Abstract
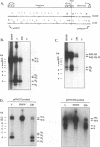
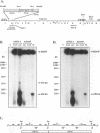
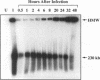
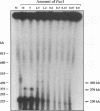
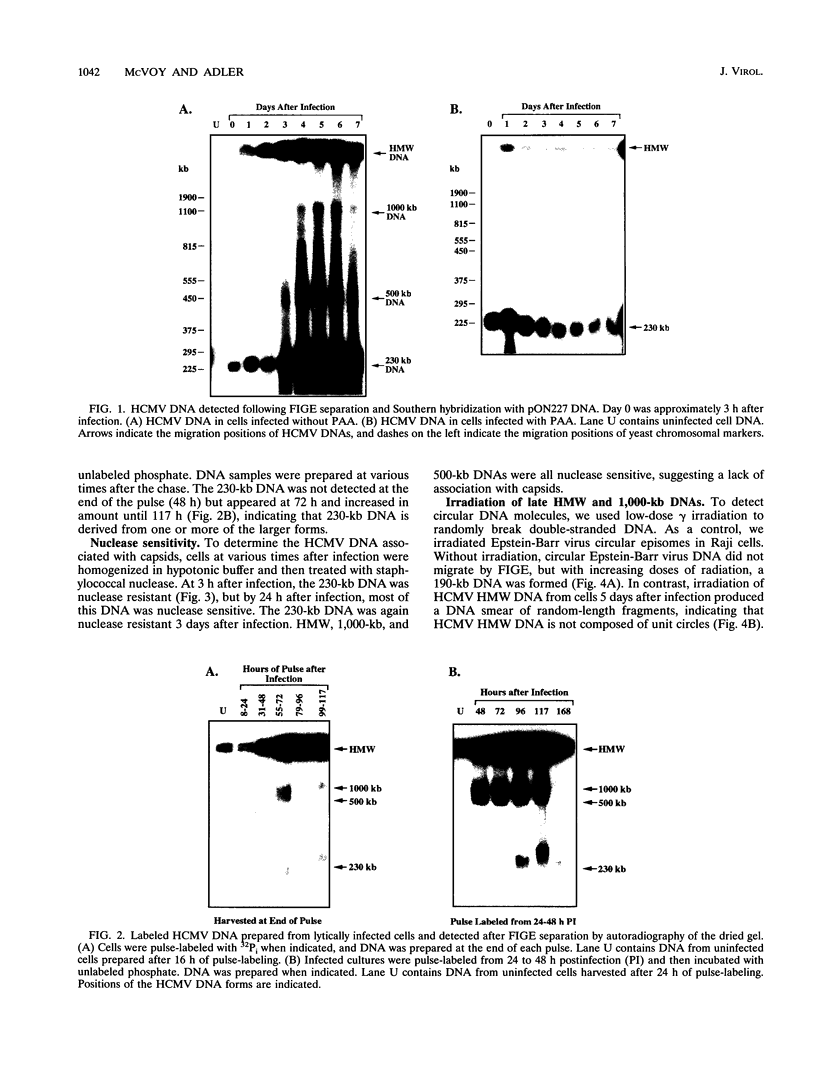
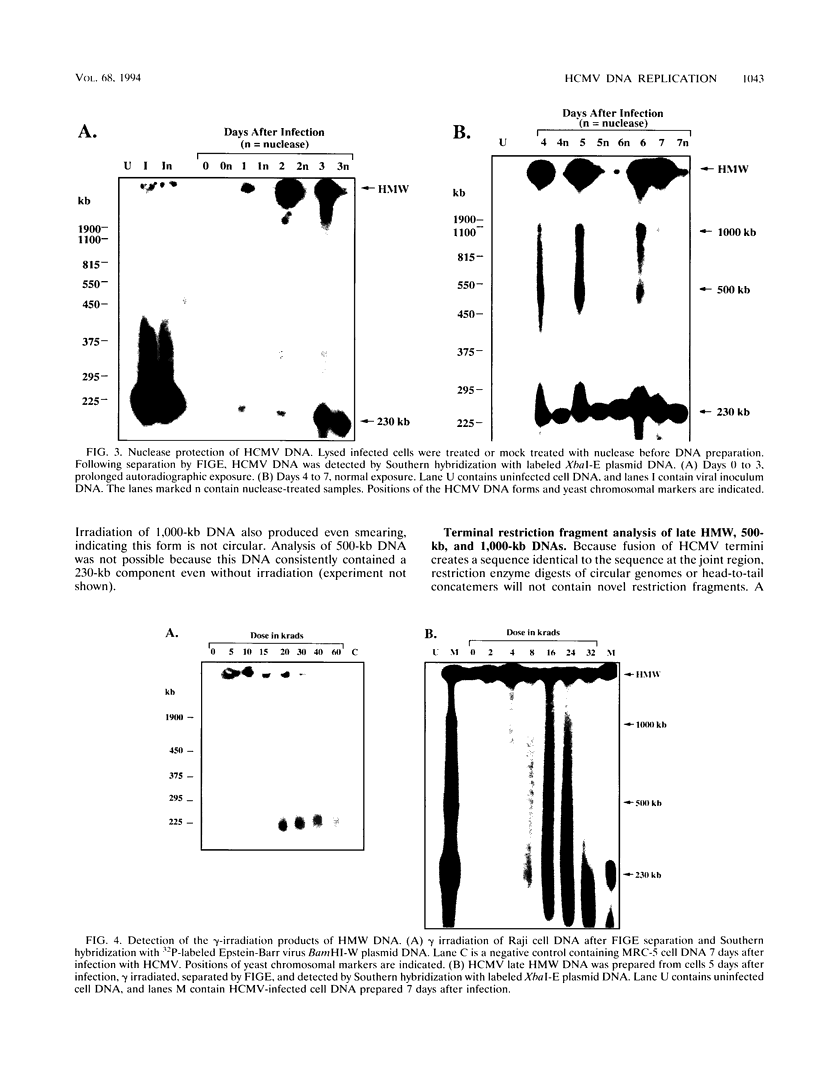
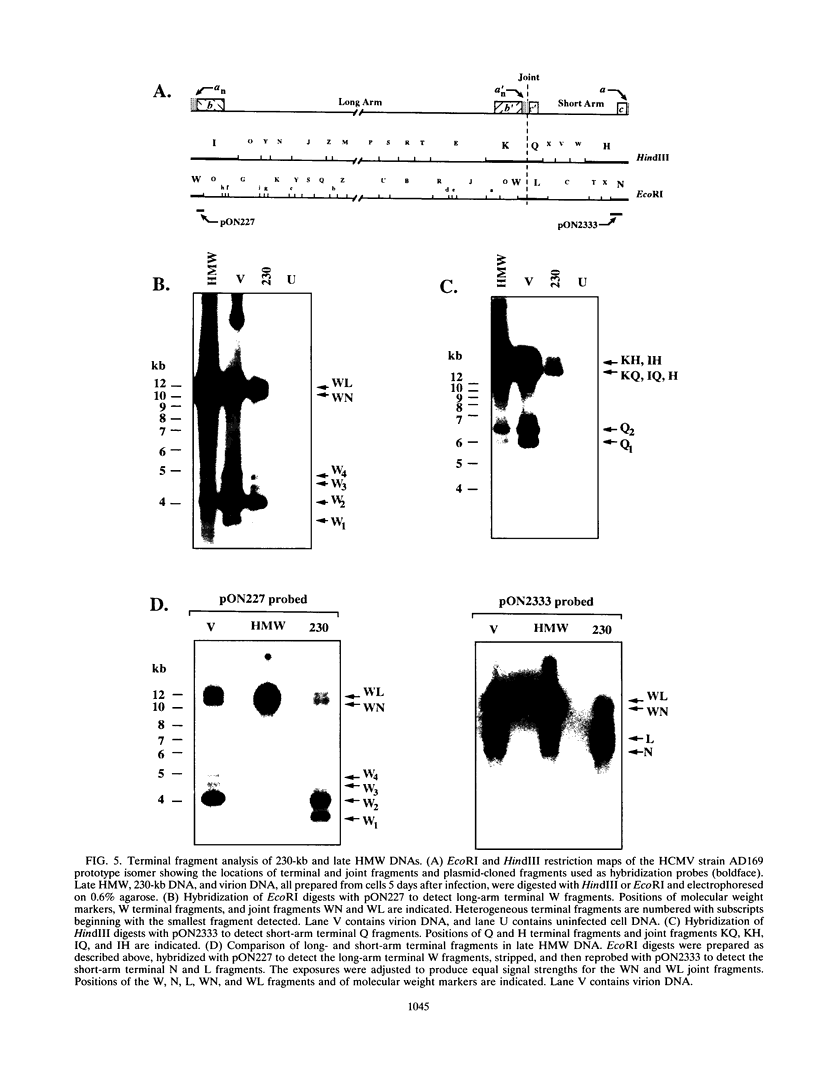
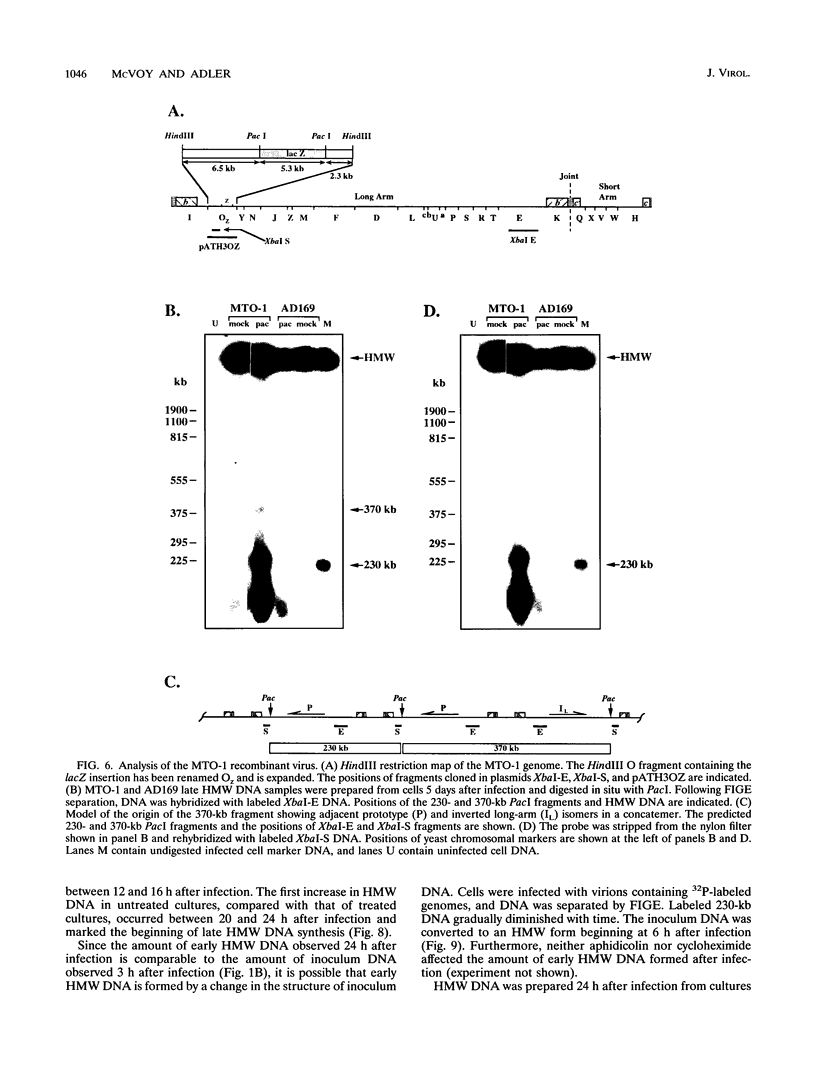
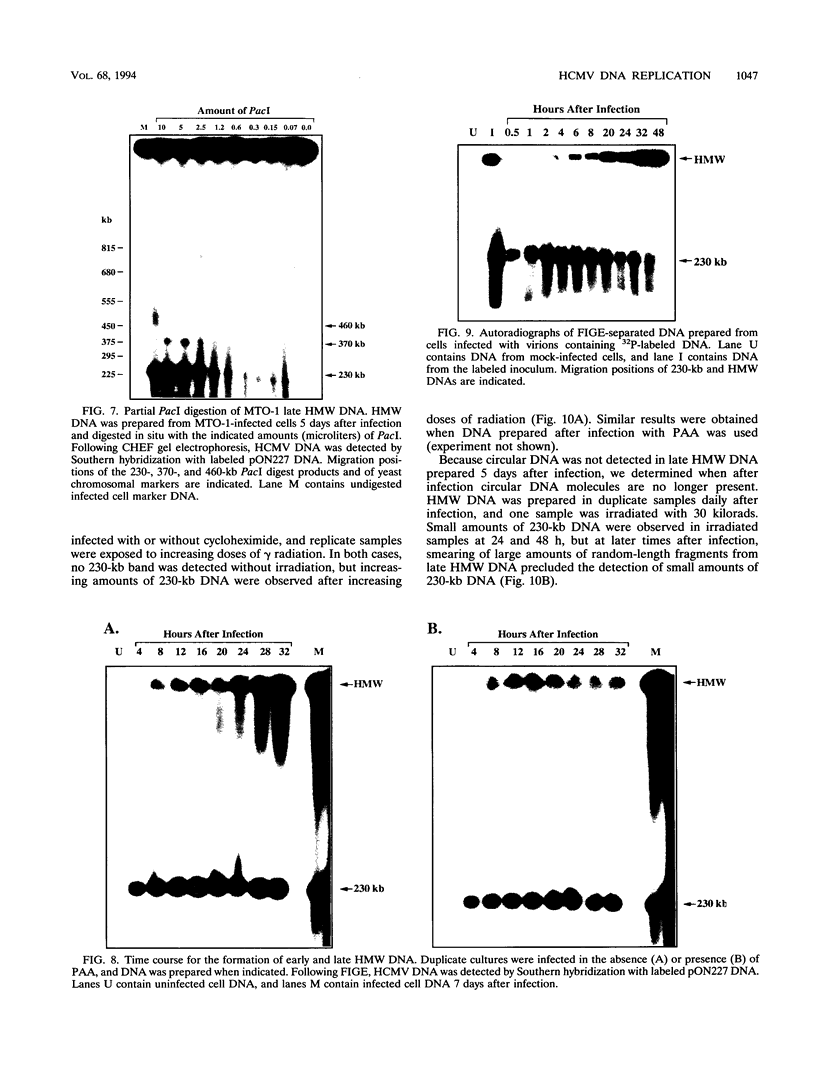
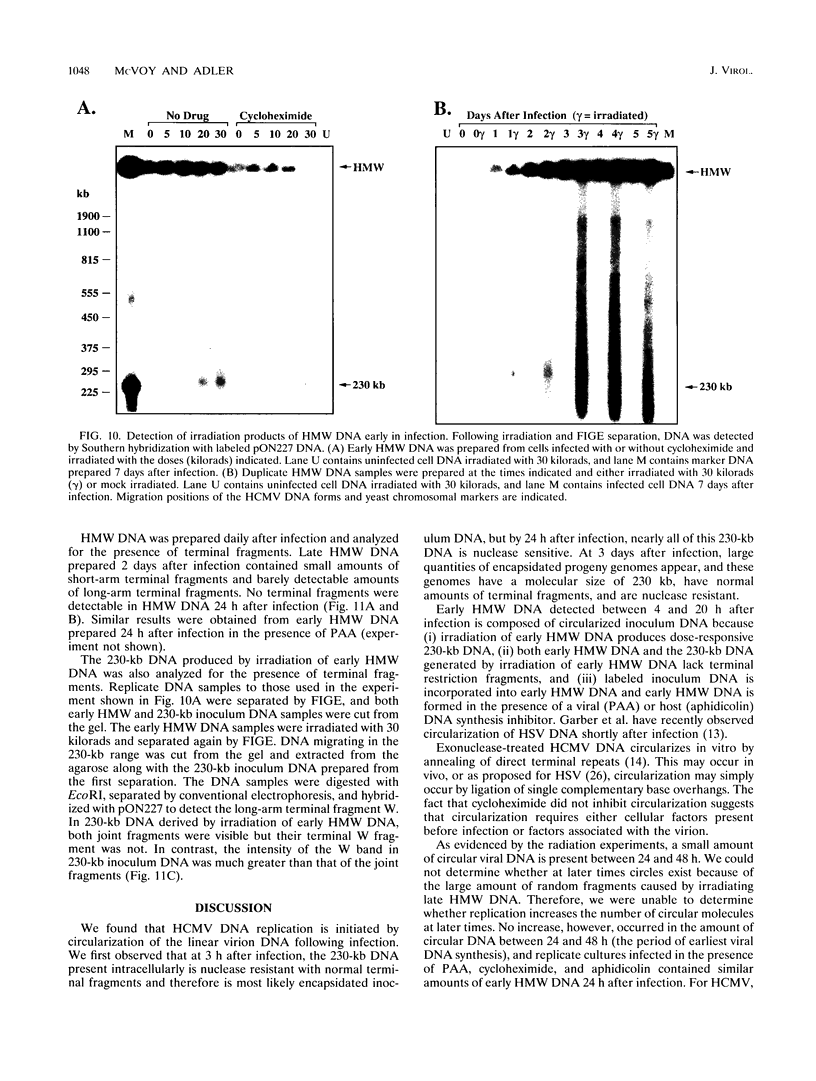
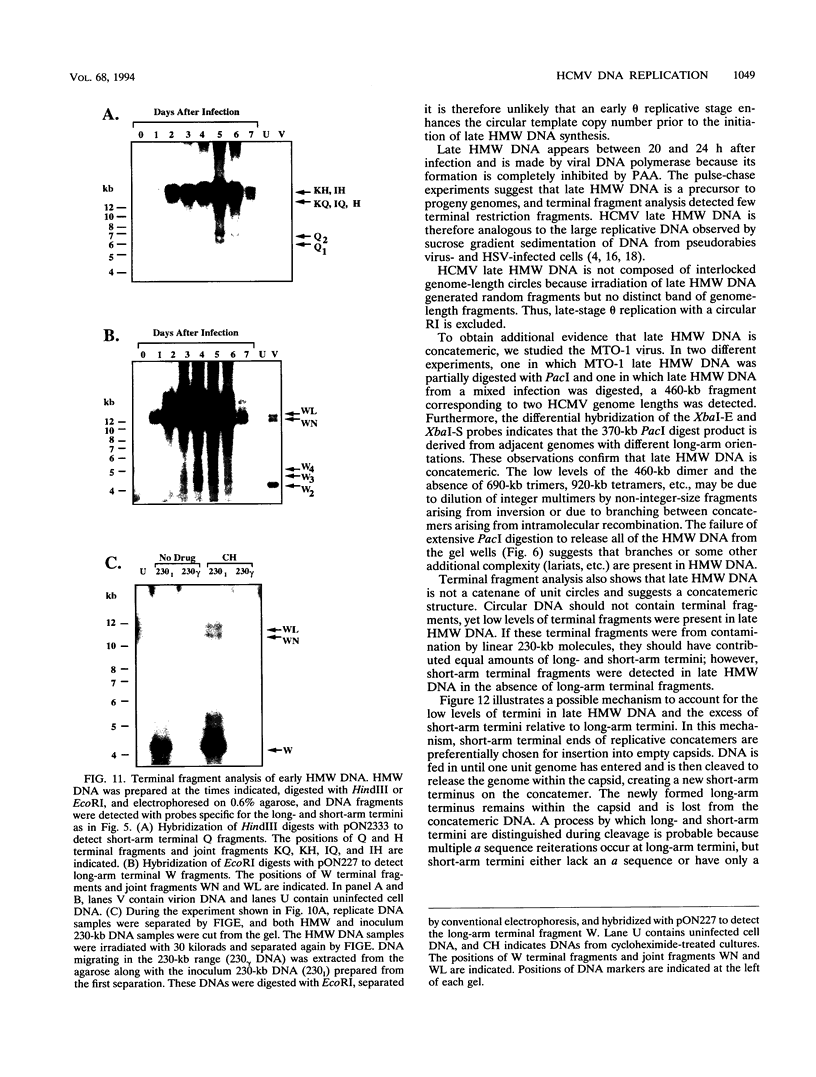
To determine the replicative mechanism for human cytomegalovirus (HCMV) DNA, field inversion gel electrophoresis was used to separate HCMV replicative DNAs during lytic infection. Unit-length circular HCMV genomes lacking terminal restriction fragments were detected starting 4 h after infection even when cells were treated with aphidicolin, phosphonoacetic acid, or cycloheximide. Viral DNA synthesis began 24 h after infection and produced large amounts of high-molecular-weight replicative DNA that was a precursor of progeny genomes. Replicative DNA contained rare terminal restriction fragments, and long-arm termini were much less frequent than short-arm termini. Replicative DNA was not composed of unit-length circles because low-dose gamma irradiation of replicative DNA generated numerous random high-molecular-weight fragments rather than unit-length molecules. PacI digestion of replicative DNA from a recombinant HCMV with two closely spaced PacI sites revealed that replicative DNA is concatemeric and genome segment inversion occurs after concatemer synthesis. These results show that after circularization of the parental genome, DNA synthesis produces concatemers and genomic inversion occurs within concatemeric DNA. The results further suggest that concatemers acquire genomic termini during the cleavage/packaging process which preferentially inserts short-arm termini into empty capsids, causing a predominance of short-arm termini on the concatemer.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler S. P. Molecular epidemiology of cytomegalovirus: evidence for viral transmission to parents from children infected at a day care center. Pediatr Infect Dis. 1986 May-Jun;5(3):315–318. [PubMed] [Google Scholar]
- Adler S. P. The molecular epidemiology of cytomegalovirus transmission among children attending a day care center. J Infect Dis. 1985 Oct;152(4):760–768. doi: 10.1093/infdis/152.4.760. [DOI] [PubMed] [Google Scholar]
- Anders D. G., Punturieri S. M. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991 Feb;65(2):931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Kaplan A. S., Stehn B., Rubenstein A. S. Concatemeric forms of intracellular herpesvirus DNA. Virology. 1976 Feb;69(2):547–560. doi: 10.1016/0042-6822(76)90484-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A. Origin of replication of the DNA of a herpesvirus (pseudorabies). Proc Natl Acad Sci U S A. 1980 Jan;77(1):172–175. doi: 10.1073/pnas.77.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch R. E., Bruckner R. C., Mocarski E. S., Lehman I. R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992 Jan;66(1):277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann A., Becker Y. Circular and circular-linear DNA molecules of herpes simplex virus. J Gen Virol. 1977 Oct;37(1):205–208. doi: 10.1099/0022-1317-37-1-205. [DOI] [PubMed] [Google Scholar]
- Friedmann A., Shlomai J., Becker Y. Electron microscopy of herpes simplex virus DNA molecules isolated from infected cells by centrifugation in CsCl density gradients. J Gen Virol. 1977 Mar;34(3):507–522. doi: 10.1099/0022-1317-34-3-507. [DOI] [PubMed] [Google Scholar]
- Garber D. A., Beverley S. M., Coen D. M. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993 Nov;197(1):459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- Hamzeh F. M., Lietman P. S., Gibson W., Hayward G. S. Identification of the lytic origin of DNA replication in human cytomegalovirus by a novel approach utilizing ganciclovir-induced chain termination. J Virol. 1990 Dec;64(12):6184–6195. doi: 10.1128/jvi.64.12.6184-6195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch I., Cabral G., Patterson M., Biswal N. Studies on the intracellular replicating DNA of herpes simplex virus type 1. Virology. 1977 Aug;81(1):48–61. doi: 10.1016/0042-6822(77)90057-5. [DOI] [PubMed] [Google Scholar]
- Hirsch I., Roubal J., Vonka V. Replicating DNA of herpes simplex virus type 1. Intervirology. 1976;7(3):155–175. doi: 10.1159/000149949. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977 Aug;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J. H., Ben-Porat T. Appearance in vivo of single-stranded complementary ends on parental herpesvirus DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2674–2678. doi: 10.1073/pnas.73.8.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J. H., Blankenship M. L., Ben-Porat T. Replication of herpesvirus DNA. I. Electron microscopic analysis of replicative structures. Virology. 1977 Jun 15;79(2):281–291. doi: 10.1016/0042-6822(77)90355-5. [DOI] [PubMed] [Google Scholar]
- Jean J. H., Yoshimura N., Furukawa T., Plotkin S. A. Intracellular forms of the parental human cytomegalovirus genome at early stages of the infective process. Virology. 1978 May 1;86(1):281–286. doi: 10.1016/0042-6822(78)90029-6. [DOI] [PubMed] [Google Scholar]
- Lai A. C., Chu Y. A rapid method for screening vaccinia virus recombinants. Biotechniques. 1991 May;10(5):564–565. [PubMed] [Google Scholar]
- MacGregor G. R., Mogg A. E., Burke J. F., Caskey C. T. Histochemical staining of clonal mammalian cell lines expressing E. coli beta galactosidase indicates heterogeneous expression of the bacterial gene. Somat Cell Mol Genet. 1987 May;13(3):253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J. The genome of herpes simplex virus: structure, replication and evolution. J Cell Sci Suppl. 1987;7:67–94. doi: 10.1242/jcs.1987.supplement_7.6. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Jacob R. J., Knipe D. M., Morse L. S., Ruyechan W. T. On the structure, functional equivalence, and replication of the four arrangements of herpes simplex virus DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):809–826. doi: 10.1101/sqb.1979.043.01.088. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Friedmann A., Becker Y. Replication intermediates of herpes simplex virus DNA. Virology. 1976 Feb;69(2):647–659. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Duncan J., Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990 Oct;64(10):5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. The alpha sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985 Jun;54(3):817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Filpula D., Friedmann T., Spector D. H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984 Nov;52(2):541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Spector D. H. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986 Sep;59(3):591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]