential distribution of mutations in several regions has been reported.37,38 We also observed clustering of mutations in three regions, but only one region (80–123) has been reported previously in DLBCLs.29 Although one report37 suggested that mutations in one cluster (420–443) were associated with increased protein expression, we did not find any correlation of BCL6 mRNA or protein expression with any of the intronic clusters, thus confirming other reports.35,36 We found that 10% of our DLBCL cases had mutations affecting exon-1, present mostly in the GCB subgroup (nine of 13; 69%). These mutations were confined to the DNase-1 hypersensitivity sites in exon-1, often affecting the two BCL6 autoregulatory sites.21 Mutations in this region may be associated with upregulation of BCL6 mRNA and protein expression, but more cases need to be studied to confirm this observation.
BCL6 translocation in the MBR encompassing the first non-coding exon and 5′ region of the first intron has been reported to occur in 20–40% of DLBCL,16,46,47 but more recent studies have found the frequency to be closer to the lower end of this range.15,17,48 Our analysis showed a frequency of 19% and, interestingly, the incidence was higher in the PM (33%) and ABC (26%) subgroups than in the GCB (10%) subgroup. In the GCB subgroup, where BCL6 expression is generally high, overexpression as a result of translocation may not be readily appreciable. In the ABC subgroup, BCL6 translocation was associated with an increase in BCL6 mRNA expression, but the level was still significantly lower than that typically found in the GCB subgroup. This relationship did not change, when ABR cases were added to the analysis, but with reduced fold change (1.4 vs 1.5). In the PM subgroup, we did not observe any appreciable increase in BCL6 mRNA or protein. There was no evidence of target gene repression in association with this translocation in DLBCL, and the functional significance of BCL6 translocation in established DLBCL is unclear. This observation does not, however, invalidate the possibility that BCL6 translocation is important in the pathogenesis of DLBCL. Translocation of the BCL6 gene involves a number of non-Ig partners, whose expression may be under the influence of the BCL6 regulatory regions,49–52 and the expression of some of these partner genes may influence the biology of the DLBCL. Many of known partner genes (5 of 8) were highly expressed in non-IgH/BCL6 translocated cases and the role of these genes in lymphomagenesis warrant further investigation.
Another breakpoint at the ABR located between 245 and 285 kb 5′ of BCL627 is preferentially associated with follicular lymphoma,28 but also occurs in a small percentage of DLBCL. We observed this rearrangement in 6% of DLBCL cases and these cases were mainly present in the ABC subgroup. There are too few cases to independently study the breakpoint at ABR but analyzing all the translocated cases showed no significant differences from the results obtained from studying only cases with a MBR translocation. Because BCL6 translocation is not associated with the repression of known BCL6 target genes, we evaluated the association of BCL6 mRNA and protein expression with target gene repression. Repression of many known BCL6 target genes was clearly observed in the GCB cases that expressed high levels of BCL6 protein or mRNA. We selected a subset of genes that were most consistently repressed in the GCB subgroup and used this set to assess target repression in the other subgroups. Repression of most of these genes was seen when there was a sufficient level of BCL6 mRNA or protein expression. However, some of the target genes are not repressed in the ABC subgroup and an even larger set were not repressed in PM-DLBCL. This observation suggests that target gene repression is partly dependent on the cellular context. Certain co-repressors may be present at a low level or not function properly in the non-GC environment. Some BCL6 targets such as MIP-1α, BCL-XL, CD44, CD80 and IP10 are also NF-κB targets (see Supplementary Reference list) and the NF-κB pathway is activated in ABC- and PM-DLBCL.53,54 It is possible that BCL6 may have other target genes in the non-GC B-cell microenvironment, and BCL6 has recently been shown to exert its function through binding to another transcription factor, MIZ.33 Therefore, BCL6 may exert significantly different functional effects in different subsets of DLCBL.
It has been shown that the p53 gene is a target of BCL6 in normal GC B cells,32 but a clear inverse relationship was not observed even in the GCB-DLBCLs. One possibility is that the level of p53 is normally low and its upregulation requires an inducing signal. Therefore, the regulatory effect of BCL6 on p53 may not be apparent. An alternative explanation is that the control of p53 in DLBCL is more complicated than in normal GC B cells and repression by BCL6 may not be consistently observed. We also did not observe a significant inverse relationship between BCL6 translocation and p53 gene abnormalities suggesting that BCL6 deregulation is not functionally equivalent to p53 deletion.
We correlated BCL6 mRNA and protein expression in the DLBCL subgroups and a significant association was seen only in the ABC subgroup. In the GCB subgroup, BCL6 mRNA and protein expression is frequently high, but does not show a significant correlation, suggesting the possibility of post-transcriptional and translational control of BCL6 protein expression.55
We demonstrated in this study that patients with higher mRNA or protein expression had significantly better OS. The relationship between BCL6 mRNA and survival was also shown previously in a study of a smaller number of cases.23 BCL6 gene expression can stratify patients into good and poor prognostic groups, even in the low and high IPI risk groups. We also observed that the mutation status of BCL6 has prognostic significance. It is possible that the better OS reflect DLBCL subgroup distinctions, because the better prognostic GCB subgroup had a significantly higher number of mutant cases and also significantly higher mRNA and protein expression. However, high expression of BCL6 in the ABC subgroup was also associated with better OS, which suggests that BCL6 could be a biomarker independent of subgroup distinction.
In conclusion, this study demonstrated significant differences in the frequency of BCL6 molecular alterations in the different subgroups of DLBCL. Translocations were more frequent in the ABC subgroup and associated with an increase in BCL6 mRNA expression. Mutations in intron-1 were more frequent in the GCB subgroup, but did not seem to have a significant influence on the expression level of BCL6. BCL6 is a target of somatic hypermutation and many mutations will be stochastic with no consequence on BCL6 expression. Exon-1 mutation was much more frequent in the GCB subgroup and may upregulate BCL6 expression when it involves the BCL6 autoregulatory sites. The set of target genes repressed by BCL6 expression is influenced by the cellular context, and hence, the effect of BCL6 expression may be unique for different subsets. Nevertheless, high BCL6 expression is associated with better survival even in the ABC subgroup. In the GCB subgroup, BCL6 may downregulate NF-κB134 and STAT3 expression (BH Ye, personal communication). The lack of activation of these two important oncogenic pathways may account for the better survival in these patients. The lack of apparent repression of p53, in contrast to normal GC B cells, may also be significant because p53 mutation/deletion is generally associated with poor survival. What then is the role of BCL6 translocation in DLBCL, as the reported target genes are not repressed? It is possible that BCL6 is important in the initial phase of transformation of these tumors, but is no longer necessary after establishment of the lymphoma. The possibility that BCL6 continues to exert its oncogenic effects in DLBCL, by repressing target genes other than those involved in normal GC B cells warrants further investigation. The contribution of translocation partners other than IgH in BCL6 translocated cases should also be investigated.
Acknowledgements
This work was supported in part by US Public Health Service grants CA36727 and CA84967 awarded by the National Cancer Institute, Department of Health and Human Services and Lymphoma Research Foundation.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Anderson JR, Armitage JO, Weisenburger DD. Epidemiology of the non-Hodgkin's lymphomas: distributions of the major subtypes differ by geographic locations. Non-Hodgkin's Lymphoma Classification Project. Ann Oncol. 1998;9:717–720. doi: 10.1023/a:1008265532487. [DOI] [PubMed] [Google Scholar]
- 2.Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatter KC, Warnke R. Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Diffuse large B-cell lymphoma. Lyon: IARC Press; World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001:171–174.
- 4.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 8.Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol. 2004;165:159–166. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 10.Baron BW, Nucifora G, McCabe N, Espinosa R, III, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci USA. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 12.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 14.Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 15.Jerkeman M, Aman P, Cavallin-Stahl E, Torlakovic E, Akerman M, Mitelman F, et al. Prognostic implications of BCL6 rearrangement in uniformly treated patients with diffuse large B-cell lymphoma – a Nordic Lymphoma Group study. Int J Oncol. 2002;20:161–165. doi: 10.3892/ijo.20.1.161. [DOI] [PubMed] [Google Scholar]
- 16.Kramer MH, Hermans J, Wijburg E, Philippo K, Geelen E, van Krieken JH, et al. Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma. Blood. 1998;92:3152–3162. [PubMed] [Google Scholar]
- 17.Barrans SL, O'Connor SJ, Evans PA, Davies FE, Owen RG, Haynes AP, et al. Rearrangement of the BCL6 locus at 3q27 is an independent poor prognostic factor in nodal diffuse large B-cell lymphoma. Br J Haematol. 2002;117:322–332. doi: 10.1046/j.1365-2141.2002.03435.x. [DOI] [PubMed] [Google Scholar]
- 18.Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, et al. LAZ3 rearrangements in non-Hodgkin's lymphoma: correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood. 1994;83:2423–2427. [PubMed] [Google Scholar]
- 19.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, et al. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitolo U, Botto B, Capello D, Vivenza D, Zagonel V, Gloghini A, et al. Point mutations of the BCL-6 gene: clinical and prognostic correlation in B-diffuse large cell lymphoma. Leukemia. 2002;16:268–275. doi: 10.1038/sj.leu.2402349. [DOI] [PubMed] [Google Scholar]
- 21.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 22.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossos IS, Jones CD, Warnke R, Natkunam Y, Kaizer H, Zehnder JL, et al. Expression of a single gene, BCL-6, strongly predicts survival in patients with diffuse large B-cell lymphoma. Blood. 2001;98:945–951. doi: 10.1182/blood.v98.4.945. [DOI] [PubMed] [Google Scholar]
- 24.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 26.Zu Y, Steinberg SM, Campo E, Hans CP, Weisenburger DD, Braziel RM, et al. Validation of tissue microarray immunohisto-chemistry staining and interpretation in diffuse large B-cell lymphoma. Leuk Lymphoma. 2005;46:693–701. doi: 10.1080/10428190500051844. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Butler M, Rao PH, Chaganti SR, Louie DC, Dalla-Favera R, et al. The t(2;3)(q21;q27) translocation in non-Hodgkin's lymphoma displays BCL6 mutations in the 5′ regulatory region and chromosomal breakpoints distant from the gene. Oncogene. 1998;17:1717–1722. doi: 10.1038/sj.onc.1202098. [DOI] [PubMed] [Google Scholar]
- 28.Bosga-Bouwer AG, Haralambieva E, Booman M, Boonstra R, van den Berg A, et al. BCL6 alternative translocation breakpoint cluster region associated with follicular lymphoma grade 3B. Genes Chromosomes Cancer. 2005;44:301–304. doi: 10.1002/gcc.20246. [DOI] [PubMed] [Google Scholar]
- 29.Greiner TC, Smith LM, Rosenwald A, Weisenburger DW, Gascoyne R, Connor J, et al. mRNA expression patterns in p53 mutant vs. wildtype subgroups of diffuse large B-cell lymphoma. Blood. 2003;102 Abstract# 1338. [Google Scholar]
- 30.Greiner TC. Enhanced detection of TP53 mutations utilizing a GC-clamp in denaturing high performance liquid chromatography. Diagn Mol Pathol. 2007;16:32–37. doi: 10.1097/PDM.0b013e31802c29de. [DOI] [PubMed] [Google Scholar]
- 31.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 32.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 33.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Wang X, Yu RY, Ding BB, Yu JJ, Dai XM, et al. BCL-6 negatively regulates expression of the NF-kappaB1 p105/p50 subunit. J Immunol. 2005;174:205–214. doi: 10.4049/jimmunol.174.1.205. [DOI] [PubMed] [Google Scholar]
- 35.Capello D, Vitolo U, Pasqualucci L, Quattrone S, Migliaretti G, Fassone L, et al. Distribution and pattern of BCL-6 mutations throughout the spectrum of B-cell neoplasia. Blood. 2000;95:651–659. [PubMed] [Google Scholar]
- 36.Lossos IS, Levy R. Mutation analysis of the 5′ noncoding regulatory region of the BCL-6 gene in non-Hodgkin lymphoma: evidence for recurrent mutations and intraclonal heterogeneity. Blood. 2000;95:1400–1405. [PubMed] [Google Scholar]
- 37.Artiga MJ, Saez AI, Romero C, Sanchez-Beato, Mateo MS, Navas C, et al. A short mutational hot spot in the first intron of BCL-6 is associated with increased BCL-6 expression and with longer overall survival in large B-cell lymphomas. Am J Pathol. 2002;160:1371–1380. doi: 10.1016/S0002-9440(10)62564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jardin F, Ruminy P, Parmentier F, Picquenot JM, Courel MN, Bertrand P, et al. Clinical and biological relevance of single-nucleotide polymorphisms and acquired somatic mutations of the BCL6 first intron in follicular lymphoma. Leukemia. 2005;19:1824–1830. doi: 10.1038/sj.leu.2403915. [DOI] [PubMed] [Google Scholar]
- 39.Takeda N, Arima M, Tsuruoka N, Okada S, Hatano M, Sakamoto A, et al. Bcl6 is a transcriptional repressor for the IL-18 gene. J Immunol. 2003;171:426–431. doi: 10.4049/jimmunol.171.1.426. [DOI] [PubMed] [Google Scholar]
- 40.Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, et al. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 41.Chen YW, Liang AC, Au WY, Chu KM, Wong KY, Hu X, et al. Multiple BCL6 translocation partners in individual cases of gastric lymphoma. Blood. 2003;102:1931–1932. doi: 10.1182/blood-2003-06-1786. author reply 1932. [DOI] [PubMed] [Google Scholar]
- 42.Xu WS, Liang RH, Srivastava G. Identification and characterization of BCL6 translocation partner genes in primary gastric high-grade B-cell lymphoma: heat shock protein 89 alpha is a novel fusion partner gene of BCL6. Genes Chromosomes Cancer. 2000;27:69–75. doi: 10.1002/(sici)1098-2264(200001)27:1<69::aid-gcc9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Akasaka T, Miura I, Takahashi N, Akasaka H, Yonetani N, Ohno H, et al. A recurring translocation, t(3;6)(q27;p21), in non-Hodgkin's lymphoma results in replacement of the 5′ regulatory region of BCL6 with a novel H4 histone gene. Cancer Res. 1997;57:7–12. [PubMed] [Google Scholar]
- 44.Baron BW, Anastasi J, Montag A, Huo D, Baron RM, Karrison T, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci USA. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malpeli G, Barbi S, Moore PS, Scardoni M, Chilosi M, Scarpa A, et al. Primary mediastinal B-cell lymphoma: hypermutation of the BCL6 gene targets motifs different from those in diffuse large B-cell and follicular lymphomas. Haematologica. 2004;89:1091–1099. [PubMed] [Google Scholar]
- 46.Miki T, Kawamata N, Hirosawa S, Aoki N. Gene involved in the 3q27 translocation associated with B-cell lymphoma, BCL5, encodes a Kruppel-like zinc-finger protein. Blood. 1994;83:26–32. [PubMed] [Google Scholar]
- 47.Offit K, Lo Coco F, Louie DC, Parsa NZ, Leung D, Portlock C, et al. Rearrangement of the bcl-6 gene as a prognostic marker in diffuse large-cell lymphoma. N Engl J Med. 1994;331:74–80. doi: 10.1056/NEJM199407143310202. [DOI] [PubMed] [Google Scholar]
- 48.Katzenberger T, Ott G, Klein T, Kalla J, Muller-Hermelink HK, Ott MM. Cytogenetic alterations affecting BCL6 are predominantly found in follicular lymphomas grade 3B with a diffuse large B-cell component. Am J Pathol. 2004;165:481–490. doi: 10.1016/S0002-9440(10)63313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, et al. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akasaka T, Ueda C, Kurata M, Akasaka H, Yamabe H, Uchiyama T, et al. Nonimmunoglobulin (non-Ig)/BCL6 gene fusion in diffuse large B-cell lymphoma results in worse prognosis than Ig/BCL6. Blood. 2000;96:2907–2909. [PubMed] [Google Scholar]
- 51.Ueda C, Uchiyama T, Ohno H. Immunoglobulin (Ig)/BCL6 versus non-Ig/BCL6 gene fusion in diffuse large B-cell lymphoma corresponds to a high- versus low-level expression of BCL6 mRNA. Blood. 2002;99:2624–2625. doi: 10.1182/blood-2001-11-0117. [DOI] [PubMed] [Google Scholar]
- 52.Ueda C, Akasaka T, Kurata M, Maesako Y, Nishikori M, Ichinohasama R, et al. The gene for interleukin-21 receptor is the partner of BCL6 in t(3;16)(q27;p11), which is recurrently observed in diffuse large B-cell lymphoma. Oncogene. 2002;21:368–376. doi: 10.1038/sj.onc.1205099. [DOI] [PubMed] [Google Scholar]
- 53.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B celllike diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feuerhake F, Kutok JL, Monti S, Chen W, LaCasce AS, Cattoretti G, et al. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood. 2005;106:1392–1399. doi: 10.1182/blood-2004-12-4901. [DOI] [PubMed] [Google Scholar]
- 55.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]


