Abstract
The PGC-1 family of regulated coactivators (PGC-1α, PGC-1β, and PRC) plays an important role in directing respiratory gene expression in response to environmental signals. Here, we show that PRC and PGC-1α differ in their interactions with nuclear hormone receptors but are highly similar in their direct binding to several nuclear transcription factors implicated in the expression of the respiratory chain. Surprisingly, neither coactivator binds NRF-2(GABP), a multisubunit transcriptional activator associated with the expression of many respiratory genes. However, the NRF-2 subunits and PRC are co-immunoprecipitated from cell extracts indicating that the two proteins exist in a complex in vivo. Several lines of evidence indicate that HCF-1 (host cell factor 1), a major chromatin component, mediates the association between PRC and NRF-2. Both PRC and NRF-2β bind HCF-1 in vitro, and the molecular determinants required for the interactions of each with HCF-1 are also required for PRC trans-activation through promoter-bound NRF-2. These determinants include a consensus HCF-1 binding site on PRC and the NRF-2 activation domain. In addition, PRC and NRF-2β can complex with HCF-1 in vivo, and all three associate with NRF-2-dependent nuclear genes that direct the expression of the mitochondrial transcription factors, TFB1M and TFB2M. Finally, short hairpin RNA-mediated knock down of PRC protein levels leads to reduced expression of TFB2M mRNA and mitochondrial transcripts for cytochrome oxidase II (COXII) and cytochrome b. These changes in gene expression coincide with a marked reduction in cytochrome oxidase activity. The results are consistent with a pathway whereby PRC regulates NRF-2-dependent genes through a multiprotein complex involving HCF-1.
Mitochondria produce the bulk of cellular energy through their oxidation of pyruvate and fatty acids. Chemical bond energy is converted to reducing equivalents that are used by the electron transport chain of the inner mitochondrial membrane to establish an electrochemical proton gradient. Dissipation of this gradient drives the synthesis of ATP and the generation of heat (1, 2). Mitochondria are semiautonomous in that they contain their own genetic system based on a multicopy mitochondrial DNA (mtDNA) genome. In vertebrates, a covalently closed circular mtDNA of ∼16.5 kilobases encodes 13 essential protein subunits of respiratory complexes I, III, IV, and V along with the 22 tRNAs and 2 rRNAs required for their translation within the mitochondrial matrix (3–5). This limited coding capacity necessitates that nuclear genes specify most of the numerous gene products required for the molecular architecture and biochemical functions of the organelle (6, 7). These include the majority of respiratory chain subunits, all of the protein constituents of the mitochondrial translation system, and all of the gene products required for the transcription and replication of mtDNA.
At the transcriptional level, nucleo-mitochondrial interactions rely upon nucleus-encoded transcription factors and transcriptional coactivators. Certain of these factors direct the transcription of mtDNA, whereas others act on nuclear genes required for the biogenesis and function of the organelle (5, 7). Among the latter are the nuclear respiratory factors, NRF-12 and NRF-2(GABP). These proteins were identified as activators of cytochrome c (8, 9) and cytochrome oxidase (10) genes and have subsequently been associated with the expression of many genes whose products contribute essential mitochondrial functions, particularly those related to the respiratory apparatus (6, 7). In addition, both factors have also been implicated in functions related to cell proliferation (11, 12), results consistent with the early embryonic lethality associated with targeted disruptions of NRF-1 (13) or NRF-2(GABP) (14) in mice.
In addition to these transcription factors, members of the PGC-1 family of inducible coactivators act as intermediaries between the environment and the transcriptional machinery specifying a number of important pathways related to cellular energetics (15, 16). PGC-1α, the founding member of the family, was originally identified for its role in adaptive thermogenesis in brown fat (17). The coactivator is induced robustly in brown fat in response to cold exposure and participates in the induction of uncoupling protein 1. In addition, PGC-1α orchestrates a program of mitochondrial biogenesis in part by serving as a trans-activator of NRF-1 and NRF-2 target genes (18). The coactivator binds NRF-1 in a manner similar to that observed for PPARγ and directs expression of respiratory subunits as well as mtDNA transcription and replication factors (18, 19). PGC-1β, a close relative of PGC-1α, also functions as a NRF-1 coactivator (20) but differs from PGC-1α in mediating biological responses in liver and muscle (21, 22).
A third PGC-1 family member was designated as PRC (PGC-1-related coactivator) (23). Although divergent from PGC-1α in overall sequence, PRC has a number of structural features that are spatially conserved including a potent amino-terminal activation domain, a central proline-rich region, an arginine/serine rich domain (R/S domain), and an RNA recognition motif. However, PRC differs from PGC-1α in that it is not induced significantly during adaptive thermogenesis but, rather, exhibits the properties of a cell growth regulator (23). PRC mRNA and protein are markedly down-regulated when cultured cells exit the cell cycle as a result of serum starvation or contact inhibition. The mRNA and protein are also rapidly induced upon serum stimulation of quiescent cells in the G0 to G1 transition. This induction is insensitive to cycloheximide and, thus, occurs in the absence of de novo protein synthesis (24). Moreover, cycloheximide treatment leads to super induction and stabilization of PRC mRNA. These properties are characteristic of the class of immediate early genes whose rapid responses to growth factors represent the earliest events in the genetic program leading to cell proliferation (25).
Like PGC-1α, PRC binds NRF-1 both in vitro and in vivo and directs the expression of NRF-1 target genes related to respiratory chain expression (19, 23). In addition, both PRC and PGC-1α are known to utilize NRF-2 binding sites to trans-activate NRF-2-dependent promoters in transfected cells (19). However, neither coactivator has been shown to interact directly with NRF-2. This suggests that PRC or PGC-1α coactivation through NRF-2 may require a third party that binds both the transcription factor and the coactivators. An ideal candidate for such a role is host cell factor-1 (HCF-1). HCF-1 is an abundant, chromatin-associated protein that was first identified through its participation in the VP16 activation of the herpes simplex virus immediate-early genes (26). A large 2035-amino acid HCF-1 precursor is cleaved autocatalytically to generate multiple amino- and carboxyl-terminal fragments that remain associated noncovalently (27, 28). HCF-1 is expressed ubiquitously and is required for cell cycle progression. A temperature-sensitive mutation in the β-propeller domain of HCF-1 results in G0/G1 arrest at the nonpermissive temperature (29). The cell cycle arrest is reversed at the permissive temperature, and the cells reenter the proliferative cycle. Moreover, specific HCF-1 subunits promote exit from mitosis and progression through G1 (30).
In addition to its interaction with VP16, HCF-1 binds NRF-2(GABP) through the transcriptional activation domain on the NRF-2β(GABPβ) subunit (31). Mutations that interfere with NRF-2(GABP) trans-activation also block binding to HCF-1, suggesting that HCF-1 functions as a NRF-2(GABP) coactivator. Here, we establish that PRC exists in a complex with HCF-1 and NRF-2β. The sequence requirements for interactions between PRC and HCF-1 and between HCF-1 and NRF-2β are the same as those required for PRC trans-activation of NRF-2-dependent transcription. Finally, chromatin immunoprecipitations coupled with loss of function experiments demonstrate that the PRC-containing complex associated with the promoter of a key mitochondrial transcription factor contributes to the expression of mitochondrial transcripts and respiratory enzyme activity. The results establish that HCF-1 is a functional intermediary in the PRC trans-activation of at least a subset of NRF-2 target genes required for mitochondrial respiratory function.
EXPERIMENTAL PROCEDURES
Plasmids—A PRC expression vector was constructed from pBSII/N-myc FL-PRC, a modified derivative of the original pBSII/FL-PRC (23), by inserting a XhoI/NotI restriction fragment containing the full-length PRC coding region into SalI/NotI-digested pSV Sport. This vector, pSV Sport/N-myc FL-PRC, was used as a template to delete the HCF-1 binding site (ΔDHDY: GACCATGACTAT) by PCR using a previously described strategy (24). The resulting DraIII/NotI PRC fragment containing the internal deletion of the codons specifying the DHDY HCF-1 binding site was then subcloned into DraIII/NotI-digested pSV Sport/N-myc FL-PRC to generate pSV Sport/N-myc PRC (ΔDHDY). The Gal4-NRF-2β fusion constructs including the full-length NRF-2β as well as those containing only the activation domain and its variants with alanine substitution mutations have been described (32, 33).
Plasmids pSG5/CREB-HA (24) and pCGN HCF(2–2035)9E10 (27) were constructed as described. The ERRα coding region used for the construction of the ERRα expression vector pSG5/ERRα-HA was generated by PCR using HeLa cDNA as template. The resulting PCR product was digested with BamHI/BglII and cloned into BamHI/BglII-digested pSG5. The NRF-2β expression vector, pSG5/NRF-2β-HA, was constructed by incorporating the hemagglutinin (HA) tag into the coding region from the original NRF-2β cDNA clone (32) by PCR. An activation domain deletion (ΔTAD) was introduced into the NRF-2β coding region by cutting the plasmid pSG5/NRF-2β-HA with PstI and re-ligating to generate pSG5/NRF-2β-HA(ΔTAD). This resulted in an in-frame 88-codon deletion encompassing NRF-2β amino acids 255–342. This deletion removed the entire NRF-2β transcriptional activation domain (ΔTAD), which was mapped previously to amino acids 258–327 (32, 33).
Coimmunoprecipitation and Immunoblotting—Immunoprecipitations were carried out using either untransfected 293FT cells or cells transfected with hemagglutinin-tagged proteins. This human cell line was used for immunological methods because our antibodies were developed against the human factors, the cells exhibit abundant constitutive expression of PRC, and they have a high transfection efficiency. Untransfected 293FT cells were grown to ∼70% confluence and harvested for the preparation of cell extract. Hemagglutinin-tagged proteins were expressed by electroporating ∼4.8 × 106 293FT cells with pCGN HCF(2–2035)9E10 (60 μg), pSG5/CREB-HA (20 μg), pSG5/ERRα-HA (50 μg), or pSG5/NRF-2β-HA (40 μg). Cells were plated in 15-cm tissue culture dishes and maintained at 37 °C for ∼48 h. Extracts from untransfected and transfected cells were prepared by suspending cells in Nonidet P-40 lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40) containing mini-complete protease inhibitor mixture (Roche Applied Science) as described (23, 24). Protein concentrations were measured by the Bradford assay (Bio-Rad). Immunoprecipitations were performed by adding 15 μl of rabbit pre-immune serum control, 15 μl of rabbit anti-PRC (1047–1379) serum (24), 2 μl of rabbit anti-HCF-1 serum (a generous gift from Winship Herr, University of Lausanne), 10 μl of rabbit anti-NRF-2α or 15 μl of rabbit anti-NRF-2β serum to 400–800 μg of whole-cell extract in a total volume of 250 μl of Nonidet P-40 lysis buffer. Reactions were incubated at 4 °C overnight on a rocking table followed by the addition of 20 μl of protein A-agarose (Roche Applied Science). After an additional 3 h of incubation at 4 °C, immunoprecipitates were centrifuged at 12,000 × g for 20 s at 4 °C, washed 4 times with 500 μl of Nonidet P-40 lysis buffer, and resuspended in 25 μl of 2× sample buffer containing β-mercaptoethanol. For detection of PRC and HCF by immunoblotting, samples were subjected to electrophoresis on 8.5% denaturing acrylamide gels and transferred to nitrocellulose membranes (Schleicher & Schuell) with high molecular weight buffer as described (24). For detection of CREB, ERRα, and NRF-2β, precipitates were subjected to electrophoresis on 12% denaturing acrylamide gels and transferred to nitrocellulose membranes using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad) with Towbin transfer buffer (23). Immunoblots were probed with either rat monoclonal high affinity (3F10) anti-HA-peroxidase antibody (Roche Applied Science), rabbit anti-HCF antibody, or rabbit anti-PRC-(1047–1379) serum. Proteins were visualized using SuperSignal West Pico chemiluminescent substrate (Pierce).
Transfections—Transient transfections of BALB/3T3 cells were performed by calcium phosphate precipitation as described (23). This cell line was utilized for transfections because conditions for PRC trans-activations were originally developed using these cells (19, 23, 24). BALB/3T3 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% calf serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). Cells were plated at a density of 2600 cells per cm2 in 6-well plates and transfected with 0.6 μg of 5×Gal4/Luc reporter and 45 ng of pRL-null control vector (Promega) together with different Gal4-NRF-2β fusion constructs. PRC trans-activations were performed by including either pSV Sport/N-myc FL-PRC or pSV Sport/N-myc PRC (ΔDHDY) lacking the HCF-1 binding site (ΔDHDY). After 5–6 h, cells were washed twice with Dulbecco's phosphate-buffered saline (Invitrogen) and grown for additional 40 h in fresh media. Cell extracts were prepared, and luciferase assays were performed using the dual luciferase reporter assay system (Promega). Firefly luciferase activity from the 5×Gal4/Luc reporter construct was normalized to Renilla luciferase luminescence from the pRL-null control vector.
S-tag Pulldown Assay—Pulldown assays were performed as described (23, 24). Binding of PRC and PGC-1α to the nuclear hormone receptors PPARγ, TRβ, and RAR was determined in the presence and absence of 1 μm receptor ligands MCC-555, tri-iodothyronine, and 9-trans-retinoic acid, respectively.
Mobility Shift Assays—NRF-2α and -β subunits were translated in vitro as performed for the S-tag pulldown assays except for the omission of radiolabeled methionine. Subunits were subjected to mobility shift assay using a 32P-labeled cytochrome oxidase subunit IV promoter oligonucleotide containing tandem NRF-2 recognition sites as described previously (19).
Chromatin Immunoprecipitation—Chromatin immunoprecipitations were performed on 293FT cells as described (24) using rabbit anti-NRF-2β, rabbit anti-PRC-(1047–1379) (24), and rabbit anti-HCF-1 antibodies (a generous gift from Winship Herr, University of Lausanne) along with rabbit IgG as a control (Sigma). Immunoprecipitated promoter fragments were quantitated by real-time PCR on the ABI PRISM 7900HT Sequence detection system with the SYBR Green PCR Mastermix (Applied Biosystems). The primers used for real-time PCR were specific for the human TFB1M and TFB2M promoter (19). Amplifications were performed in triplicate in each chromatin immunoprecipitation experiment, and the results were quantitated using the ΔΔCt method (34) and expressed as the average of three independent experiments ± S.E.
Histochemistry—For histochemical staining of cytochrome c oxidase activity (35), cells grown on glass coverslips were air-dried for 1 h at room temperature and then preincubated with 1 mm CoCl2 in 50 mm Tris-HCl, pH 7.6, containing 10% sucrose for 15 min at room temperature. After rinsing with 0.1 m sodium phosphate, pH 7.6, containing 10% sucrose, the cells were incubated for 6 h at 37 °C in incubation medium (10 mg of cytochrome c, 10 mg 3,3′-diaminobenzidine hydrochloride, 2 mg of catalase, 10% sucrose in 0.1 m sodium phosphate, pH 7.6). The coverslips were rinsed in 0.1 m sodium phosphate, pH 7.6, containing 10% sucrose and mounted in VectaMount AQ (Vector Laboratories).
Real-time Quantitative Reverse Transcription-PCR—Transcript levels were quantitated by real-time reverse transcription-PCR by extracting total RNA using Trizol (Invitrogen) from U2OS cells washed in phosphate-buffered saline. RNA samples were then DNase-treated with the Turbo DNA-free kit (Ambion) and reverse-transcribed with random hexamer primers and the TaqMan reverse transcription reagents kit (Applied Biosystems) according to the manufacturer's instructions. The reverse-transcribed RNA was then amplified by real-time PCR using the ABI PRISM 7900HT Sequence detection system with the Power SYBR Green PCR Mastermix (Applied Biosystems). The primers used for real-time were specific for the following genes: PRC (hPRC sybr sense (S), AGTGGTTGGGGAAGTCGAAG; hPRC sybr antisense (AS), CCTGCCGAGAGAGACTGAC), TFB1M (hTFB1 sybr S, CCTCCGTTGCCCACGATTC; hTFB1 sybr AS, GCCCACTTCGTAAACATAAGCAT), TFB2M (hTFB2 sybr S, CGCCAAGGAAGGCGTCTAAG; hTFB2 sybr AS, CTTTCGAGCGCAACCACTTTG), COXII (hCOXII sybr S, ACAGATGCAATTCCCGGACGTCTA; hCOXII sybr AS, GGCATGAAACTGTGGTTTGCTCCA), hcytochrome b (hcytb sybr S, AATTCTCCGATCCGTCCCTA; hcytb sybr AS, GGAGGATGGGGATTATTGCT), and β-actin (hβ-actin S, CATGTACGTTGCTATCCAGGC; hβ-actin AS, CTCCTTAATGTCACGCACGAT). Reactions were carried out using the following conditions: an initial step of 2 min at 50 °C and 10 min at 95 °C followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C. The results were analyzed using the Relative Quantification Study program with SDS 2.1 software (Applied Biosystems). Samples were analyzed in triplicate, and mRNA quantities were normalized to 18 S RNA. Relative gene expression levels were determined by the comparative Ct method and expressed as the average of at least three separate determinations ± S.E.
Generation of Lentivirus Transductants Expressing shRNA—Double-stranded oligonucleotides targeting the PRC gene (PRCsh#1S, CACCGCCATCAGGACATCACCATCACGAATGATGGTGATGTCCTGATGGC; PRCsh#1AS, AAAAGCCATCAGGACATCACCATCATTCGTGATGGTGATGTCCTGATGGC) and a negative control sequence derived from the MISSION nontarget shRNA control vector (Sigma) (control shS, CACCCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG; control shAS, AAAACAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG) were ligated into the pENTR/U6 vector using the BLOCK-iT U6 RNAi Entry Vector kit (Invitrogen). The control hairpin contains four base pair mismatches to any known human or mouse gene (36). The resulting entry vectors were designated pENTR/PRCshRNA#1 and pENTR/control. The lentiviral expression vectors pLenti/PRCshRNA#1 and pLenti/control and pLenti-GW/U6-LaminshRNA were generated by transferring the U6-PRC and U6-control and U6-Lamin RNA-mediated interference cassettes into the pLenti6/BLOCK-iT DEST vector using the LR recombination reaction. Lentiviral particles of these constructs were generated in 293FT cells using the BLOCK-iT Lentiviral RNAi Expression system according to the manufacturer's protocol (Invitrogen). U2OS cells were transduced with each lentiviral construct at a multiplicity of infection of 10, and stable shRNA-expressing clones were selected with blasticidin. U2OS cells were used because they are a human cell line that exhibits regulated expression of PRC (24). Clones were cultured, and cell lysates were prepared and analyzed by immunoblotting using an anti-lamin A/C antibody (a generous gift from Robert Goldman, Northwestern University) and rabbit anti-PRC-(1047–1379) (24).
RESULTS
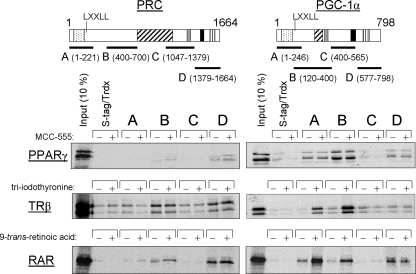
Similarities and Differences in Transcription Factor Recognition by PRC and PGC-1α—PRC is similar to PGC-1α in both its structure and in its ability to trans-activate NRF target genes. Here, we compare the two coactivators for their ability to interact with relevant transcription factors using a thioredoxin pulldown assay. As shown in Fig. 1, PRC differs from PGC-1α in its interaction with several nuclear hormone receptors. PRC shows little if any specific interaction with PPARγ under conditions where specific binding of PPARγ to PGC-1α subfragments A, B, and D is evident. The results also confirm that the interaction between PGC-1α and PPARγ is ligand-independent since MCC-555, a thiazolidinedione ligand for PPARγ, fails to enhance the signal. In contrast to PRC, PGC-1α engages in ligand-dependent binding to both TRβ and RAR through a domain containing the LXXLL coactivator signature motifs (17). This result is confirmed in Fig. 1, which shows ligand-dependent binding of PGC-1α subfragments A and B to both TRβ and RAR. Under similar conditions, only weak ligand-independent binding is observed to PRC subfragments B and D, neither of which contains the LXXLL motif. A PRC fragment bounded by amino acids 1–700 spanning fragments A and B also exhibited a weak ligand-independent interaction with both nuclear hormone receptors (not shown). These results are suggestive of functional differences between the two coactivators in their interactions with nuclear hormone receptors.
FIGURE 1.
Comparison of nuclear hormone receptor binding to PRC and PGC-1α. The in vitro binding of PRC and PGC-1α subfragments to the nuclear hormone receptors PPARγ, TRβ, and RAR was determined by S-tag pulldown assay as described under “Experimental Procedures.” Trdx, thioredoxin. Schematic representation of PRC and PGC-1α is shown above with the various functional domains indicated (stippled, activation domain; cross-hatched, proline-rich region; gray-shaded, consensus recognition site (DHDY) for host cell factor (HCF); solid, R/S domain; vertical-hatched, RNA recognition motif). Subfragments of each coactivator denoted as A, B, C, or D with their amino acid coordinates shown in parentheses were used in S-tag pulldown assays with 35S-labeled nuclear hormone receptor. Binding of the various subfragments to each 35S-radiolabeled receptor was compared with that of S-tagged thioredoxin as a negative control. Ligand-dependent binding was determined by inclusion of the indicated receptor ligand in the binding reaction as described under “Experimental Procedures.” Bound proteins were eluted from the S-protein-agarose and visualized by autoradiography.
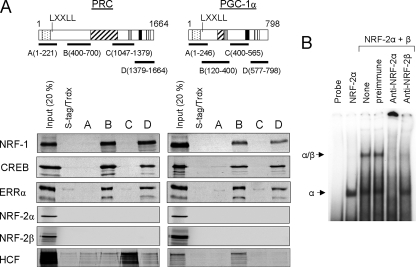
PRC and PGC-1α were also compared for their ability to bind transcription factors implicated in the expression of the mitochondrial respiratory chain. As shown in Fig. 2A, both PRC and PGC-1α bind NRF-1, CREB, and ERRα specifically through their respective subfragments B and D (24). The binding specificity is demonstrated by the fact that neither the thioredoxin control nor other subfragments (A or C) of either coactivator bind any of these transcription factors. CREB has been associated with the trans-activation of cytochrome c expression by PRC and is known to bind the same sites as NRF-1 within PRC subfragments B and D (24). The orphan nuclear hormone receptor ERRα is a target for PGC-1α-directed mitochondrial biogenesis (37). Notably, both PRC and PGC-1α bind ERRα through the same subfragments used for their interactions with NRF-1 and CREB (Fig. 2A).
FIGURE 2.
Comparison of transcription factor binding to PRC and PGC-1α. A, the in vitro binding of PRC and PGC-1α subfragments to transcription factors linked to respiratory chain expression (NRF-1, CREB, ERRα, NRF-2α, and NRF-2β) and HCF-1 was determined by S-tag pulldown assay as described under “Experimental Procedures.” Schematic representation of PRC and PGC-1α is as shown in Fig. 1. Subfragments of each coactivator denoted as A, B, C, or D with their amino acid coordinates shown in parentheses were used in S-tag pulldown assays with 35S-labeled transcription factor. Binding of the various subfragments to each 35S-radiolabeled factor was compared with that of S-tagged thioredoxin (Trdx) as a negative control. Bound proteins were eluted from the S-protein-agarose and visualized by autoradiography. B, NRF-2α and -β subunits were translated in vitro as done for the pulldown assay, except radiolabeled methionine was omitted from the reaction mixtures. NRF-2α or a mixture of NRF-2α and -β subunits was subjected to mobility shift assay using a radiolabeled cytochrome oxidase subunit IV promoter fragment containing tandem NRF-2 recognition sites. Either 1 μl of preimmune serum as a negative control or 1 μl of rabbit anti-NRF-2α or anti-NRF-2β serum was added to the binding reactions as indicated.
Surprisingly, neither coactivator binds either the α or β subunit of NRF-2 (Fig. 2A) despite the fact that the expressed NRF-2 subunits have been shown to interact with each other to produce a functional heterotetrameric complex (19). This is confirmed here by a mobility shift experiment showing that the in vitro translated NRF-2α and -β subunits used in the pulldown assay are capable of forming the expected heteromeric complexes. As shown in Fig. 2B, in vitro translated NRF-2α binds a radiolabeled cytochrome oxidase subunit IV promoter fragment containing tandem NRF-2 recognition sites. The addition of the in vitro translated NRF-2β subunit results in the appearance of a slower migrating complex consistent with the formation of the NRF-2α2/β2 heterotetramer (19). Both complexes are supershifted using anti-NRF-2α serum demonstrating that α is present in both. However, only the heteromeric complex containing NRF-2β is supershifted with anti-NRF-2β serum. This confirms the identity of these complexes and demonstrates that the in vitro translated subunits can interact. Thus, the failure of these subunits to bind PRC or PGC-1α is unlikely to be explained by their inability to engage in biologically relevant interactions. The results demonstrate that the trans-activation of NRF-2 target genes by PGC-1α and PRC occurs in the absence of a direct interaction with this transcription factor.
Specific in Vitro and in Vivo Binding of HCF-1 to PRC and NRF-2β—Although neither PGC-1 coactivator engages in a direct interaction with NRF-2, they may exist in a complex with NRF-2 through interaction with a third party that binds both the coactivator and the transcription factor. HCF-1 is an ideal candidate for such a function. HCF-1 acts as a NRF-2(GABP) coactivator (31) and also binds PGC-1α and -β through a protein-protein interaction motif defined by the amino acid sequence DHDY (20). The data in Fig. 1 confirm the in vitro interaction of HCF-1 with PGC-1α subfragment B and also demonstrates specific HCF-1 binding to PRC subfragment C. In each case, the subfragment that binds HCF-1 is the only one containing the DHDY HCF-1 binding motif.
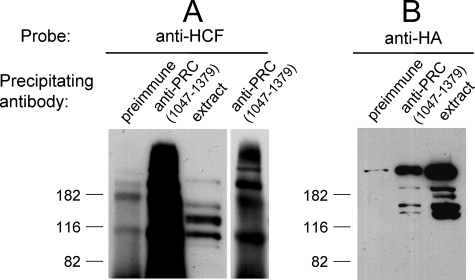
The focus of our work is on PRC as it relates to the regulation of mitochondrial biogenesis and cell growth. Because of the proposed role of PRC and HCF-1 as cell growth regulators, it was of interest to determine whether PRC and HCF-1 exist in a complex in vivo. To this end, PRC was immunoprecipitated from whole cell extracts using anti-PRC serum. The immunoprecipitates were electrophoresed on denaturing gels, and coprecipitation of HCF-1 was assayed by immunoblotting using anti-HCF-1 serum. As shown in Fig. 3A, copious amounts of anti-HCF-1 reactive material was detected in the anti-PRC immunoprecipitates under conditions where the preimmune control showed only a weak signal. The observed HCF-1 heterogeneity is expected because the full-length 2035-amino acid HCF-1 precursor is cleaved autocatalytically into several amino- and carboxyl-terminal fragments that remain associated in vivo (27, 28). The identity of the precipitated protein as HCF-1 was further verified by expressing HA-tagged HCF-1 from a transfected vector and then assaying for HA-tagged HCF-1 in anti-PRC immunoprecipitates with anti-HA antibody. As shown in Fig. 3B, the immunoprecipitates contained a major anti-HA reactive protein corresponding to the full-length HCF-1 expressed in cell extracts. In addition, several minor species likely representing autocatalytic products were also observed. Although the relative abundance of each species differed substantially between the immunoprecipitated endogenous (Fig. 3A) and transfected (Fig. 3B) HCF-1, there was generally good correspondence between the masses of the protein species represented. The exception was a major species migrating below the 116-kDa standard that was present in immunoprecipitates of endogenous but not transfected protein. This is almost certainly a carboxyl-terminal cleavage product that would not be detected in the transfected extracts because the HA tag is expressed on the amino terminus of HCF-1. These results support the conclusion that PRC and HCF-1 exist in a complex in vivo.
FIGURE 3.
In vivo interaction between PRC and HCF-1. A, cell extracts from human 293FT cells were immunoprecipitated with either rabbit preimmune serum as a negative control or rabbit anti-PRC-(1047–1379). Immune complexes were brought down with protein A-agarose, washed, and run on an SDS-10% PAGE gel. For comparison, cell extract was run in the indicated lane. After transfer, the immunoblot was probed with rabbit anti-HCF-1 antibody. A lighter exposure of the anti-PRC-(1047–1379) lane is shown in the adjacent panel. B, HA-tagged HCF-1 was expressed in 293FT cells after electroporation with pCGN HCF(2–2035)9E10. 293FT cell extracts were immunoprecipitated with either rabbit preimmune serum as a negative control or anti-PRC-(1047–1379). Immune complexes were precipitated and electroblotted as in A. For comparison, total cell extract was run in the indicated lane. After transfer, the immunoblot was probed with mouse anti-HA monoclonal antibody. Molecular mass standards in kilodaltons are indicated at the left in each panel.
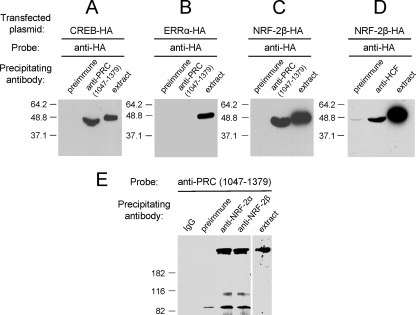
If NRF-2 exists in a ternary complex with PRC and HCF-1, one would expect that NRF-2 would be immunoprecipitated with antibodies directed against either PRC or HCF-1. This was investigated by expressing an HA-tagged derivative of the NRF-2β subunit and immunoprecipitating the cell extracts with anti-PRC or anti-HCF-1 antibodies. In this experiment CREB serves as a positive control because its in vitro and in vivo interaction with PRC has already been demonstrated (24). The results in Fig. 4A confirm the immunoprecipitation of CREB with anti-PRC serum. Interestingly, ERRα serves as negative control because, despite the fact that it interacts specifically with PRC in the in vitro assay (Fig. 2A), antibodies to PRC failed to immunoprecipitate the expressed protein from cell extracts (Fig. 4B). Under these conditions, a robust and specific immunoprecipitation of NRF-2β is detected using anti-PRC serum (Fig. 4C). The slightly increased migration observed after immunoprecipitation is likely the result of a spurious gel artifact rather than a specific modification because it affects both CREB and NRF-2β similarly.
FIGURE 4.
In vivo binding of NRF-2 to PRC and HCF-1. A, as a positive control, HA-tagged CREB was expressed in 293FT cells after electroporation with pSG5/CREB-HA. Cell extracts were immunoprecipitated with rabbit preimmune serum as a negative control or anti-PRC-(1047–1379). B, as a negative control, HA-tagged ERRα was expressed in 293FT cells after electroporation with pSG5/ERRα-HA. Cell extracts were immunoprecipitated as in panel A. C, HA-tagged NRF-2β was expressed in 293FT cells after electroporation with pSG5/NRF-2β-HA. Cell extracts were immunoprecipitated as in panels A and B. D, HA-tagged NRF-2β was expressed in 293FT cells after electroporation with pSG5/NRF-2β-HA. Cell extracts were immunoprecipitated with rabbit preimmune serum as a negative control or with rabbit anti-HCF-1 antibody. For panels A–D, the immunoblot was probed with mouse anti-HA monoclonal antibody. E, cell extract from untransfected cells was subject to immunoprecipitation with anti-NRF-2α, anti-NRF-2β, or the controls rabbit IgG or preimmune serum. The immunoblot was probed with rabbit anti-PRC-(1047–1379). For each panel, cell extract was run in the indicated lane with molecular mass standards in kilodaltons indicated at the left.
The PRC-NRF-2 interaction was further established using untransfected cells by immunoprecipitating cell extracts with anti-NRF-2α or anti-NRF-2β sera and probing immunoblots with anti-PRC-(1047–1379). As shown in Fig. 4E, antibodies directed against either the NRF-2α or β subunits can immunoprecipitate PRC from cell extracts under conditions where the IgG or preimmune serum controls do not. The formation of a complex between endogenously expressed proteins demonstrates that the interaction is not dependent on the expression of NRF-2 as a tagged protein from a transfected vector. Thus, although NRF-2 fails to bind PRC in vitro, it exists in a complex with PRC in cell extracts. In addition, NRF-2β is also immunoprecipitated with anti-HCF-1 antibody (Fig. 4D), confirming the previous findings of others that GABPβ, the mouse homologue of human NRF-2β, interacts directly with HCF-1 (31). Because PRC and HCF-1 exist in a complex (Fig. 3) and NRF-2 does not bind PRC directly (Fig. 2A), these data are consistent with the interpretation that NRF-2β enters into a complex with PRC through its interaction with HCF-1.
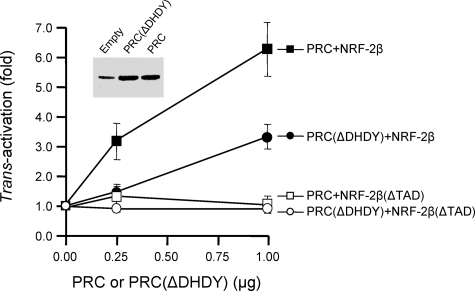
PRC Trans-activation through NRF-2 Requires Both the HCF-1 Binding Site on PRC and Essential Hydrophobic Residues within the NRF-2 Activation Domain—If the in vivo interactions among PRC, HCF-1, and NRF-2 observed by co-immunoprecipitation are functionally significant, the sequence motifs required for these interactions should play a role in the PRC mediated trans-activation through NRF-2. As demonstrated (Fig. 2A), PRC binds HCF-1 through a subfragment containing the DHDY HCF-1 binding site. In addition, it has been established that HCF-1 binding to GABPβ(NRF-2β) requires the same amino acid residues within the NRF-2β activation domain that are also required for transcriptional activation by NRF-2 (31, 33). The requirement for these motifs was tested by measuring the PRC-dependent trans-activation of a Gal4-luciferase reporter in the presence of a Gal4-NRF-2β fusion protein. In this system, PRC trans-activates the reporter to a level ∼6–7-fold above that achieved with the Gal4-NRF-2β fusion protein alone (Fig. 5). Site-directed deletion of the HCF-1 binding site on PRC (ΔDHDY) inhibits this activity significantly. The inhibition does not result from differences in expression from the transfected vectors because PRC (ΔDHDY) and PRC are expressed at similar steady-state levels (Fig. 5). The observed partial inhibition may reflect a requirement for more than a single contact. For example, PRC may be bound to the complex via DHDY but also through its interactions with other coactivators via its activation domain. Deletion of the NRF-2β activation domain (ΔTAD) completely abolishes trans-activation of the reporter by PRC in both the presence and absence of the DHDY motif (Fig. 5). These results establish that the HCF-1 interaction domains on both the coactivator (PRC) and the transcription factor (NRF-2β) are essential for maximal trans-activation by PRC.
FIGURE 5.
Molecular determinants of PRC trans-activation through promoter bound NRF-2β. PRC trans-activations were carried out using a Gal4/luc reporter plasmid. Nearly full-length NRF-2β1 wild type or the same protein containing a deletion in the activation domain (ΔTAD) was directed to the luc promoter through their expression as fusions to Gal4-(1–147) as described under “Experimental Procedures.” The fold trans-activation by either PRC (filled and open squares) or a mutated derivative lacking the DHDY HCF-1 consensus binding site (ΔDHDY) (filled and open circles) was determined by measuring luciferase activity after cotransfection with either 0.25 or 1.0 μg of plasmid expressing each construct. Values were normalized to Renilla luciferase to correct for differences in transfection efficiency. The inset panel shows the steady-state PRC expression in cells transfected with pSV Sport (empty vector), pSV Sport/N-myc PRC(ΔDHDY), or pSV Sport/N-myc PRC.
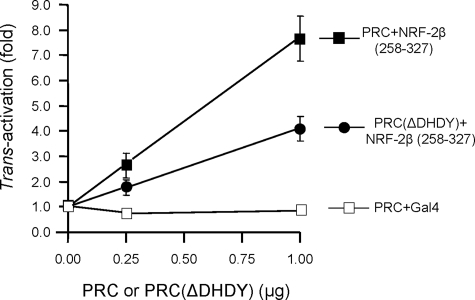
To investigate whether the NRF-2 activation domain is sufficient for trans-activation by PRC, a Gal4 fusion containing only the essential region of the NRF-2β activation domain bounded by amino acids 258–327 (33) was constructed. This construct was trans-activated by PRC to a degree similar (7–8-fold) to that achieved using the full-length Gal4-NRF-2β fusion (Fig. 6). Gal4 alone gave no activity, whereas trans-activation by PRC(ΔDHDY) was significantly reduced. Thus, the NRF-2β activation domain alone is sufficient for PRC-dependent trans-activation of the reporter.
FIGURE 6.
The NRF-2β activation domain is sufficient for trans-activation by PRC. PRC trans-activations were carried out using a Gal4/luc reporter plasmid as in Fig. 5. In this case a fragment containing only the NRF-2β1 activation domain (NRF-2β1/258–327) was directed to the luc promoter through its expression as a fusion to Gal4-(1–147) as described under “Experimental Procedures.” The fold trans-activation by either PRC (filled squares) or a mutated derivative lacking the DHDY HCF-1 consensus binding site (ΔDHDY) (filled circles) was determined as in Fig. 5 and compared with that derived from PRC and Gal4-(1–147) as a negative control (open squares).
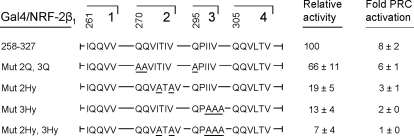
Clusters of hydrophobic amino acid residues within NRF-2β activation domain are essential for NRF-2 transcriptional activity (33). These same residues are also required for interaction between GABPβ(NRF-2β) and HCF-1 (31). In fact, those residues that contribute most to transcriptional activation are also the major contributors to HCF-1 binding to NRF-2β. Thus, if PRC trans-activation occurs through a complex containing PRC, HCF-1, and NRF-2, one would expect that trans-activation by PRC would require the same residues necessary for the NRF-2β-HCF-1 interaction and for NRF-2β-mediated transcription. This was tested using a series of NRF-2β activation domain mutants where clusters of amino acid residues containing either glutamines or hydrophobic residues were converted to alanines by site-directed mutagenesis (33). As shown in Fig. 7, conversion of glutamines within clusters 2 (Gln-270 and -271) and 3 (Gln-295) of the NRF-2β activation domain reduced transcriptional activity by about 34% and had a similar effect on the fold trans-activation by PRC. By contrast, conversion of hydrophobic residues within clusters 2 (Ile-274 and -76) or 3 (Ile-297 and -298 and Val-299) to alanines had a much larger effect on NRF-2β transcription and a proportionately larger effect on trans-activation by PRC. Combined mutations in clusters 2 and 3 reduced NRF-2β transcription by >90% and completely abolished trans-activation by PRC. These results establish that key amino acids required for both transcription by NRF-2β and for NRF-2β interaction with HCF-1 are also required for PRC-dependent trans-activation through NRF-2.
FIGURE 7.
The same NRF-2β activation domain hydrophobic residues are required for interaction with HCF-1 and for trans-activation by PRC. Either glutamines or hydrophobic amino acids within glutamine-containing hydrophobic clusters 2 and 3 of the NRF-2β activation domain were converted to alanines (underlined). Gal4 fusion constructs containing the wild type activation domain (NRF-2β1/258–327) or the alanine substitution mutants were assayed for their activation of the Gal4/luc reporter (Relative activity) and for their ability to support the trans-activation of the same reporter by PRC (Fold PRC activation). Values were normalized to Renilla luciferase to correct for differences in transfection efficiency and represent the average ± S.E. for five separate determinations.
In Vivo Occupancy of NRF-2-dependent Promoters by NRF-2β, PRC, and HCF—If a complex containing NRF-2, PRC, and HCF-1 is physiologically significant, one might expect that all three components occupy NRF-2-dependent promoters in vivo. In a previous study, we established that the promoters from both isoforms of mitochondrial transcription factor B designated as TFB1M and TFB2M (38, 39) were dependent on functional NRF-2 recognition sites for both their basal activity and for their trans-activation by PRC (19). In addition, chromatin immunoprecipitations revealed that NRF-2α was bound to both promoters in vivo. Based on these results, it was of interest to determine whether NRF-2β, PRC, and HCF-1 were also localized to the TFB promoters in vivo. To this end chromatin immunoprecipitations were carried out using antibodies specific for each of these factors. As shown in Table 1, significant occupancy of both TFB promoters by NRF-2β, PRC, and HCF-1 was detected. The signal is less robust for PRC compared with the other two factors possibly because of the low level of PRC expression or because of masking of the 1047–1379 epitope by protein-protein interactions within the chromatin-bound complex. Nevertheless, the results are consistent with the in vitro experiments showing a functional association among NRF-2β, PRC, and HCF-1 and support the conclusion that all three factors can associate with NRF-2-dependent promoters in vivo.
TABLE 1.
Chromatin immunoprecipitation analysis of mitochondrial transcription factor B promoter occupancy by NRF-2β, PRC, and HCF
|
Precipitating antibody
|
Promoter
occupancya
|
|
|---|---|---|
| TFB1M | TFB2M | |
| Rabbit IgG | 1.0 | 1.0 |
| Anti-NRF-2β | 31.6 ± 7.9 | 59.8 ± 19.2 |
| Anti-PRC-(1047-1379) | 4.5 ± 0.5 | 4.4 ± 0.8 |
| Anti-HCF-1 | 44.2 ± 4.7 | 96.2 ± 12.1 |
Values of relative promoter occupancy represent the average ± S.E. for three separate determinations.
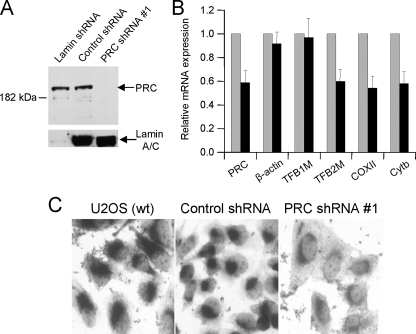
Effects of shRNA-mediated PRC Knockdown on TFB and COX Expression—The results presented are consistent with a pathway whereby PRC activates the expression of the TFBs and possibly other NRF-2 target genes through its interaction with an NRF-2·HCF-1 complex. One prediction of this model is that reduced PRC expression might lead to diminished mitochondrial transcript levels and the consequent reduction in respiratory enzyme activities. This was examined by constructing a lentivirus transductant of U2OS cells that expresses a small hairpin RNA designed to knock down the expression of PRC. The U2OS cell line was chosen because it is a contact inhibited human line that displays regulated cell-cycle expression of PRC (24). As shown in Fig. 8A, one of the transductants tested (PRC shRNA#1) showed specific shRNA-mediated reduction in PRC protein expression. This transductant showed the largest reduction in PRC protein levels among 20 individual isolates tested. The inhibition was specific to the PRC shRNA because a transductant expressing a hairpin with a negative control sequence showed no reduction in PRC. Moreover, a lentivirus transductant expressing a lamin-specific control hairpin displayed markedly reduced lamin expression and no change in the steady-state level of PRC. The knockdown of PRC protein in these cells was accompanied by reduced PRC mRNA expression as measured by quantitative real time reverse transcription-PCR (Fig. 8B). This coincided with diminished levels of TFB2M mRNA and two different mitochondrial transcripts encoding COXII and cytochrome b. Thus, reduced PRC expression is associated with the down-regulation of transcripts encoding a key mitochondrial transcription factor (TFB2M) and mitochondrial respiratory chain subunits. Surprisingly, TFB1M mRNA was not diminished significantly in the PRC shRNA transductant and was expressed at levels equivalent to the β-actin negative control. This suggests that the effects of PRC likely depend on promoter context or unknown compensatory interactions. The downstream effect of these changes in gene expression on respiratory activity was assessed by staining cells for cytochrome oxidase activity. As shown in Fig. 8C, the PRC shRNA transductant displayed diminished COX staining compared with the robust staining observed in wild type U2OS and transductants expressing the negative control hairpin. The results are consistent with a pathway whereby the PRC-dependent expression of NRF-2 target genes can mediate changes in the expression of a respiratory enzyme complex.
FIGURE 8.
Down-regulation of TFB2M, mitochondrial transcripts, and cytochrome oxidase activity associated with stable shRNA-mediated knock down of PRC expression. A, lentivirus transductants of U2OS cells expressing shRNAs directed against a lamin shRNA control, PRC shRNA#1 or a negative control oligonucleotide. Cell extracts of each were subjected to immunoblotting with antibodies directed against lamin and PRC. B, total RNA was isolated from transductants expressing either the control sequence or PRC shRNA#1. Quantitative real time reverse transcription-PCR was carried out with primers specific for PRC, β-actin, TFB1M, TFB2M, COXII, and cytochrome b (Cytb), and the transcript levels for each in the PRC shRNA#1 transductant are expressed relative to those of the negative control. C, cytochrome oxidase activity staining was performed as described under “Experimental Procedures” on U2OS wild type (wt) cells and on transductants expressing the negative control oligonucleotide and PRC shRNA#1.
DISCUSSION
The PGC-1 family of regulated coactivators functions in the relay of environmental cues to the transcriptional machinery (7, 16, 40). This is accomplished partly through interactions with transcription factor targets that act on an array of genes governing programs of cellular energetics and differentiation. PGC-1α exhibits a broad range of transcription factor interactions that include a host of nuclear hormone receptors as well as transcription factors implicated in mitochondrial biogenesis, muscle fiber type switching, and many other biological processes (6, 16, 40). The induction of PGC-1α by cAMP-dependent transcription and its post-translational modification are important means of its regulation by extracellular signaling events (41, 42). PRC is defined as a member of the PGC-1 family by conservation of structural domains and by its ability to interact with NRF-1 in the activation of NRF-1 target genes involved in the expression of the respiratory chain (19, 23). However, PRC expression differs from that of PGC-1α in that it is not induced during thermogenesis but, rather, responds to signals regulating cell proliferation (23, 24). Here, we show that PRC also differs from PGC-1α in its interaction with nuclear hormone receptors. It shows only a weak interaction with PPARγ as well as ligand-independent binding to TRβ and RAR. These results along with its inability to respond to thermogenic signals likely reflect significant divergence between PRC and PGC-1α in signaling via nuclear hormone receptor pathways.
In contrast to these differences in nuclear hormone receptor interactions, PRC and PGC-1α are virtually identical in their binding to an array of transcription factors that have been implicated in the expression of the respiratory chain. In particular, strong interactions by both coactivators with NRF-1 and ERRα are consistent with significant similarities between the two factors in their ability to trans-activate the promoters of target genes that specify respiratory chain subunits and mitochondrial transcription factors. Surprisingly, neither coactivator engages in a direct interaction with NRF-2(GABP) despite the fact that both have been associated with NRF-2-dependent gene expression (18, 19). Here, we demonstrate that HCF-1 serves as an important intermediary between PRC and NRF-2 target genes by binding both PRC and the NRF-2β subunit. Significant inhibition of PRC trans-activation function can be achieved by mutation of the DHDY HCF-1 consensus binding site on PRC. This agrees with both the in vitro pulldown assays showing direct binding of HCF-1 to the DHDY-containing PRC subfragment and with immunoprecipitations showing that HCF-1 is precipitated from cell extracts using antibodies directed against PRC. The data also show that the NRF-2β transcriptional activation domain is both absolutely required and sufficient for PRC-directed transcriptional activation. This function is associated with key hydrophobic amino acid residues in the NRF-2β activation domain. This result is consistent with previous findings showing that the same hydrophobic residues are essential for binding of HCF-1 to NRF-2β(GABPβ), thus implicating HCF-1 as a coactivator of this transcription factor (31). The finding that all three proteins occupy NRF-2-dependent TFB promoters as demonstrated by chromatin immunoprecipitations reinforces the physiological significance of these interactions.
In addition to its structural and functional similarities with PGC-1α (18, 19, 23), a role for PRC in the expression of the respiratory chain is supported by the finding that a dominant negative PRC allele consisting of the NRF-1/CREB binding site inhibits respiratory growth on galactose when expressed in trans (24). Here, we show that PRC loss of function through shRNA-mediated knock down is associated with the down-regulation of the TFB2M mRNA encoding a key mitochondrial transcription factor. This coincides with reductions in mitochondrial transcripts for respiratory subunits, one of which encodes COXII, an essential subunit of the cytochrome oxidase complex. The down-regulation of COXII mRNA in the PRC shRNA transductant is accompanied by reduced COX enzyme activity demonstrating the physiological consequences of these changes in gene expression. However, the normal level of TFB1M expression in the transductant indicates that PRC is not limiting for all NRF-2-dependent genes. This might be explained by unknown differences in promoter context or by promoter-specific compensatory interactions. It remains to be determined to what extent PRC selectively mediates the coordinate expression of the family of NRF target genes.
It is notable that the TFB1M and TFB2M isoforms have distinct biological functions. The TFB1M isoform is transiently down-regulated relative to that of the TFB2M isoform in serum-stimulated quiescent fibroblasts, suggesting that the latter is favored in the transition to proliferative growth (19). RNA-mediated interference knockdown of the Drosophila B2 isoform results in reduced mtDNA transcription and copy number (43). This contrasts with RNA-mediated interference knockdown of the B1 isoform, which has no effect on mtDNA transcription or replication but does result in reduced mitochondrial translation (44). This is consistent with the finding that overexpression of Drosophila TFB2M but not TFB1M increases mtDNA copy number. These results match those obtained in human cells where overexpression of human TFB2M but not TFB1M enhances mitochondrial transcription and transcription-primed replication (45). Thus, it is not surprising that we observe a decrease in mitochondrial transcript levels in the PRC shRNA transductant where only the TFB2M mRNA is down-regulated. This appears sufficient to mediate changes in the mitochondrial transcriptional machinery in both Drosophila and human systems.
Our previous work has implicated PRC as a potential regulator of cell proliferation (23, 24). It of interest in this context that PRC exists in a complex with HCF-1 and NRF-2 and that the molecular determinants of these interactions are required for maximal trans-activation by PRC. HCF-1 and GABP(NRF-2) were both originally described as cellular factors required for the expression of herpes simplex virus immediate early genes (27, 46, 47). Subsequently, HCF-1 was found to interact with a number of transcription factors, including VP16 and GABPβ(NRF-2β), as well as chromatin-remodeling cofactors (27, 31). HCF-1 is an important component of a molecular switch that triggers immediate early gene expression by interacting with the VP16·Oct1 transcription factor complex (26). Moreover, genetic evidence supports an essential role for HCF-1 in progression beyond G1 of the cell cycle, suggesting that it may serve as transcriptional coactivator for cell cycle regulated genes (29). This is especially interesting in light of the recent finding that GABP(NRF-2) can direct a D-cyclin-independent pathway of entry to the cell cycle (12). The association between HCF-1 and NRF-2(GABP) may serve to integrate the cell proliferative cycle with components of the mitochondrial biogenesis program related to the expression of the respiratory chain. PRC appears to be a regulated moiety of this complex that functions to enhance the basal expression of essential genes in preparation for cell division. Although antibodies directed against PRC can immunoprecipitate both HCF-1 and NRF-2β, its association with chromatin-bound complexes may be transient. A transient association might facilitate a regulatory function and is consistent with the immediate early expression of PRC, its relatively rapid turnover, and its low abundance (23, 24). With the current results, it is now clear that HCF-1 binds all three members of the PGC-1 coactivator family. It interacts with both PGC-1α and -β and enhances their transcriptional activities in vitro (20). Moreover, phosphorylation of both PGC-1α and GABPβ (NRF-2β) augments their ability to enter into a complex with HCF-1 in the regulation of neuromuscular gene expression (48). Although the three family members are differentially regulated, their association with HCF-1 appears to be fundamental to their ability to activate transcription through NRF-2 and possibly other transcription factors.
Acknowledgments
We are grateful to Dr. Robert Goldman (Northwestern University) and Dr. Winship Herr (University of Lausanne) for the gift of antibodies. We thank Raymond A. Pasko for expert technical assistance.
This work was supported United States Public Health Service National Institutes of Health Grant GM32525-25. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NRF, nuclear respiratory factor; PRC, PGC-1-related coactivator; PGC, peroxisome proliferator-activated receptor γ coactivator; GABP, GA-binding protein; HCF, host cell factor; COX, cytochrome oxidase; HA, hemagglutinin; TFB, transcription factor B; CREB, cyclic AMP response element-binding protein; ERR, estrogen-related receptor; TR, thyroid receptor; RAR, retinoic acid receptor; PPAR, peroxisome proliferator-activated receptor; TAD, trans-activation domain; shRNA, short hairpin RNA; FL, full length.
References
- 1.Hatefi, Y. (1985) Annu. Rev. Biochem. 54 1015–1069 [DOI] [PubMed] [Google Scholar]
- 2.Cannon, B., and Nedergaard, J. (2004) Physiol. Rev. 84 277–359 [DOI] [PubMed] [Google Scholar]
- 3.Scarpulla, R. C. (2004) in Oxidative Phosphorylation in Health and Disease (Smeitink, J. A. M., Sengers, R. C. A., and Trij, J. M. F., eds) pp. 28–42, Landes Bioscience, New York
- 4.Wallace, D. C. (2005) Annu. Rev. Genet. 39 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonawitz, N. D., Clayton, D. A., and Shadel, G. S. (2006) Mol. Cell 24 813–825 [DOI] [PubMed] [Google Scholar]
- 6.Kelly, D. P., and Scarpulla, R. C. (2004) Genes Dev. 18 357–368 [DOI] [PubMed] [Google Scholar]
- 7.Scarpulla, R. C. (2006) J. Cell. Biochem. 97 673–683 [DOI] [PubMed] [Google Scholar]
- 8.Evans, M. J., and Scarpulla, R. C. (1989) J. Biol. Chem. 264 14361–14368 [PubMed] [Google Scholar]
- 9.Evans, M. J., and Scarpulla, R. C. (1990) Genes Dev. 4 1023–1034 [DOI] [PubMed] [Google Scholar]
- 10.Virbasius, J. V., Virbasius, C. A., and Scarpulla, R. C. (1993) Genes Dev. 7 380–392 [DOI] [PubMed] [Google Scholar]
- 11.Cam, H., Balciunaite, E., Blias, A., Spektor, A., Scarpulla, R. C., Young, R., Kluger, Y., and Dynlacht, B. D. (2004) Mol. Cell 16 399–411 [DOI] [PubMed] [Google Scholar]
- 12.Yang, Z. F., Mott, S., and Rosmarin, A. G. (2007) Nat. Cell Biol. 9 339–346 [DOI] [PubMed] [Google Scholar]
- 13.Huo, L., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ristevski, S., O'Leary, D. A., Thornell, A. P., Owen, M. J., Kola, I., and Hertzog, P. J. (2004) Mol. Cell. Biol. 24 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, J., Handschin, C., and Spiegelman, B. M. (2005) Cell Metab. 1 361–370 [DOI] [PubMed] [Google Scholar]
- 16.Finck, B. N., and Kelly, D. P. (2006) J. Clin. Investig. 116 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M., and Spiegelman, B. M. (1998) Cell 92 829–839 [DOI] [PubMed] [Google Scholar]
- 18.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., and Spiegelman, B. M. (1999) Cell 98 115–124 [DOI] [PubMed] [Google Scholar]
- 19.Gleyzer, N., Vercauteren, K., and Scarpulla, R. C. (2005) Mol. Cell. Biol. 25 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, J., Puigserver, P., Donovan, J., Tarr, P., and Spiegelman, B. M. (2002) J. Biol. Chem. 277 1645–1648 [DOI] [PubMed] [Google Scholar]
- 21.Lin, J. D., Tarr, P. T., Yang, R. J., Rhee, J., Puigserver, P., Newgard, C. B., and Spiegelman, B. M. (2003) J. Biol. Chem. 278 30843–30848 [DOI] [PubMed] [Google Scholar]
- 22.Arany, Z., Lebrasseur, N., Morris, C., Smith, E., Yang, W., Ma, Y., Chin, S., and Spiegelman, B. M. (2007) Cell Metab. 5 35–46 [DOI] [PubMed] [Google Scholar]
- 23.Andersson, U., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vercauteren, K., Pasko, R. A., Gleyzer, N., Marino, V. M., and Scarpulla, R. C. (2006) Mol. Cell. Biol. 26 7409–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkles, J. A. (1998) Prog. Nucleic Acid Res. Mol. Biol. 58 41–78 [DOI] [PubMed] [Google Scholar]
- 26.Wysocka, J., and Herr, W. (2003) Trends Biochem. Sci. 28 294–304 [DOI] [PubMed] [Google Scholar]
- 27.Wilson, A. C., LaMarco, K., Peterson, M. G., and Herr, W. (1993) Cell 74 115–125 [DOI] [PubMed] [Google Scholar]
- 28.Vogel, J. L., and Kristie, T. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9425–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goto, H., Motomura, S., Wilson, A. C., Freiman, R. N., Nakabeppu, Y., Fukushima, M., Herr, W., and Nishimoto, T. (1997) Genes Dev. 11 726–737 [DOI] [PubMed] [Google Scholar]
- 30.Julien, E., and Herr, W. (2003) EMBO J. 22 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel, J. L., and Kristie, T. M. (2000) EMBO J. 19 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gugneja, S., Virbasius, J. V., and Scarpulla, R. C. (1995) Mol. Cell. Biol. 15 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gugneja, S., Virbasius, C. A., and Scarpulla, R. C. (1996) Mol. Cell. Biol. 16 5708–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak, K. J., and Schmittgen, T. D. (2001) Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- 35.Salviati, L., Hernandez-Rosa, E., Walker, W. F., Sacconi, S., DiMauro, S., Schon, E. A., and Davidson, M. M. (2002) Biochem. J. 363 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G., Piqani, B., Eisenhaure, T. M., Luo, B., Grenier, J. K., Carpenter, A. E., Foo, S. Y., Stewart, S. A., Stockwell, B. R., Hacohen, N., Hahn, W. C., Lander, E. S., Sabatini, D. M., and Root, D. E. (2006) Cell 124 1283–1298 [DOI] [PubMed] [Google Scholar]
- 37.Mootha, V. K., Handschin, C., Arlow, D., Xie, X. H., St Pierre, J., Sihag, S., Yang, W. L., Altshuler, D., Puigserver, P., Patterson, N., Willy, P. J., Schulman, I. G., Heyman, R. A., Lander, E. S., and Spiegelman, B. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCulloch, V., Seidel-Rogol, B. L., and Shadel, G. S. (2002) Mol. Cell. Biol. 22 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falkenberg, M., Gaspari, M., Rantanen, A., Trifunovic, A., Larsson, N.-G., and Gustafsson, C. M. (2002) Nat. Genet. 31 289–294 [DOI] [PubMed] [Google Scholar]
- 40.Handschin, C., and Spiegelman, B. M. (2006) Endocr. Rev. 27 728–735 [DOI] [PubMed] [Google Scholar]
- 41.Herzig, S., Long, F. X., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., Spiegelman, B., and Montminy, M. (2001) Nature 413 179–183 [DOI] [PubMed] [Google Scholar]
- 42.Puigserver, P., Rhee, J., Lin, J., Wu, Z., Yoon, J. C., Zhang, C. Y., Krauss, S., Mootha, V. K., Lowell, B. B., and Spiegelman, B. M. (2001) Mol. Cell 8 971–982 [DOI] [PubMed] [Google Scholar]
- 43.Matsushima, Y., Garesse, R., and Kaguni, L. S. (2004) J. Biol. Chem. 279 26900–26905 [DOI] [PubMed] [Google Scholar]
- 44.Matsushima, Y., Adan, C., Garesse, R., and Kaguni, L. S. (2005) J. Biol. Chem. 280 16815–16820 [DOI] [PubMed] [Google Scholar]
- 45.Cotney, J., Wang, Z., and Shadel, G. S. (2007) Nucleic Acids Res. 35 4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaMarco, K. L., and McKnight, S. L. (1989) Genes Dev. 3 1372–1383 [DOI] [PubMed] [Google Scholar]
- 47.Kristie, T. M., and Sharp, P. A. (1990) Genes Dev. 4 2383–2396 [DOI] [PubMed] [Google Scholar]
- 48.Handschin, C., Kobayashi, Y. M., Chin, S., Seale, P., Campbell, K. P., and Spiegelman, B. M. (2007) Genes Dev. 21 770–783 [DOI] [PMC free article] [PubMed] [Google Scholar]