Abstract
Pairing between the hexamer seed region of a small interfering RNA (siRNA) guide strand (nucleotides 2–7) and complementary sequences in the 3′ UTR of mature transcripts has been implicated as an important element in off-target gene regulation and false positive phenotypes. To better understand the association between seed sequences and off-target profiles we performed an analysis of all possible (4096) hexamers and identified a nonuniform distribution of hexamer frequencies across the 3′ UTR transcriptome. Subsequent microarray analysis of cells transfected with siRNAs having seeds with low, medium, or high seed complement frequencies (SCFs) revealed that duplexes with low SCFs generally induced fewer off-targets and off-target phenotypes than molecules with more abundant 3′ UTR complements. These findings provide the first experimentally validated strategy for designing siRNAs with enhanced specificity and allow for more accurate interpretation of high throughput screening data generated with existing siRNA/shRNA collections.
Keywords: seed complement frequencies (SCFs), RNA interference (RNAi), small interfering RNAs (siRNA), microRNAs (miRNAs), RNA induced silencing complex (RISC)
INTRODUCTION
RNA interference (RNAi) is a near-ubiquitous post-transcriptional gene regulatory pathway that is mediated by microRNAs (miRNAs) and other small noncoding RNAs (Fig. 1; Zamore and Haley 2005). In mammals, the miRNA-loaded RNA-induced silencing complex (RISC) modulates expression of genes by one of two mechanisms: direct cleavage (in the case of close to perfect complementarity between the miRNA guide strand and the mRNA target) or translational attenuation and mRNA degradation with P-body relocation (in cases of partial complementarity between the miRNA guide strand and the 3′ UTR of target genes) (Doench et al. 2003; Doench and Sharp 2004; Jakymiw et al. 2005; Liu et al. 2005a,b; Sen and Blau 2005).
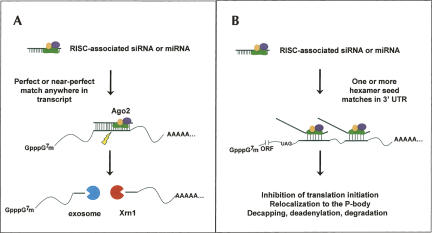
FIGURE 1.
Degradation of message RNA can occur by two separate pathways in RNAi. (A) Perfect or near-perfect complementarity of the siRNA guide strand with a target site leads to RISC-mediated endonucleolytic cleavage and degradation of the message. (B) The seed region (nucleotides ∼2–7) of the siRNA guide strand binds to the 3′ UTR of the mRNA and inhibits translation and/or promotes mRNA degradation.
Synthetic, 19-base-pair (bp) small interfering RNAs (siRNA) can enter the RNAi pathway and target genes for cleavage (Elbashir et al. 2001). Though this technique has been widely adopted as a tool for functional genomics (Bartz and Jackson 2005; Chatterjee-Kishore and Miller 2005; Collins et al. 2006; Neumann et al. 2006), studies have revealed that siRNA specificity is not as stringent as originally anticipated (Jackson et al. 2003) and that off-target gene knockdown induced by partial complementarity between the sense or antisense of the duplex and unintended targets is widespread. As recent investigations have revealed that (1) off-targeting can induce false positives during RNAi-based phenotypic screening (Lin et al. 2005; Fedorov et al. 2006), and (2) identity based algorithms such as BLASTn and Smith–Waterman fail to enhance siRNA specificity (Birmingham et al. 2006), novel bioinformatic methods that enhance siRNA specificity are urgently needed.
Microarray profiling studies have demonstrated that different siRNAs induce widely disparate numbers of off-targets. Given that the mechanism of siRNA off-targeting appears to be similar to that of miRNA on-targeting and requires pairing between a distinct hexamer sequence of the guide strand (the seed region, nucleotides 2–7) and complementary sequences in the 3′ UTR of the off-targeted gene (Lim et al. 2005; Birmingham et al. 2006; Jackson et al. 2006b; Valencia-Sanchez et al. 2006), the variations in off-target signature size might be explained by differences in the frequency at which seed complements appear in the 3′ UTR transcriptome. For these reasons, we decided to experimentally test the impact of low seed complement frequency on (1) off-target signature size and (2) overall siRNA functionality. To this end, bioinformatic studies designed to examine the frequency of all possible hexamers were first performed and revealed a distribution of 3′ UTR hexamer frequencies that spanned two orders of magnitude. This allowed the design and testing of siRNAs having seeds with low, medium, and high seed complement frequencies (SCFs). Studies that employed both microarray analysis and phenotypic assays showed that siRNAs with low 3′ UTR SCFs generated fewer off-targets and false positive phenotypes than duplexes with seeds having moderate to high SCFs. This study provides the first experimental demonstration that bioinformatic principles based on seed complement frequencies can be utilized to minimize off-target signatures and the frequency at which false positives occur in RNAi-based phenotypic screens. In addition, these findings will greatly aid in the stratification of hits from high throughput siRNA screens. Specifically, siRNAs that (1) induce measurable phenotypes and (2) have low SCFs have a higher probability of representing true target knockdown due to the lower likelihood of inducing extensive off-targeting signatures.
RESULTS
A broad hexamer frequency range exists in the 3′ UTR transcriptome
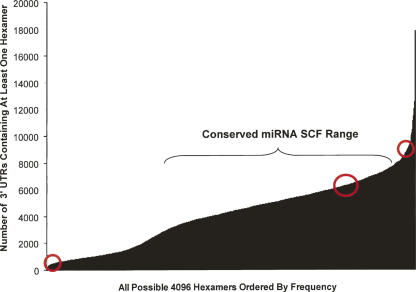
For the SCF to play a role in establishing the magnitude of the off-target signature, a range of SCFs is expected to be present in the 3′ UTR genome. To investigate this, the list of all possible (4096) 6-nucleotide (nt) sequences was mapped to the collection of transcripts that are (1) present in the NCBI human RefSeq 15 database or (2) expressed in HeLa cells/detectable by Agilent Human 1A expression arrays (see Materials and Methods), to determine the number of 3′ UTRs containing at least one copy of any given hexamer. For the RefSeq 15 database, a broad (approximately two orders of magnitude) and nonuniform distribution of frequencies was observed with the lowest and highest occurring hexamer motifs appearing in 142 and 13,662 3′ UTRs, respectively (Fig. 2; Supplemental Table 1; data excludes AAAAAA [17,121 occurrences] and AATAAA [17,844 occurrences] motifs). Plotting the number of hexamers versus the number of 3′ UTRs in which they occur (at least one time) identified a bimodal distribution (resulting from the paucity of CG dinucleotides in the mammalian genome) containing a population of hexamers that occurred in a relatively low number of 3′ UTRs (see Supplemental Fig. 1). This pattern was reiterated in the HeLa cell transcriptome, albeit in a compressed form due to the smaller number of expressed genes in this cell type (distribution spanning 66 to ∼6000 3′ UTRs out of ∼11,000 detectable XM or NM transcripts). When the complements of the 62 seed families present in well-conserved vertebrate miRNAs (Lewis et al. 2005) were mapped to the RefSeq 15 and HeLa distributions, the number of potentially targeted 3′ UTRs predominantly clustered in the mid- to high-frequency range (∼4500–6500 and 2000–3000 3′ UTRs in the RefSeq 15 and HeLa transcriptomes, respectively). As miRNAs are predicted to target large numbers of genes (Brennecke et al. 2005; Farh et al. 2005; Krek et al. 2005; Lewis et al. 2005; Stark et al. 2005; Xie et al. 2005) and the mechanism of miRNA on-targeting and siRNA off-targeting are believed to be similar, mid- to high and low SCF ranges were predicted to be associated with large and modest off-target signatures, respectively.
FIGURE 2.
3′ UTR hexamer frequency is widely distributed in the human genome. A plot of all possible 4096 hexamers (X-axis) versus the number of 3′ UTRs that contain a given hexamer (Y-axis) is presented. 3′ UTR sequences were derived from Refseq 15. The approximate seed frequency ranges from which low, medium, and high-frequency seeds were chosen is indicated by red circles.
siRNAs with low SCFs induce smaller off-target signatures
Previous studies have utilized microarray profiling to document the extent of siRNA off-target effects (Chi et al. 2003; Jackson et al. 2003; Semizarov et al. 2003). To test whether a relationship exists between SCF and off-target signatures, 29 unique siRNAs targeting two housekeeping genes, PPIB and GAPDH, and having high (>3800), medium (∼2500–2800), or low (<350) SCFs in the HeLa transcriptome (∼10 siRNAs for each group) were transfected into HeLa cells and assessed for off-target signatures by microarray expression analysis. Though the size and position of these windows are somewhat arbitrary in nature, these distinct groupings were chosen to sample sufficient numbers of duplexes with SCFs that were less frequent (low), representative of (medium), or more frequent (high) than SCFs found in conserved miRNA seeds. Additionally, for each experiment the window size was constrained by the composition of all possible seeds found in the targeted gene sequence as well as the need to identify multiple sequences that are highly functional (against a given target gene) within our collection. All of the siRNAs used in this study exhibited ≥ 84% silencing of the primary target mRNA (see Supplemental Table 2). This criterion demonstrated that functionality was not associated with siRNAs having particular SCFs and ensured that the off-target signature was predominantly the result of a single (antisense/guide) strand (Khvorova et al. 2003).
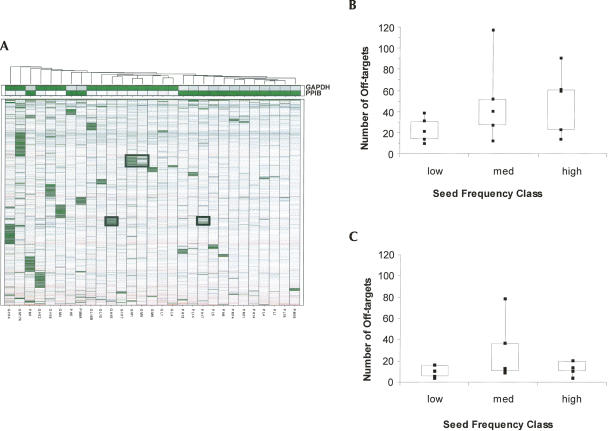
The off-target signatures of siRNA-transfected cultures revealed a remarkable bias; the siRNAs having the most extensive off-target signatures all had seeds with medium or high SCFs (Fig. 3). While low seed frequency was not essential for small off-target effects (some siRNAs with seeds that had medium or high SCFs induced only modest signatures), box-plots summarizing the overall distribution of off-targets for each siRNA identified a clear relationship between SCFs and the size of the off-target signature (Fig. 3B,C). The differences in off-target distributions between the low and medium SCF classes were statistically significant (P-value of 0.09 between low and medium distribution in the GAPDH studies, Fig. 3B, and P-value of 0.009 between low and medium distributions in the PPIB studies, Fig. 3C). At the same time, off-target distributions between medium and high groups across the two datasets were indistinguishable.
FIGURE 3.
Microarray signatures of GAPDH- and PPIB-targeting siRNAs identify a relationship between seed complement frequency and the magnitude of off-target signatures. (A) Heatmaps of off-target signatures generated by PPIB- and GAPDH-targeting siRNA having low (L), medium (M), or high (H) seed complement frequencies. Level of target knockdown is represented by zoomed-in bars above the heatmap. Dashed black boxes outline the nearly identical signatures of PPIB H17 and GAPDH H15, two siRNAs targeting unrelated genes but having identical seeds. A solid black box outlines signatures of GAPDH M1 and M8, two siRNAs that differ by a 1-nt shift in the target site. (A) Saturation on the color scale reflects fourfold up (red) and fourfold down (green) with respect to mock lipid transfection. (B,C) Box plots quantitating the number of off-targets induced by each class of siRNA targeting (B) GAPDH, and (C) PPIB.
These studies reiterate the importance of the seed region in determining the identity of off-targeted genes (Birmingham et al. 2006; Jackson et al. 2006b). Two distinct siRNAs targeting unrelated genes yet having identical seeds (GAPDH H15 sense: 5′-GAAGUAUGACAACAGCCUC-3′; PPIB H17 sense: 5′-CGACAGUCAAGACAGCCUG-3′, seed complement sequence underlined) generated comparable off-target signatures (Fig. 3A, dashed black boxes; Supplemental Table 2). Likewise, two GAPDH-targeting siRNAs that differed by a single nucleotide shift in the target site (GAPDH M1 sense: 5′-GGCUCACAACGGGAAGCUU-3′; GAPDH M8 sense: 5′-GCUCACAACGGGAAGCUUG-3′) also exhibit very similar signatures (Fig. 3A, solid black box). These findings have been observed with two additional sequences (data not shown) and (1) reaffirm the importance of the seed in determining off-target identity and (2) support previous claims by Birmingham and others (Lewis et al. 2005; Lim et al. 2005; Birmingham et al. 2006) that the boundaries of the seed region are not static. In addition, the frequency at which one or more hexamer seed complement matches were found in the 3′ UTRs of off-targets was significantly enriched over that observed in untargeted (control) mRNA populations with comparable 3′ UTR lengths (Supplemental Fig. 2). The elevated occurrence of seed complements was present across all three SCF classes (low, medium, and high, p < 0.01), whereas an identical test for matches between the 3′ UTRs and sense-strand seed sequences failed to identify a significant difference between the off-targeted and the untargeted gene sets. Overall, these findings support previous conclusions that complementarity between the 3′ UTR of the off-targeted gene and the seed region of the siRNA guide strand is critical for off-targeting (Jackson et al. 2003; Lim et al. 2005; Birmingham et al. 2006) and validate that SCF can be used as a predictor for siRNA specificity.
The seed region can determine off-target signature size
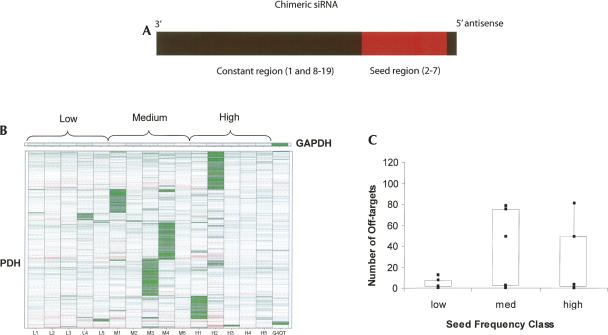
To determine whether the off-target signature size bias observed in the GAPDH and PPIB experiments could be attributed solely to the seed sequence, chimeric siRNAs having seeds with high, medium, or low SCFs were synthesized with an invariable GAPDH-targeting scaffold region [sense strand: 5′-UGGUUUACAUGU(6-nt seed complement)A-3′, see Supplemental Table 2; Fig. 4A]. All of the duplexes incorporated 2′-O-methyl modifications on nucleotides 1 and 2 of the sense strand. This modification pattern has been shown to minimize sense strand entry into RISC, thus limiting the off-target signature to genes targeted by the antisense (guide) strand (Jackson et al. 2006a). In addition, while none of the chimeric sequences have endogenous targets, all of the duplexes were shown to be functional (>75% silencing) against a perfectly matched target site inserted into the 3′ UTR of a luciferase reporter gene (see Supplemental Table 2).
FIGURE 4.
Chimeric siRNAs demonstrate that the seed plays a dominant role in determining off-target signature size. (A) Diagram of the chimeric siRNA having seeds with low (L), medium (M), or high (H) SCFs associated with constant regions (position 1 and positions 8–19) targeting GAPDH. (B) Heatmaps generated from HeLa cells transfected with chimeric siRNAs. The knockdown and signature of the siRNA from which the scaffold was taken (G4 OT) is in the last lane. (C) Box plot showing the relationship between off-target signature size and SCFs.
The off-target signature bias found in the original PPIB and GAPDH studies was similarly observed in the chimeric siRNA experiments. Chimeric siRNAs having low SCFs generated fewer off-targets than medium and high SCF counterparts (Fig. 4B,C, P value between low and medium SCF class = 0.002). From this we conclude that the SCF alone can explain the observed bias and that siRNA sequences outside the seed region play little to no role in determining the extent of the off-target signature. Taken together, these studies are the first demonstration that the SCF is a critical siRNA design feature that can be manipulated to minimize off-target effects.
siRNAs with low SCFs generate fewer false positives in an RNAi phenotypic screen
Previous studies by Lin (Lin et al. 2005), Fedorov (Fedorov et al. 2006), and others demonstrated that off-target effects can induce measurable phenotypes. As siRNAs with low SCFs induced fewer off-targets, we predicted that (as a class) these duplexes should generate fewer off-target mediated false positives. To test this, 144 GAPDH, PPIB, and ACTB-targeting siRNAs having a preferred RISC-entry strand (as determined by thermodynamic end stability calculations described by Khvorova et al. 2003) and containing seeds with low, medium, or high SCFs (in the HeLa-expressed set) were transfected into HeLa cells and tested in the Apo-ONE (caspase 3/7 activity and apoptosis) assay. None of the target genes used in these studies have documented roles in apoptosis, therefore Apo-ONE phenotypes (defined as 1.5-fold or higher caspase activity over background) that result from siRNA delivery are assumed to have off-target origins. Data were plotted in five SCF groups (∼29 sequences per group) to investigate the false positive rates of siRNA with different SCFs in detail.
siRNAs with the lowest SCFs generated fewer Apo-One off-target phenotypes than duplexes with higher SCFs. Of the group of sequences having the lowest SCFs (between 91 and 588) only 10% generated off-target phenotypes (Fig. 5; Supplemental Table 3). The groups containing siRNAs with SCFs ranging from 613–2382 and 2385–2657 generated 21% and 31% false positive rates, respectively, while the remaining two groupings (SCFs between 2660–3301 and 3313–5377) exhibited a 21% and 14% false positive rate, correspondingly. Thus, in this study, the false positive rate of siRNAs with low (91–588) SCFs was two- to threefold lower than that observed with siRNAs having SCFs ranging from 613 to 3301. The false positive rates of siRNAs with SCFs in the range of 91–588 were statistically indistinguishable from siRNAs having SCFs in the range of 3313–5377. These trends were present regardless of whether data were binned according to equal numbers of siRNAs in each bin or equal-sized SCF ranges. Moreover, these same patterns were similarly observed in a cell viability screen (Fig. 5B), indicating that the described phenomenon is not assay dependent. These findings strongly support the notion that seed complement frequency can be employed in siRNA design algorithms to minimize false positives in RNAi-based phenotypic screening.
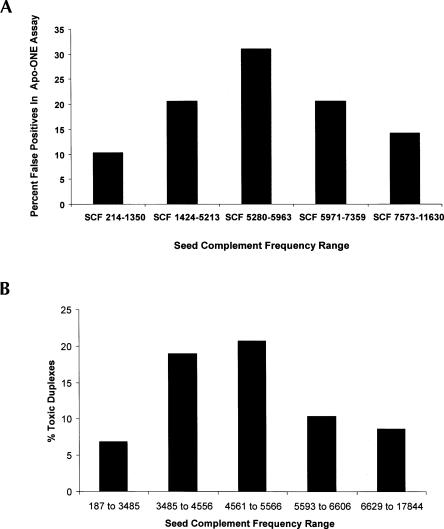
FIGURE 5.
A strong association exists between siRNA SCF and off-target induced phenotypes. (A) One hundred forty-four duplexes targeting PPIB, GAPDH, or ACTB were transfected into HeLa cells and assessed at 48 h using the APO-One assay. Sequences were divided into five seed complement frequency (SCF) ranges (91–588, 613–2382, 2385–2657, 2660–3301, and 3313–5377, 29 siRNAs per SCF range group with the exception of the highest group, which had 28 siRNAs) and plotted as the fraction of siRNAs in each group that induce positive phenotypes (scored as 1.5-fold or higher caspase induction over background). (B) Two hundred ninety duplexes targeting GAPDH or PPIB were individually transfected into cells as described by Federov et al. (2006). Following transfection, cell viability was assessed by alamarBlue according to the manufacturer's instructions. All measurements were performed in triplicate. Sequences were divided into five SCF ranges (187–3485, 3485–4556, 4561–5566, 5593–6606, and 6629–17844, 58 siRNAs per SCF group) and plotted as the fraction of siRNAs in each group that induced positive phenotypes (scored as inducing 50% or greater reduction in culture viability). SCF ranges used in both studies are derived from RefSeq 15 distributions.
A Web-based tool for identification of highly functional and specific siRNAs
The studies presented above identify 3′ UTR SCFs as a leading contributor to the size of off-target signatures and the nature of off-target induced phenotypes. While more detailed studies will be necessary to determine whether a stringent breakpoint exists for preferred SCFs, these findings represent a significant leap in our understanding of the off-target phenomenon and identify a critical parameter that can be incorporated into siRNA design algorithms to enhance specificity and minimize false positive phenotypes during gene functional analysis studies that employ RNAi-mediated silencing.
The value of the SCF specificity parameter is highlighted by the fact that functional siRNA are comparably distributed throughout all seed complement frequencies (data not shown), thus allowing the identification of functional siRNAs with low seed complement frequencies. For this reason, we have added seed-based specificity criteria to a publicly available siRNA evaluation Web tool (http://www.dharmacon.com/DesignCenter/).
DISCUSSION
Recent phenotypic screens using RNAi technologies have yielded widely variable false positive rates. In many cases, false positives have been tied to siRNA induced off-target effects, highlighting the need for highly specific assays and methods to reduce off-targets and off-target generated phenotypes. The studies presented above identify seed complement frequency as a contributing factor to the off-target signature size. The association of SCF and off-target profile size observed in this study is similarly present in previously reported off-target data sets (see Supplemental Table 4, based on data presented by Kittler et al. 2007), thus providing further support that SCF can be used to minimize off-target gene knockdown and the false positive phenotypes associated with them.
While the studies presented above have identified a critical attribute of off-target signatures, we have noted that the relationship between the SCF and off-target signature size is not strictly linear. In several cases the size of the off-target signature was observed to decrease at the higher SCFs. Though the reasons behind this phenomenon are not fully understood, we speculate that in the context of high-frequency seed complements, the off-target effects of the loaded RISC are widely dispersed, making the concentration of any single RISC-mRNA below the threshold required for pronounced gene down-regulation. While further studies will be necessary to dissect these phenomena, this model is consistent with the observed concentration dependence of off-target effects. In a separate issue not addressed by these studies, we and others have noted that regardless of the SCF, only a small fraction of possible targets having 3′ UTR seed matches are actually down-regulated (Bartel 2004; Farh et al. 2005; Lim et al. 2005; Birmingham et al. 2006). Why some genes containing 3′ UTR seed matches are off-targeted by a given siRNA while others are not is still unexplained. It may be (as suggested by Jackson et al. 2006b) that for genes to be off-targeted, they must also contain seed matches for endogenous miRNA regulators or that cooperatively between siRNA and miRNA seeds requires the positioning of sites within a certain distance of each other. While the importance of miRNA target sites and other sequences that play a role in mRNA stability remains to be explained, it is likely future studies in this field will identify additional points of overlap between siRNA off-targeting and endogenous post-transcriptional gene regulatory mechanisms.
The study presented here is based on a large database of experimentally confirmed off-targets (∼700 genes). While microarray is a robust and reproducible tool for off-target detection, caveats to this experimental approach should be noted. First, given (1) the similarities between siRNA off-targeting and miRNA targeting and (2) that the mechanism of miRNA targeting is believed (in many cases) to be translation attenuation, a substantially large off-target signature may also exist at the protein level. With this in mind, it should be noted that the mRNA signature acquired by microarray profiling may not be comprehensive, but instead is indicative of overall specificity of any individual duplex. Furthermore, it is possible that some fraction of the changes observed in the gene expression patterns could be attributed to biological responses resulting from minute discrepancies in either the concentration, activity, or stability of each siRNA. Though future studies designed to test these theories will require a rigorous proteomics study, (possibly at the level of single cell resolution) such analyses are expected to broaden our understanding of the specific and nonspecific cellular effects induced by introduction of siRNA.
While the low-frequency seed complement principle can be applied to all genes in the human genome, merging this attribute with the large numbers of functional design criteria can be challenging. In cases where only a small population of highly functional siRNAs are available for gene targeting, additional methodologies (including chemical modifications [Jackson et al. 2006a] and pooling strategies) are available to reduce off-target signatures. Also, a limited number of off-targets identified in this study contain no 3′ UTR seed matches. This may be an indication of (1) a seed-independent pathway for off-target generation; (2) the tolerance of GU wobble or mismatches in the seed region requiring compensatory binding elsewhere; or (3) genuine, downstream events triggered by on-target gene modulation. While future studies designed to understand these seed-independent gene knockdown events will likely lead to further enhancements in siRNA specificity and a deeper understanding of miRNA-mediated gene regulation, the demonstration that siRNA seed complement frequency can be used to enhance specificity provides a critical missing element in the optimization of siRNA design.
The utility of this experimentally confirmed criterion is twofold. First, new collections of silencing reagents (both synthetic siRNAs and expressed short interfering hairpins) can incorporate the concept of SCF to develop a new generation of tools that are both functional and highly specific. In addition, the importance of SCFs can be exploited to interpret data generated from preexisting siRNA collections. Depending upon the specificity of the assay, phenotypes generated by siRNAs with low SCFs may have a higher probability of resulting from knockdown of the intended target than those generated by siRNAs with medium SCFs. As such, a hit list generated by single siRNA duplexes could be weighted to give hits generated by low SCF duplexes the greatest priority. Thus, while a seed frequency evaluation tool is not a substitute for independent confirmation of phenotypes with multiple siRNAs, it may offer a quick and convenient strategy for primary high throughput hit stratification required for detailed follow-up studies.
MATERIALS AND METHODS
siRNA synthesis
Modified and unmodified 19-bp duplex siRNAs were synthesized using 2′-ACE chemistry (Hartsel et al. 2004) and contain 3′-UU overhangs on both strands, which are generally omitted from sequence lists and in the text. siRNA sequences used in this study are available in Supplemental Table 2.
Cell culture, transfection, and cellular assays
HeLa cells (ATCC) were cultured under standard conditions (37°C, 5% CO2, Dulbecco's Modified Eagle medium [HyClone], 10% fetal bovine serum, 2 nM L-glutamine, supplemented with penicillin [100 U/mL] and streptomycin [100 mg/mL]). For transfections, 10 × 104 cells were seeded per well (in a 96-well plate) 24 h before the experiment in antibiotic-free medium. Cells were transfected with siRNA (100 nM) using the cationic lipid DharmaFECT 1 (0.20 μL/well; ThermoFisher Scientific). Twenty-four hours following transfection, knockdown of the intended target was assessed by branched DNA assay (Quantigene, Panomics) using GAPDH (NM_002046) or PPIB (NM_000942) as a housekeeping reference. Cellular viability was monitored by alamarBlue reagent (Biosource)—25 μL per well, 2 h incubation, 37°C, 5% CO2. The effect of transfection on cell viability (alamarBlue, BioSource, Intl.) was less than 20% in all experiments. The Apo-ONE assay (Promega) was performed 48 h post-transfection according to the published protocol with the exception that the growth media was replaced upon addition of the Apo-ONE reagent. (Apo-One reagent is diluted 2× in fresh cell growth media. After aspiration of growth media from the plate, 100 μm of this diluted reagent is added to each well.) Chimeric reporters were constructed by inserting the 19-nt, fully complementary target site for each chimeric siRNA (see Supplemental Table 2) into the psiCHECK-2 vector (Promega). Dual luciferase activity was measured 48 h after co-transfection with the Dual-Glo Luciferase Assay System (Promega) and normalized to the empty psiCHECK-2 vector as described in the published protocol.
Protocols described by Federov et al. (2006) were used to determine the effects of SCF on cell viability. Briefly, 72 h (HeLa) or 144 h (MCF7, DU145) after transfection with siRNAs, 25 μL of Alamar Blue dye was added to wells containing cells in 100 μL of media. Cultures were then incubated 0.5 h (HeLa) or 2 h (MCF7 and DU145) at 37°C in a humidified atmosphere with 5% CO2. The data presented are an average of nine data points coming from three independent experiments performed in triplicate on different days. For the purpose of this study, siRNAs were defined as toxic when the average from nine different experiments (taking into account standard deviations) showed cell viability below 50%.
Microarray experiments
For each sample, 650 ng of total RNA isolated from siRNA-treated HeLa cells (collected 24 h post-transfection) was amplified, labeled with Cy5 (for experimental channel) or Cy3 (for control, reference channel) (Cy-5 and Cy-3 CTP, Perkin Elmer) using the Low Input RNA Fluorescent Linear Amplification Kit (Agilent) without the use of spike-ins. The hybridization reference was Cy3-labeled RNA derived from lipid-treated (control) samples, or Cy-3 labeled RNA from untreated cells in control arrays. Hybridizations were performed on Human 1A (V2) Oligo Microarrays (Agilent; >21,000 unique probes) for 17–20 h at 60°C, according to the published protocol (http://www.chem.agilent.com). Slides were washed using 6× and 0.06× saline–sodium phosphate–EDTA (SSPE) buffers (Amresco, each with 0.025% N-lauroylsarcosine), dried using a nonaqueous drying and stabilization solution (Agilent), and scanned (Agilent microarray scanner, model G2505B). The raw image was processed using Feature Extraction software (v8.5). Further analysis was performed using Spotfire Decision Site 8.1 software and the Spotfire Functional Genomics Module. Outlier flagging was not employed. Due to the large number of samples, only one array was processed per sample. This limitation does pose some constraints on the quality of the data generated by each individual duplex, as multiple replicates would give a range for the signature size. However, previous experiments have demonstrated excellent concurrence between biological replicates when transfections were performed under controlled conditions as described above (Birmingham et al. 2006; data not shown). A set of ∼15,000 records (the “HeLa-expressed set”) were selected for log ratio analysis based on their combined red and green processed signal being over 631 intensity units (log > 2.8) in the average measurement from three separate, biological replicate, self–self arrays (these arrays were performed and analyzed previously to all arrays reported in this study and are listed in the ArrayExpress data submission as HeLa self-self control #1, etc.). This cutoff was chosen because very few records in this set (typically, 10–30) exhibit changes over twofold in any of the self–self arrays due to either low signal, technical “noise,” or dye bias. From this set, off-targets were identified as genes that were down-regulated by twofold or more (log ratio more negative than −0.3) by any given siRNA but were not modulated by other functionally equivalent siRNAs targeting the same gene. Samples/arrays were processed in three batches based on target (PPIB-targeting, GAPDH-targeting, and chimeric siRNAs) with mock–mock self arrays and mock–untransfected arrays as controls for each batch. Records that exhibited significant log ratio differences in the mock self–self array or which displayed spurious log ratio differences across all of the arrays (∼50 records), were removed from the analysis, as well as several records in the GAPDH experiment that were identified as true downstream effects of GAPDH knockdown.
Computational methods
The frequency of hexamer occurrences in the human transcriptome was calculated by computationally enumerating all existing six-base strings in the 3′ UTRs of well-annotated RefSeq 15 human mRNAs (those with accession numbers prefixed with “NM”).
3′ UTRs were assumed to start at the first base position after the end of the coding sequence (as defined in each RefSeq record's feature table) and to run through the end of the RefSeq mRNA sequence. Hexamers containing degenerate bases were enumerated but removed from further analysis. These data were combined to determine the number of 3′ UTRs containing at least one of each possible cannonical hexamer. For calculation of hexamer SCF in 3′ UTRs of the HeLa-expressed set, the 15,000 records identified by microarray analysis described above were further abridged to only include well-annotated transcripts (resulting in ∼11,000 records).
The seed frequency analysis was performed on a subset of the off-target data for which adequate 3′ UTR annotation was available (853 distinct mRNAs). For each of these, we computationally identified a control mRNA from the set of well-annotated RefSeq 15 human mRNAs that had discernable signal and were not down-regulated by any of the tested siRNAs. The control was required to have a 3′ UTR length that was ± 55 nt in length relative to that of the off-targeted mRNA; three off-targets were discounted from further analysis due to the inability to find a control within this length-based parameter. The analysis counted the number of occurrences of exact substrings (identical to positions 13–18 inclusive) of an siRNA sense strand to the 3′ UTRs of each of its target and controls (when the same mRNA was off-targeted by multiple siRNAs, the same control was used for each case.) The significance of association between off-targeting and the presence of at least one hexamer match was calculated using a chi-square test for independence. The significance was assessed for each individual experiment (GAPDH, PPIB, chimera) and SCF class (low, medium, high) as well as for the combined set of experiments. All results were significant at less than p < 0.01 with the exception of the GAPDH low SCF test, which exhibited the same tendency at a slightly lower significance value (p = 0.01). An identical analysis was performed using positions 2–7 inclusive of sense strand, and no significant associations were identified.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org. Array data were deposited at ArrayExpress (E-MEXP-1402).
ACKNOWLEDGMENTS
We thank Queta Smith for helpful discussions and Cara Kotas for assistance in preparation of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.704708.
REFERENCES
- Bartel, D. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartz, S., Jackson, A.L. How will RNAi facilitate drug development? Sci. STKE. 2005;2005:pe39. doi: 10.1126/stke.2952005pe39. [DOI] [PubMed] [Google Scholar]
- Birmingham, A., Anderson, E.M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Stark, A., Russell, R.B., Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee-Kishore, M., Miller, C.P. Exploring the sounds of silence: RNAi-mediated gene silencing for target identification and validation. Drug Discov. Today. 2005;10:1559–1565. doi: 10.1016/S1359-6446(05)03623-8. [DOI] [PubMed] [Google Scholar]
- Chi, J.T., Chang, H.Y., Wang, N.N., Chang, D.S., Dunphy, N., Brown, P.O. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl. Acad. Sci. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, C.S., Hong, J., Sapinoso, L., Zhou, Y., Liu, Z., Micklash, K., Schultz, P.G., Hampton, G.M. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc. Natl. Acad. Sci. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., Sharp, P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Farh, K.K.-H., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fedorov, Y., Anderson, E.M., Birmingham, A., Reynolds, A., Karpilow, J., Robinson, K., Leake, D., Marshall, W.S., Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsel, S.A., Kitchen, D., Scaringe, S.A., Marshall, W.S. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. In: Herdewijn P., editor. Methods in molecular biology. Humana Press; Totowa, NJ: 2004. pp. 33–49. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J.M., Lim, L., Karpilow, J., Nichols, K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Schelter, J., Chau, B.N., Cleary, M., Lim, L., Linsley, P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw, A., Lian, S., Eystathioy, T., Li, S., Satoh, M., Hamel, J.C., Fritzler, M.J., Chan, E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kittler, R., Surendranath, V., Heninger, A.K., Slabicki, M., Theis, M., Putz, G., Franke, K., Caldareli, A., Grabner, H., Kozak, K., et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Krek, A., Grun, D., Poy, M., Wolf, R., Rosenberg, L., Epstein, E., MacMenamin, P., da Piedade, I., Gunsalus, K., Stoffel, M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis, B., Burge, C., Bartel, D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim, L., Lau, N., Garrett-Engele, P., Grimson, A., Schelter, J., Castle, J., Bartel, D., Linsley, P., Johnson, J. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin, X., Ruan, X., Anderson, M.G., McDowell, J.A., Kroeger, P.E., Fesik, S.W., Shen, Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Rivas, F.V., Wohlschlegel, J., Yates J.R., 3rd, Parker, R., Hannon, G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1161–1166. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, B., Held, M., Liebel, U., Erfle, H., Rogers, P., Pepperkok, R., Ellenberg, J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D.N., Fesik, S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L., Blau, H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R., Cohen, S. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′ UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez, M.A., Liu, J., Hannon, G.J., Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Xie, X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., Kellis, M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Haley, B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]