Abstract
A C-terminally truncated Y145Stop variant of the human prion protein (huPrP23–144) is associated with a hereditary amyloid disease known as PrP cerebral amyloid angiopathy. Previous studies have shown that recombinant huPrP23–144 can be efficiently converted in vitro to the fibrillar amyloid state, and that residues 138 and 139 play a critical role in the amyloidogenic properties of this protein. Here, we have used magic-angle spinning solid-state NMR spectroscopy to provide high-resolution insight into the protein backbone conformation and dynamics in fibrils formed by 13C,15N-labeled huPrP23–144. Surprisingly, we find that signals from ≈100 residues (i.e., ≈80% of the sequence) are not detected above approximately −20°C in conventional solid-state NMR spectra. Sequential resonance assignments revealed that signals, which are observed, arise exclusively from residues in the region 112–141. These resonances are remarkably narrow, exhibiting average 13C and 15N linewidths of ≈0.6 and 1 ppm, respectively. Altogether, the present findings indicate the existence of a compact, highly ordered core of huPrP23–144 amyloid encompassing residues 112–141. Analysis of 13C secondary chemical shifts identified likely β-strand segments within this core region, including β-strand 130–139 containing critical residues 138 and 139. In contrast to this relatively rigid, β-sheet-rich amyloid core, the remaining residues in huPrP23–144 amyloid fibrils under physiologically relevant conditions are largely unordered, displaying significant conformational dynamics.
Keywords: protein structure, solid-state NMR, protein misfolding, transmissible spongiform encephalopathies
Polymerization of normally soluble proteins into amyloid deposits underlies a number of dreaded human disorders, including Alzheimer's, Parkinson's, and Huntington's diseases, and type II diabetes (1). Regardless of the amino acid sequence and three-dimensional structure of amyloidogenic precursor proteins, mature amyloid fibrils share many physicochemical properties including elongated morphology, the ability to bind Congo red dye in a manner leading to characteristic apple-green birefringence, and a so-called “cross-β” fiber diffraction pattern indicative of the presence of β-sheets with individual β-strands arranged perpendicular to the fibril axis (2). With the notable exception of extensively studied Aβ peptide (3, 4), however, higher-resolution data for these biologically important structures are very scarce and incomplete. This is largely due to difficulties in applying classical methods of structural biology to study high-molecular-weight protein aggregates such as amyloid fibrils. Recent crystallographic studies have provided atomic-level insight into amyloid-like microcrystals formed by a number of short (4–7 residues) peptides, revealing a common “steric zipper” motif where pairs of β-sheets form a dry interface with interdigitation of side chains (5). Remarkably, these basic steric-zippers can be organized in distinct molecular architectures, suggesting structural complexity of amyloids formed by larger, biologically relevant proteins. Direct crystallographic studies for such larger proteins are, however, not yet feasible.
The conversion of a monomeric protein to highly ordered, amyloid-like aggregates is also associated with the pathogenesis of transmissible spongiform encephalopathies (TSEs), a group of neurodegenerative disorders such as Creutzfeldt–Jakob disease (CJD) in humans, scrapie in sheep, and bovine spongiform encephalopathy in cattle (6–8). The protein of interest here is an ≈209-aa glycoprotein, dubbed prion protein (PrP). Most remarkable, the aggregates formed upon conversion of the normal form of mammalian prion protein (PrPC) to the misfolded conformation (PrPSc) are believed to be infectious, accounting for the transmissibility of TSEs (6–8). Once highly controversial, the hypothesis that a disease can be transmitted by a “protein-only” mechanism is now gaining acceptance, especially in view of recent progress in generating infectious PrPSc in vitro (9–11). Furthermore, the notion that proteins alone can be infectious is strongly supported by a plethora of findings regarding protein conformation-based inheritance in fungi (12, 13).
Despite recent advances, our understanding of the biophysical mechanisms and structural basis of mammalian prion protein conversion to the amyloid state is relatively poor and of low resolution. Recently, we and others have shown that certain aspects of this process can be conveniently studied by using the recombinant Y145Stop mutant of the human prion protein, huPrP23–144 (14, 15). This C-truncated protein, associated with prion protein hereditary amyloidosis known as PrP cerebral amyloid angiopathy (16), undergoes very efficient autocatalytic conformational conversion under physiologically relevant buffer conditions (14, 15); a similar reaction for recombinant full-length prion protein or PrP90–231 has been reported only in the presence of relatively high concentrations of chemical denaturants (17, 18). Most importantly, using this experimentally tractable model, we were able to reproduce in vitro some of the most fundamental aspects of prion propagation, including the phenomena of “species barrier” and prion strains (19, 20). One of the main lessons from these studies was that both the seeding specificity and the strain variability of PrP amyloid are fully encoded in the fibril conformation. However, the techniques used in previous conformational analysis were of intrinsically low resolution, precluding molecular-level description of PrP23–144 amyloid structure and the structural basis of strain specificity.
Here, we have analyzed the molecular conformation and dynamics of huPrP23–144 amyloid fibrils using high-resolution magic-angle spinning (MAS) solid-state NMR (SSNMR) spectroscopy. MAS SSNMR is one of very few techniques that can provide site-specific information about proteins in noncrystalline solid state; therefore, this approach is uniquely suited for structural studies of amyloid fibrils. Indeed, some of the most successful recent applications of SSNMR have been aimed at the atomic-level characterization of amyloidogenic peptides and proteins, including Aβ (3), α-synuclein (21, 22), fungal prion proteins HET-s, Sup35p and Ure2p (23–25), and peptide fragments of β2-microglobulin (26) and transthyretin (27, 28). Our SSNMR measurements performed by using uniformly 13C, 15N-labeled protein allowed us to identify a compact (≈28 residues), rigid core of the huPrP23–144 amyloid, as well as to determine the location of β-strands within this core region.
Results
SSNMR Spectra Identify a Highly Ordered, Compact Amyloid Core with a Predominantly β-Strand Conformation.
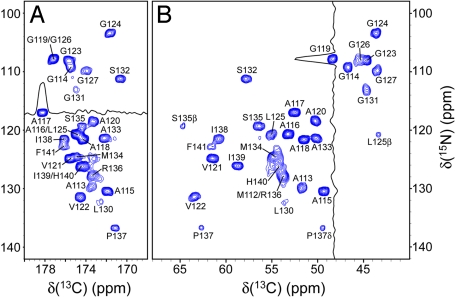
The fingerprint 2D 15N-13C′ (NCO), 15N-13Cα (NCA), and 13C-13C (CC) chemical-shift correlation spectra of uniformly 13C,15N-labeled huPrP23–144 amyloid fibrils at 0°C are shown in Fig. 1 and supporting information (SI) Fig. S1. Essentially identical spectra (i.e., |Δδ|avg < 0.2 ppm) were obtained for two independently prepared fibril samples, indicating high reproducibility of the experiments. Remarkably, only ≈20% of the cross-peaks (i.e., 28 of a total of 125 or 126) expected in 2D NCO, NCA, or CC spectra (C′–Cα region) are observed under the present experimental conditions (note that huPrP23–144 used in this study contains 126 residues, including a 4-residue N-terminal extension). Given that cross-polarization (CP), which requires the presence of substantial through-space magnetic dipole–dipole couplings (29, 30), was used to transfer magnetization between 1H, 13C, and 15N nuclei, this clearly indicates the presence of a compact (≈28 residues), relatively rigid “core” region of the amyloid. In contrast to this rigid core region, the remainder of the protein backbone in huPrP23–144 fibrils shows a high degree of conformational flexibility (see Variable Temperature NMR Studies of Conformational Dynamics in huPrP23–144 Amyloid). The existence of such rigid and dynamic segments has been previously reported for amyloid fibrils formed by other proteins, including α-synuclein (21, 22) and HET-s (23, 31).
Fig. 1.
Two-dimensional NCO (A) and NCA (B) spectra of huPrP23–144 fibrils, acquired at a 11.111-kHz MAS rate and 0°C. Each spectrum was recorded as a 768* (t1, 15N) × 1,400* (t2, 13C) data matrix with acquisition times of 15.3 ms (t1) and 28 ms (t2), and a measurement time of 8.5 h. Cross-peaks are drawn with the lowest contour at ≈10 times the root-mean-square noise level and labeled by residue number according to the 15N frequency. Also shown are representative 13C and 15N 1D traces for residues A117 (A) and G119 (B).
Site-specific 13C and 15N NMR linewidth measurements further reaffirm that the amyloid core of huPrP23–144 fibrils is highly ordered on the atomic level. Indeed, as demonstrated by representative 13C and 15N 1D traces shown in Fig. 1, for most residues 2D NCO and NCA spectra display relatively narrow and symmetric resonances. Quantitative measurements for a set of 20 well resolved peaks in the 2D NCA spectrum (processed with no apodization) gave the average 15N and 13C widths at half-height of 0.98 ± 0.18 and 0.63 ± 0.13 ppm, respectively. These linewidths, comparable with those recently reported for amyloids formed by a synthetic fragment of transthyretin (27, 28) and HET-s protein (23), approach the limit of narrowness observed for peptides and globular proteins in a highly ordered microcrystalline state (32). Moreover, a 2D NCO spectrum recorded in the presence of 1H and 15N spin decoupling during 13C detection shows that for residues distributed throughout the amyloid core, the 13C spectral resolution is to a large extent limited by the presence of 13C′-13Cα J-couplings (see Fig. S2). This further confirms high degree of order in this part of huPrP23–144.
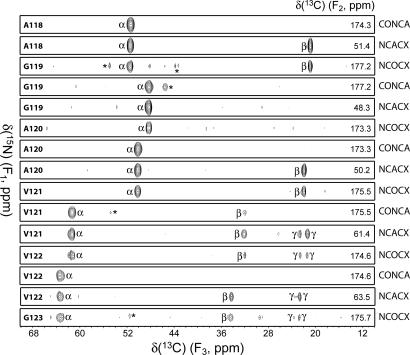
To obtain the sequential 13C and 15N backbone and side-chain resonance assignments for the amyloid core region, a set of 2D and 3D chemical-shift correlation spectra was recorded at 0°C (see Materials and Methods). Representative strips from 3D 13C′-15N-13Cα (CONCA), 15N-13Cα-13CX (NCACX), and 15N-13C′-13CX (NCOCX) spectra (Fig. 2) show sequential backbone connectivity for residues A118–V122. The resonance assignments are listed in Table S1 and shown in the 2D spectra in Fig. 1 and Fig. S1. Importantly, no signals from residues 23–111 and 142–144 were observed in any of the experiments, clearly indicating that the amyloid core of huPrP23–144 maps to the region encompassing residues 112–141. Resonances for residues 112, 130, and 141 exhibited notably reduced intensities, and no cross-peaks were observed for residues 128–129 (except M129 13C′, via a weak correlation to L130 15N in the NCO spectrum). This points to the presence within the 112–141 core of regions characterized by varying degrees of conformational flexibility, with the short segment Y128–M129 being especially highly dynamic. Note that the identification of residues within the 112–141 core region is unambiguous. This unambiguousness of assignment, confirmed by multiple 2D and 3D NMR spectra, was facilitated by the fact that several amino acid types with unique 13C chemical-shift patterns (i.e., Ala, Ile, Val) are present only in this part of huPrP23–144.
Fig. 2.
Representative strips from 3D CONCA, NCACX, and NCOCX spectra of huPrP23–144 fibrils showing sequential backbone connectivity for residues A118–V122. Spectra were recorded at a 11.111-kHz MAS rate and 0°C. Acquisition parameters were as follows. CONCA: 24* (t1, 15N) × 24* (t2, 13C′) × 1,000* (t3, 13C) data matrix with acquisition times of 8.3 ms (t1), 8.3 ms (t2), and 20 ms (t3), measurement time of 38.5 h; NCACX: 26* (t1, 15N) × 28* (t2, 13Cα) × 1,400* (t3, 13C) data matrix with acquisition times of 9.0 ms (t1), 4.9 ms (t2), and 28 ms (t3), measurement time of 48.5 h; NCOCX: 26* (t1, 15N) × 24* (t2, 13C′) × 1,400* (t3, 13C) data matrix with acquisition times of 9.0 ms (t1), 8.3 ms (t2), and 28 ms (t3), measurement time of 41.5 h. Cross-peaks are drawn with the lowest contour at ≈8 times the root-mean-square noise level. Strips are labeled by residue number according to the 15N (F1) frequency, 13C F2 frequencies are given inside each strip on the right, and the major cross-peaks corresponding to α, β, and γ carbons, are identified (minor peaks from neighboring spectral planes are marked by asterisks).
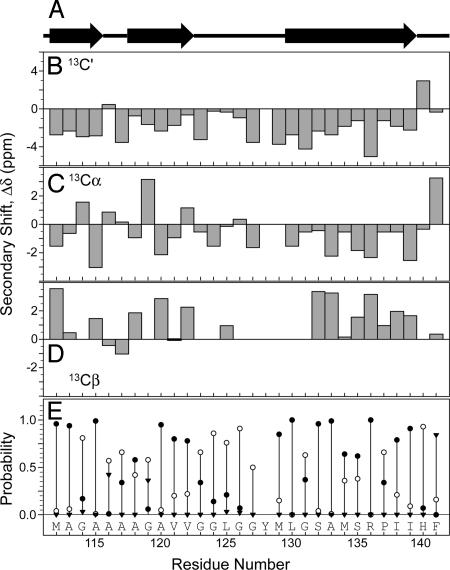
It is well established that NMR chemical shifts are highly sensitive reporters of protein secondary structure, both in solution and the solid phase. Here, the 13C and 15N shifts were used to predict the backbone conformation of the huPrP23–144 amyloid core region. The 13C′, 13Cα, and 13Cβ secondary shifts, Δδ (i.e., the differences between the experimental chemical shifts and the corresponding random-coil values), are plotted as a function of the residue number in Fig. 3. The inspection of these plots reveals that the 13C′ secondary shifts for all but two residues are negative, with an average value of Δδavg = −1.9 ± 1.6 ppm. Furthermore, most 13Cα and 13Cβ secondary shifts are negative (Δδavg = −0.6 ± 1.5 ppm) and positive (Δδavg = 1.5 ± 1.4 ppm), respectively. This immediately indicates that the majority of residues in the huPrP23–144 amyloid core region adopt a β-strand conformation. To further probe this issue, a semiquantitative conformational analysis was performed by using the program PSSI (33). The normalized probabilities of different secondary structure types (i.e., α-helix, β-strand, and coil) and the secondary structure classification are shown in Fig. 3. Altogether, this analysis clearly indicates that most residues between 112 and 122, and 130 and 139 are in a β-strand conformation, whereas unordered structure is predicted for residues 140–141 at the edge of the amyloid core, as well as residues 123–129. The latter region contains four glycines and the highly flexible Y128–M129 segment.
Fig. 3.
Secondary structure of the amyloid core in huPrP23–144 fibrils. (A) Prediction by PSSI. (B–D) Secondary chemical shifts for 13C′ (B), 13Cα (C), and 13Cβ (D). (E) Normalized PSSI probabilities of the secondary structure types: β-strand, filled circles; α-helix, filled inverted triangles; coil, open circles.
Variable Temperature NMR Studies of Conformational Dynamics in huPrP23–144 Amyloid.
As noted above, most of the NMR signals, including those corresponding to the entire N-terminal segment 23–111, were not observed in 0°C experiments with cross-polarization. This suggests that large segments of huPrP23–144 in the amyloid state remain highly dynamic, attenuating nuclear dipole–dipole couplings required for efficient cross-polarization. To confirm the presence of this segmental dynamics, we performed systematic SSNMR studies as a function of temperature between −30 and 30°C. Fig. S3 shows a series of 1D 13C MAS spectra, where the initial 13C magnetization was generated by either 1H-13C CP (i.e., the conditions that “filter out” flexible parts of the protein) or direct polarization (DP) (i.e., a single 90° 13C pulse, which excites with equal efficiency nuclei in both rigid and flexible regions). These experiments may be summarized as follows. First, whereas the CP spectra between −20 and 30°C show only modest temperature-dependent differences, the −30°C spectrum is markedly different, displaying substantially broader signals and an ≈2-fold increase in the total integrated intensity as compared with the spectrum recorded at −20°C. Second, the linewidths in the DP series of spectra progressively decrease as the temperature increases; a particularly striking example is the region corresponding to side chains of His, Phe, Trp, and Tyr (≈100–140 ppm). Third, for all CP/DP pairs recorded between −20 and 30°C, significant differences in peak widths and intensities (i.e., apart from a uniform intensity scaling of the entire spectrum) are observed at each temperature. Only for the −30°C CP/DP pair is the CP spectrum an essentially 2-fold enhanced version of the DP spectrum, as would be expected in the rigid-lattice limit (29). Altogether, these variable-temperature studies confirm the presence of nonuniform backbone and side-chain dynamics of huPrP23–144 molecules in the amyloid fibrils. Although the amyloid core remains highly ordered at all temperatures probed, the entire region between the N terminus and residue 112 exhibits significant conformational dynamics. This segmental flexibility is attenuated at low temperatures, becoming effectively “frozen out” at approximately −30°C.
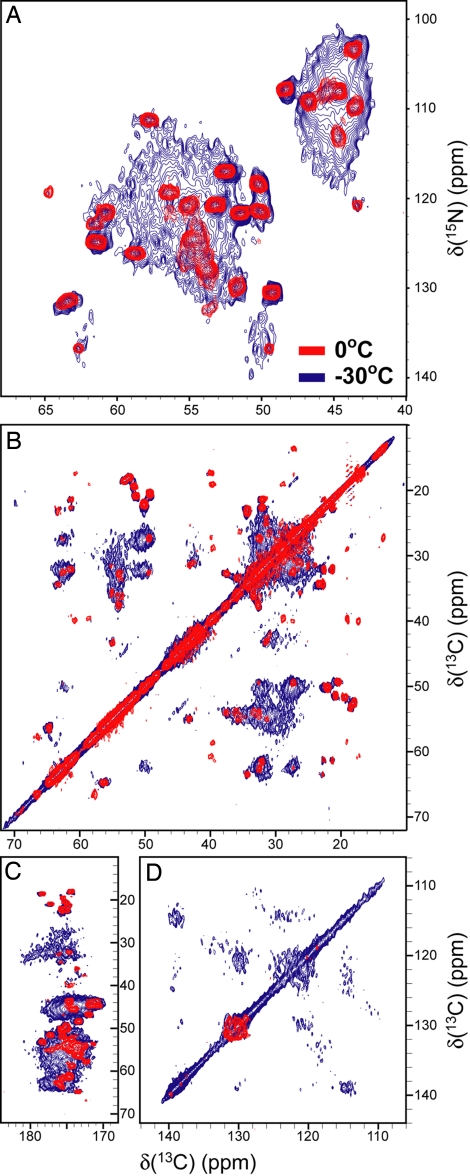
To gain higher-resolution insight into temperature-dependent changes in the segmental flexibility of huPrP23–144 fibrils, we recorded 2D NCA and CC spectra at −30°C, 0°C, and 20°C (Fig. 4 and Fig. S4). Although the 0°C and 20°C spectra are similar to each other, major changes in peak intensities and widths are evident at −30°C. In addition to the amyloid core signals, numerous additional peaks appear at −30°C in all spectral regions associated with backbone and side-chain 15N-13C and 13C-13C correlations. The appearance of these additional resonances indicates that at −30°C parts of the protein outside the amyloid core region become sufficiently rigid to enable efficient dipolar magnetization transfers between 1H, 13C, and 15N nuclei, in complete agreement with the 1D 13C spectra discussed above. In fact, quantitative analysis of the 2D NCA spectra reveals that most residues in huPrP23–144 become rigid at −30°C. Using the integrated cross-peak volumes, it can be estimated that ≈83–120 total residues and ≈35–47 glycines contribute to the −30°C spectrum. Given the approximate nature of such an analysis, these estimates are in reasonable agreement with the expectation that 125 residues in huPrP23–144, including 40 glycines, should contribute to the spectrum in the rigid-lattice limit. Finally, it is notable that, in contrast to the amyloid core resonances, additional cross-peaks that appear at −30°C exhibit substantial inhomogeneous linebroadening. This suggests that the flexible N-terminal region of huPrP23–144 in amyloid fibrils likely exists as an ensemble of largely disordered states characterized by a random-coil-like conformation. This conclusion is further supported by characteristic random-coil chemical shifts observed for all Gly 13Cα and Thr 13Cβ resonances that can be uniquely identified in the 1D 13C DP spectrum recorded at 30°C (Fig. S3I).
Fig. 4.
Two-dimensional NMR spectra of huPrP23–144 fibrils recorded at 0°C (red contours) and −30°C (blue contours). Small regions from NCA (A) and CC (B–D) spectra corresponding to aliphatic–aliphatic (B), aliphatic–carbonyl (C), and aromatic–aromatic (D) correlations (cf. Fig. 1, Fig. S1, and Fig. S4).
Discussion
In addition to cases arising sporadically and those acquired by infection, prion diseases may also have a genetic origin. Over 20 mutations in the PRNP gene have been identified to date that correspond to Gerstmann–Sträussler–Scheinker disease (GSS), familial CJD, and fatal familial insomnia. Unlike in some other forms of prion diseases, a consistent feature of GSS syndrome is extensive accumulation of amyloid fibrils. Systematic biochemical studies have revealed that the major component of this amyloid is the ≈7-kDa PrP fragment spanning residues ≈88 to ≈146, although the exact position of N and C termini can vary between residues 81–88 and 144–153, respectively (34, 35). A similar fragment is also found in amyloid deposits associated with cerebral PrP amyloid angiopathy in individuals with the Y145Stop mutation (16). Furthermore, the region encompassing residues ≈89–140 is an integral part of the minimal sequence shown to sustain prion replication in cell culture and transgenic mice (36, 37), suggesting that it plays an essential role in the PrPC to PrPSc conversion.
The apparent importance of the ≈89–144 region of PrP in the pathogenesis of different forms of prion diseases has prompted numerous biophysical and structural studies with synthetic peptides encompassing this sequence (38–41). However, despite considerable effort, structural characterization of amyloid fibrils formed by these peptides is incomplete, remaining at a relatively low resolution. To bridge this gap, we have recently initiated systematic structural studies of PrP amyloid using MAS SSNMR spectroscopy. Instead of resorting to short synthetic peptide fragments, our present work was performed by using recombinant protein encompassing the entire sequence of a disease-associated human prion protein variant Y145Stop (huPrP23–144). Apart from facilitating studies with a longer polypeptide of obvious biological relevance, another advantage of a recombinant protein is that it provides inexpensive means for the incorporation of 13C,15N-labeled amino acids. SSNMR spectra of fibrils formed by uniformly 13C,15N-labeled huPrP23–144 were of surprisingly high resolution, allowing an unambiguous assignment of resonances corresponding to all amino acid residues showing a high degree of immobilization.
The key finding of the present study is that the relatively rigid and highly ordered core of huPrP23–144 amyloid maps to the C-terminal region encompassing residues 112–141, whereas the entire N-terminal sequence 23–111 and a short segment 142–144 are highly flexible, lacking any ordered organization. Chemical-shift analysis indicates that the core region contains β-strands between residues 112–122 and 130–139, with residues 123–129 likely forming a partially flexible loop. The 112–122 segment may not consist of a single continuous strand but rather two shorter strands (residues 112–115 and 118–122) separated by a short, rigid non-β segment 116–117 (Fig. 3). Interestingly, whereas in the native structure of the full-length human prion protein monomer residues 128 and 129 are part of β-strand 128–131 (42), in the huPrP23–144 amyloid structure these residues appear to be in a less ordered conformation. Although the present data are insufficient to deduce a high-resolution structural model, the general folding motif for huPrP23–144 in amyloid fibrils appears similar to the β-strand–bend–β-strand motif first identified in Aβ(1–40) amyloid (3) and later found in many other amyloid fibrils (23–26).
Previously, SSNMR was used to probe the structure of a 55-residue synthetic peptide corresponding to mouse PrP segment 89–143 with P101L mutation (38). The authors concluded that upon aggregation the peptide “appears to convert completely to an extended, β-sheet-like form.” However, this conclusion was based on relatively limited chemical-shift data for only six residues (corresponding to G94, A115, V121, M129, A132, and G142 in the sequence of human PrP) and, thus, was rather general in nature. A subsequent, more detailed study focused specifically on residues 112–124 (113–125 in the human sequence), indicating β-sheet conformation in this region (41). A similar region has been also found to comprise the β-sheet core in fibrils formed by a short PrP fragment 106–126 (39). Although the present study with huPrP23–144 amyloid supports the existence of a β-strand(s) between residues 112 and 122, our detailed spectroscopic data are clearly at odds with the notion that essentially the entire PrP90–144 sequence is part of an extended β-sheet structure. In fact, there is conclusive evidence that the regions spanning residues 23–111 (including G94) and 141–144 (including G142) are highly flexible, falling outside the ordered β-sheet core of the amyloid. This apparent discrepancy between the present data and conclusions reached in the previous SSNMR study with mouse P101L PrP89–143 may be due to intrinsically different conformational preferences of residues ≈90–111 in the shorter peptide and PrP23–144 containing the entire N-terminal sequence, different sample preparation methods (P101L PrP89–143 aggregates were obtained by precipitation from an acetonitrile:acetate-buffered saline mixture and the solid material was completely dried before NMR measurements, whereas the present study was performed by using highly homogenous population of fully hydrated fibrils prepared by using physiologically relevant buffer conditions), or very limited direct experimental data in the previous study for the region N-terminal to residue 113. The last possibility is especially plausible given that the presence of β-structure in this region in P101L PrP89–143 was proposed solely on the basis of a single Cα chemical-shift measurement for G94, with no information for other residues (38). Furthermore, chemical-shift-based analysis of secondary structure for glycines tends to be less reliable than for other residues. One could also argue that relatively small species-dependent sequence differences and/or the P101L (P102L in the human sequence) mutation dramatically affect conformational preferences of the 90–111 region of the prion protein. This is, however, highly unlikely given essentially identical amide I infrared spectra of amyloid fibrils for human, mouse, and the P102L variant of human PrP23–144 (20, 43).
The PK-resistant core of the infectious PrPSc starts at residue ≈90. Previous modeling efforts based on electron microscopic data for 2D crystals proposed that the entire sequence 90–175 forms left-handed β-helices (44). A fundamentally different picture was deduced from molecular dynamics simulations at low pH, postulating a PrPSc model consisting of six short β-strands between residues 90–165 (five within the 90–144 region), with the entire α-helical domain of PrPC being preserved (45). The structure experimentally observed in the present study for huPrP23–144 fibrils is at odds with the motifs predicted for the 90–144 region by both of these theoretical models. Most intriguingly, recent studies with amyloid fibrils formed spontaneously by the recombinant full-length prion protein (PrP23–231) or PrP90–231 consistently indicate that the β-sheet core in these fibrils maps to the C-terminal region starting at residue ≈160, with the entire N-terminal portion of the protein lacking significant PK-resistance (46, 47) and showing no evidence for β-structure in direct biophysical experiments (47, 48). Although further studies are needed to explain the conundrum of apparently different structural preferences for residues 90–144 in fibrils formed by C-terminally truncated huPrP23–144 and PrP variants containing the entire C-terminal domain, the present observations call for great caution in extrapolating data for fragments of amyloidogenic proteins to infer structural properties of same sequences within fibrils formed by respective full-length proteins. Given these caveats, as well as obvious questions as to whether the high-resolution structures of amyloid fibrils formed in vitro are the same as those found in diseased brain, our study falls short of resolving the ongoing controversy regarding the structure of PrPSc. Nevertheless, detailed structural data for huPrP23–144 amyloid are of major interest in the context of familial prion diseases such as GSS syndrome and familial cerebral angiopathy with 145Y mutation, in which cases the C-truncated PrP fragments are of direct relevance to the disease pathogenesis.
Apart from its central role in some familial prion diseases, PrP23–144 has been recently shown to provide an invaluable model for studying basic mechanistic principles underlying the phenomenon of prion strains, as well as the molecular basis of barriers in prion propagation (19, 20). In particular, previous studies have demonstrated that the seeding specificity and strain characteristics of PrP23–144 are fully encoded in fibril conformation, and that these properties depend on the nature of amino acid residues at positions 138 and 139. However, the conformational correlates established in these studies were of intrinsically low resolution. The present work reveals that residues 138 and 139 in huPrP23–144 are located just at the edge of the C-terminal strand of the β-sheet core, a strategic location that may explain the role of these two residues in controlling fibril conformation as well their capacity for species- and strain-selective recruitment of the protein monomer. Further extension of this work to interatomic distance measurements—together with inclusion of PrP23–144 amyloid variants used in the previous study (20)—should allow the construction of atomic-resolution structural models, providing a full understanding of the structural basis of PrP23–144 amyloid strains.
Materials and Methods
Expression and Purification of huPrP23–144.
The plasmid encoding huPrP23–144 with N-terminal linker containing His6-tag and a thrombin cleavage site is described in ref. 49. Uniformly 13C,15N-labeled huPrP23–144 was expressed in Escherichia coli by using a minimal medium with 13C-glucose (3 g/liter) and 15NH4Cl (1 g/liter) (Cambridge Isotope Laboratories) as the sole carbon and nitrogen sources, respectively, and purified by using a nickel-nitrilotriacetic acid agarose resin (19). The His6-tag was cleaved by using biotinylated thrombin (Novagen), the thrombin was sequestered by using streptavidin-agarose beads, and the residual His6-tag was removed by dialysis against ultrapure water. The purified 126-residue protein consisted of the huPrP23–144 sequence with an N-terminal Gly-Ser-Asp-Pro extension. The protein was stored as a lyophilized powder and had a final purity of >95% as determined by SDS/PAGE.
Preparation of huPrP23–144 Amyloid Fibrils.
Lyophilized huPrP23–144 was dissolved in ultrapure water at a concentration of 400 μM, and fibrilization was initiated as described previously (14, 19) by addition of potassium phosphate buffer (pH 6.4) to a final concentration of 50 mM. Fibrils were allowed to form at 25°C under the conditions of gentle rotation at 8 rpm. Before NMR analysis, amyloid fibril samples were characterized by atomic force microscopy (Fig. S5; see refs. 14 and 20 for experimental details). Fibrils (≈12 mg of 13C,15N protein) were pelleted by centrifugation, washed several times with 50 mM potassium phosphate buffer (pH 6.4), and packed by using low-speed centrifugation into a 3.2-mm, limited-speed, 36-μl Varian SSNMR rotor. The rotor was sealed by using custom-made spacers to prevent sample dehydration during experiments.
SSNMR Spectroscopy.
Spectra were recorded by using a three-channel, 500-MHz Varian spectrometer equipped with a 3.2-mm BioMAS probe in the 1H–13C–15N configuration. The sample temperature was controlled by a stream of compressed air delivered to the sample via a variable-temperature (VT) stack. All temperature values listed in the article consistently refer to the temperature of the VT gas at the sample; under the experimental conditions used, the average sample temperatures were 5 ± 2°C higher than the VT gas temperature, as determined by lead nitrate calibration.
The pulse schemes used in this work are analogous to those used in previous SSNMR studies of proteins (32). The following 2D and 3D experiments were used to establish sequential resonance assignments of the huPrP23–144 amyloid core region at 0°C: 2D 13C-13C (CC), 2D 15N-13Cα (NCA), 2D 15N-13C′ (NCO), 2D 15N-(13Cα)-13CX (N(CA)CX), 2D 15N-(13C′)-13CX (N(CO)CX), 3D 13C′-15N-13Cα (CONCA), 3D 15N-13Cα-13CX (NCACX), and 3D 15N-13C′-13CX (NCOCX). Where applicable, the 15N-13Cα/13C′ correlations were established by using adiabatic SPECIFIC CP (30) (τmix,NCA/τmix,NCO = 3.5/6.0 ms), and 13C-13C magnetization transfer was achieved by using dipolar assisted rotational resonance (50) (τmix = 10 ms for 2D CC and 15 ms otherwise). Two-pulse phase modulated proton decoupling (51) at a field strength of ≈70 kHz was used during all chemical-shift evolution periods. Data were processed by using NMRPipe (52).
Supplementary Material
Acknowledgments.
This work was supported by the Ohio State University (startup funds to C.P.J.) and National Institutes of Health Grants NS 44158 and NS 38604 (to W.K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711716105/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Tycko R. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr Opin Struct Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid-state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luhrs T, et al. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawaya MR, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 8.Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 9.Legname G, et al. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 10.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 13.Wickner RB, et al. Prions: Proteins as genes and infectious entities. Genes Dev. 2004;18:470–485. doi: 10.1101/gad.1177104. [DOI] [PubMed] [Google Scholar]
- 14.Kundu B, et al. Nucleation-dependent conformational conversion of the Y145Stop variant of human prion protein: Structural clues for prion propagation. Proc Natl Acad Sci USA. 2003;100:12069–12074. doi: 10.1073/pnas.2033281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore RA, et al. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci. 2006;15:609–619. doi: 10.1110/ps.051822606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghetti B, et al. Vascular variant of prion protein cerebral amyloidosis with τ-positive neurofibrillary tangles: The phenotype of the stop codon 145 mutation in PRNP. Proc Natl Acad Sci USA. 1996;93:744–748. doi: 10.1073/pnas.93.2.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrPSc. J Mol Biol. 2005;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 18.Apetri AC, Vanik DL, Surewicz WK. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90–231. Biochemistry. 2005;44:15880–15888. doi: 10.1021/bi051455+. [DOI] [PubMed] [Google Scholar]
- 19.Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell. 2004;14:139–145. doi: 10.1016/s1097-2765(04)00155-8. [DOI] [PubMed] [Google Scholar]
- 20.Jones EM, Surewicz WK. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Heise H, et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloepper KD, et al. Temperature-dependent sensitivity enhancement of solid-state NMR spectra of α-synuclein fibrils. J Biomol NMR. 2007;39:197–211. doi: 10.1007/s10858-007-9189-z. [DOI] [PubMed] [Google Scholar]
- 23.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxa U, et al. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 26.Iwata K, et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc Natl Acad Sci USA. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaroniec CP, et al. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 30.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- 31.Siemer AB, et al. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 32.McDermott AE. Structural and dynamic studies of proteins by solid-state NMR spectroscopy: Rapid movement forward. Curr Opin Struct Biol. 2004;14:554–561. doi: 10.1016/j.sbi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Wang YJ, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagliavini F, et al. Amyloid fibrils in Gerstmann–Sträussler–Scheinker disease (Indiana and Swedish kindreds) express only PrP peptides encoded by the mutant allele. Cell. 1994;79:695–703. doi: 10.1016/0092-8674(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 35.Tagliavini F, et al. A 7-kDa prion protein (PrP) fragment, an integral component of the PrP region required for infectivity, is the major amyloid protein in Gerstmann–Sträussler–Scheinker disease A117V. J Biol Chem. 2001;276:6009–6015. doi: 10.1074/jbc.M007062200. [DOI] [PubMed] [Google Scholar]
- 36.Muramoto T, Scott M, Cohen FE, Prusiner SB. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Supattapone S, et al. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 38.Laws DD, et al. Solid-state NMR studies of the secondary structure of a mutant prion protein fragment of 55 residues that induces neurodegeneration. Proc Natl Acad Sci USA. 2001;98:11686–11690. doi: 10.1073/pnas.201404298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuwata K, et al. NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106–126. Proc Natl Acad Sci USA. 2003;100:14790–14795. doi: 10.1073/pnas.2433563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmona M, et al. Structural properties of Gerstmann–Sträussler–Scheinker disease amyloid protein. J Biol Chem. 2003;278:48146–48153. doi: 10.1074/jbc.M307295200. [DOI] [PubMed] [Google Scholar]
- 41.Lim KH, et al. Solid-state NMR structural studies of the fibril form of a mutant mouse prion peptide PrP89–143(P101L) Solid State Nucl Magn Reson. 2006;29:183–190. doi: 10.1016/j.ssnmr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Zahn R, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones EM, Surewicz K, Surewicz WK. Role of N-terminal familial mutations in prion protein fibrillization and prion amyloid propagation in vitro. J Biol Chem. 2006;281:8190–8196. doi: 10.1074/jbc.M513417200. [DOI] [PubMed] [Google Scholar]
- 44.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed β-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeMarco ML, Daggett V. Molecular mechanism for low pH triggered misfolding of the human prion protein. Biochemistry. 2007;46:3045–3054. doi: 10.1021/bi0619066. [DOI] [PubMed] [Google Scholar]
- 46.Bocharova OV, Breydo L, Salnikov VV, Gill AC, Baskakov IV. Synthetic prions generated in vitro are similar to a newly identified subpopulation of PrPSc from sporadic Creutzfeldt–Jakob Disease. Protein Sci. 2005;14:1222–1232. doi: 10.1110/ps.041186605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu X, Wintrode PL, Surewicz WK. β-Sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci USA. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morillas M, Swietnicki W, Gambetti P, Surewicz WK. Membrane environment alters the conformational structure of the recombinant prion protein. J Biol Chem. 1999;274:36859–36865. doi: 10.1074/jbc.274.52.36859. [DOI] [PubMed] [Google Scholar]
- 50.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 51.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6957. [Google Scholar]
- 52.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. 0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.