Abstract
Proper normalization is a critical but often an underappreciated aspect of quantitative gene expression analysis. This study describes the identification and characterization of appropriate reference RNA targets for the normalization of microRNA (miRNA) quantitative RT-PCR data. miRNA microarray data from dozens of normal and disease human tissues revealed ubiquitous and stably expressed normalization candidates for evaluation by qRT-PCR. miR-191 and miR-103, among others, were found to be highly consistent in their expression across 13 normal tissues and five pair of distinct tumor/normal adjacent tissues. These miRNAs were statistically superior to the most commonly used reference RNAs used in miRNA qRT-PCR experiments, such as 5S rRNA, U6 snRNA, or total RNA. The most stable normalizers were also highly conserved across flash-frozen and formalin-fixed paraffin-embedded lung cancer tumor/NAT sample sets, resulting in the confirmation of one well-documented oncomir (let-7a), as well as the identification of novel oncomirs. These findings constitute the first report describing the rigorous normalization of miRNA qRT-PCR data and have important implications for proper experimental design and accurate data interpretation.
Keywords: miRNA, normalization, RT-PCR, biomarker, lung cancer, oncomir
INTRODUCTION
MicroRNA represents an important new class of regulatory biomolecules, with roles in processes as diverse as early development, cell proliferation and differentiation, apoptosis, fat metabolism, and oncogenesis (for reviews, see Ambros 2004; Bartel 2004; Esquela-Kerscher and Slack 2006; Calin and Croce 2006a). Indeed, tumor-associated, differentially expressed miRNAs, termed “oncomirs,” have been found in many different cancers and the number of known oncomirs is growing rapidly (Calin and Croce 2006a; Esquela-Kerscher and Slack 2006). Moreover, miRNAs show promise in both diagnostic and therapeutic applications (Wu et al. 2006; Szafranska et al. 2007). Although the small size of miRNA (17–25 nucleotides [nt]) creates challenges for their detection, recent innovative adaptations of existing technologies for gene expression profiling, such as microarrays and quantitative RT-PCR (qRT-PCR), have emerged that make large-scale characterizations of miRNA expression patterns possible (Krichevsky et al. 2003; Nelson et al. 2004; Thomson et al. 2004). The accuracy of these methods for expression analysis, however, is critically dependent on proper normalization of the data inasmuch as inappropriate normalization of qRT-PCR data can lead to incorrect conclusions (Tricarico et al. 2002; Bas et al. 2004).
The goal of many miRNA qRT-PCR expression experiments is to identify differences between two groups of samples, often a “normal” control and a “disease” specimen. Thus, the purpose of normalization is to remove as much variation as possible between groups except for that difference that is a consequence of the disease state itself. Yet there are many other sources of variation in such experiments aside from disease-specific differential expression of a particular RNA. These sources may be technical, such as differences in sample procurement, stabilization, RNA extraction, and target quantification, or biological, reflecting sample-to-sample inconsistencies in cellular subpopulations or even differences in bulk transcriptional activity. Ideally, a normalizer is a single nucleic acid that exhibits invariant expression across all samples, is expressed along with the target in the cells of interest, and demonstrates equivalent storage stability, extraction, and quantification efficiency as the target of interest. In reality, such a normalizer does not exist (Vandesompele et al. 2002). In the case of mRNA, multiple published reports have argued for combinations of transcripts or ribosomal RNA (rRNA) normalizers as part of an empirical strategy to minimize unwanted variation (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004; Szabo et al. 2004).
The identification of RNA normalizers is not possible without first normalizing the data that is establishing a reference baseline of expression. This paradox has been addressed using pairwise measures (Vandesompele et al. 2002) and model-based estimates of expression variation (Andersen et al. 2004), among other approaches (Pfaffl et al. 2004; Szabo et al. 2004). Here we describe the use of two of these statistical methods to identify appropriate reference RNA species for miRNA qRT-PCR studies. As a class, miRNAs pose a significant challenge for normalization. First, miRNAs represent perhaps only 0.01% of the mass amount of total RNA in a sample, and this fraction can vary significantly across different samples (Liang et al. 2007). Thus, the normalizer of choice should mirror any wholesale changes in the global miRNA population. Further, a normalizer should have similar purification properties as miRNA. However, the extraction efficiency of miRNA from samples is very different than for much longer RNAs. Therefore, we chose to evaluate a panel of small RNAs—miRNA, ribosomal RNA, and small nuclear RNA (snRNA)—in pursuit of the most stable species in both normal and diseased human tissues. In all cases, methods for RNA extraction were used that are known to efficiently recover both small and large RNA populations. Analyses of microarray data of RNA isolated from dozens of tissues revealed a set of reference miRNA candidates that were further assessed across 13 distinct human solid tissues, and, separately, five pair of tumor and normal adjacent tissue (NAT). This analysis was then extended to include 12 pair of frozen lung cancer (LuCa) and NAT as well as 16 pair of formalin-fixed paraffin-embedded (FFPE) LuCa/NAT.
RESULTS
A global strategy for the selection of candidate RNA normalizers
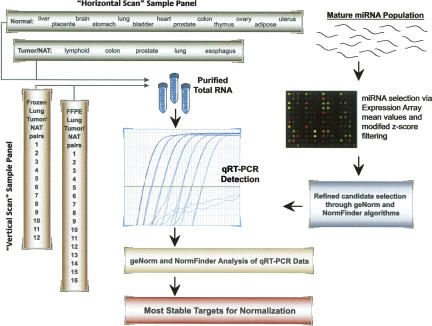
Our first goal was to develop a process for identifying the most stable targets as normalizers in miRNA qRT-PCR studies (Fig. 1). Beginning with miRNA microarray expression data, four criteria were used to select candidate normalizer miRNAs in the tissues of interest: (1) the miRNA must be highly expressed in most, if not all, of the samples; (2) the miRNA must be consistently expressed, as measured by the modified z-score (Supplemental Table 1); (3) only one representative from a given miRNA family should be considered; and (4) the miRNA must be a target of a commercially available qRT-PCR assay at the time of the work. Using these filters, data from the microarray content representing 287 human miRNAs resulted in the selection of 10–15 miRNAs for evaluation by qRT-PCR, depending on the sample set (see Supplemental Table 1 for more information). In addition, 5S rRNA (121 nt) and U6 snRNA (RNU6B, 45 nt) were included in some cases owing to their purported expression stability and use in several published miRNA qRT-PCR studies (Takamizawa et al. 2004; Choong et al. 2007; Corney et al. 2007; Pineles et al. 2007).
FIGURE 1.
Strategy for the identification of stable targets for normalizing microRNA qRT-PCR data.
Determination of the most stable miRNA normalizers in normal human tissues
Ideally, normalizers should be empirically validated for each sample type planned for qRT-PCR. The conventional wisdom is to procure 10–20 representatives of the sample(s) of interest and evaluate the stability of a set of possible normalizers within the relevant sample group(s). We refer to this approach as a “vertical scan” (Fig. 1), since the same panel of candidate normalizers is analyzed “down” multiple representatives of the same sample type. This strategy is contrasted with a “horizontal scan,” which is the evaluation of global stability of putative normalizers “across” many different sample types. We initiated our studies with a “horizontal scan” of normal human solid tissues to determine if miRNA, rRNA (5S), or snRNA (U6) demonstrated uniform expression stability across very different biological backgrounds. A plot of the raw Ct values by normal tissue type for each of the 12 RNA targets selected is shown in Supplemental Figure 1, and the raw data file for this and all other experiments described in this study are provided in the supplemental data.
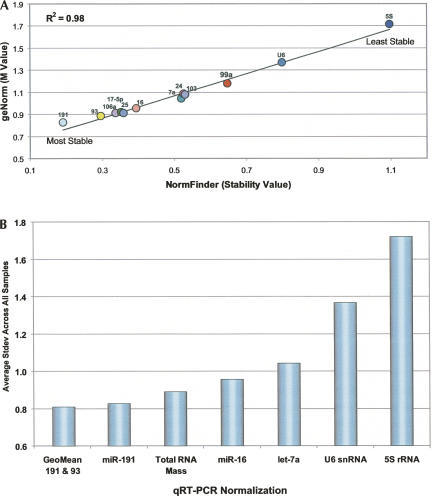
Two different algorithms were used to assess the variance in expression levels: geNorm (Vandesompele et al. 2002) and NormFinder (Andersen et al. 2004). As shown in Figure 2A, both statistical methods ranked the RNA targets similarly from most to least stable, with an excellent correlation in raw stability values. miR-191 was the most consistently expressed miRNA, followed by miR-93, miR-106a, miR-17–5p, and miR-25. In contrast, U6 and 5S, two commonly used normalizers for miRNA qRT-PCR experiments (Takamizawa et al. 2004; Pineles et al. 2007; Corney et al. 2007; Choong et al. 2007), were the least stable (Fig. 2; Supplemental Table 2). Indeed, the difference in stability between miR-191 and 5S was a standard deviation of nearly ±1Ct (cycle threshold), or a difference of ±2-fold. Normalization to total RNA mass was also evaluated, but this reference approach ranked behind miR-191 and miR-93 in stability (Fig. 2B).
FIGURE 2.
geNorm and NormFinder analyses of qRT-PCR data from a “horizontal scan” of 13 normal human tissues. (A) Correlation of the geNorm M value and the NormFinder stability value for the 12 RNA targets evaluated. (B) The average standard deviation across all tissue samples when normalized to (1) geometric mean (GeoMean) of miR-191 and miR-93, (2) miR-191, (3) total RNA mass, (4) miR-16, (5) let-7a, (6) U6 snRNA, and (7) 5S rRNA.
Determination of the most stable miRNA normalizers in a panel of cancer and NAT samples
qRT-PCR experimental designs typically include comparisons of normal and diseased tissue. As a result, we performed a “horizontal scan” of five sets of tumor and NAT sample RNA (lymphoid, colon, prostate, lung, and esophagus) (see also Supplemental Table 3) to evaluate the expression stabilities of a set of 12 miRNAs selected using the microarray gating criteria described above. The subsequent qRT-PCR data were analyzed both as two discrete groups (tumor and NAT), and one combined group (tumor + NAT). Interestingly, miR-191 was ranked as the most stable RNA by geNorm, and the second most stable by NormFinder for the NAT group, but not for the tumor group (Supplemental Table 4). miR-17–5p and miR-25 also ranked among the four most stable targets by both algorithms for the NAT group. These results were consistent with the finding that miR-191, miR-17–5p, and miR-25 were among the most stable miRNAs in the normal tissue “horizontal scan” (Supplemental Table 2), even though the NAT group was a unique sample set. In contrast, miR-103 was the most stable RNA in the tumor group. When the two sample groups were combined, both geNorm and NormFinder identified miR-191 and miR-25 as the most stable pair of normalizers. One important difference between the geNorm and NormFinder methods is that NormFinder allows for the designation of sample groups, and thus determination of both intragroup and intergroup (i.e., tumor versus NAT) variation. In this specific case, however, the intragroup variation—the only type of variation that geNorm measures—dominated the total observed variation, and thus both algorithms converged to the same pair of normalizers (here, miR-191 and miR-25).
Determination of the most stable miRNA normalizers in a panel of LuCa and NAT samples
The finding that specific miRNAs could be relatively stably expressed across different normal and cancerous tissue prompted us to evaluate miRNA normalizers in a “vertical scan” of LuCa paired with NAT samples. Two independent sample sets were evaluated: a panel of 12 flash-frozen LuCa tumor/NAT pairs (Supplemental Table 5), and a discrete collection of 16 FFPE LuCa tumor/NAT pairs (Supplemental Table 6). The rationale for this experimental design was twofold: (1) Assess the robustness of normalizer selection to different sample preparation methods using the same tissue type. (2) Determine the reproducibility in quantifying differential miRNA expression across the two sample sets following normalization using the independently determined most stable RNAs. Again, the miRNA targets were selected from array data using the gating criteria described above, with the exception that 5S was also included as a comparative reference. For both frozen and FFPE sample sets, NormFinder alone was used to identify the most stable RNA targets, given its capability to assess overall stability using distinct intragroup and intergroup measures of variability.
In the frozen LuCa sample pairs, miR-191 was determined to be the most stable single miRNA (Supplemental Table 7). The most stable combination of RNAs was miR-103 and let-7a. Closer inspection of the data reveals why miR-191 was not included in the combination of the two best normalizers: miR-191, though uniformly expressed in the respective tumor and NAT groups, was slightly underexpressed in the tumor set, whereas the combination of miR-103 and let-7a offered similarly low intragroup variability, but opposing intergroup differences that summed to essentially zero. Thus, consistent with the results presented above, miR-191 and miR-103 were among the most invariant reference RNAs (out of 16 total targets) across the 12 pairs of tumor and NAT tissues. 5S, by comparison, was one of the most unstable targets (ranked 13th out of 16 targets).
A subset of the most stable miRNAs from the frozen LuCa experiment was then evaluated in 16 pairs of FFPE LuCa tumor and NAT samples. The most stable RNA was miR-103, followed by miR-191 (Supplemental Table 8). The most stable pair of targets was miR-17–5p and miR-24. Strikingly, six of the seven miRNAs common to the frozen and FFPE LuCa datasets followed the same rank order of stability (Supplemental Table 9). The lone exception was miR-103, which ranked first in the FFPE LuCa set (out of eight total targets), but fourth in the frozen LuCa set (out of 16 total targets).
A case study for normalized differential expression measurements by miRNA qRT-PCR: Quantification of let-7a in nonsmall cell LuCa
An additional goal of this study was to determine how the independent selection of most stable normalizers would impact the quantification of miRNAs in normal versus tumor group comparisons. We selected let-7a as a case study for the following reasons: (1) several independent studies have reported that let-7a is down-regulated in LuCa (Takamizawa et al. 2004; Johnson et al. 2005; Yanaihara et al. 2006; Inamura et al. 2007); (2) a published study has measured let-7a expression in LuCa using the same qRT-PCR assay format described here (Inamura et al. 2007), permitting a quantitative comparison; and (3) a plausible biological model to support the consequences of let-7a down-regulation in LuCa tumors has been successfully tested (Johnson et al. 2005).
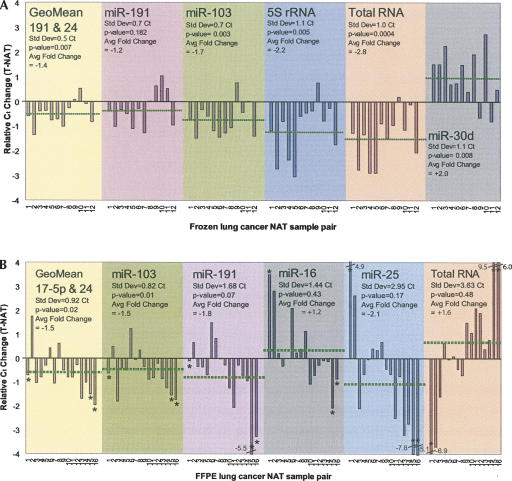
Consequently, we evaluated let-7a expression in both the frozen and FFPE LuCa sample sets when normalized to RNA targets representing both relatively stable and unstable expression. As shown in Figure 3A, let-7a expression was 1.2-fold lower in tumors than it was in NAT when normalized to miR-191, although this modest change was not statistically significant (P = 0.182). When let-7a was normalized to the most stable pair that did not include let-7a (that is, miR-191 and miR-24), the fold change was comparable (−1.4-fold), but also statistically significant (P = 0.007). In fact, this degree of reduced expression of let-7a in tumors was similar to that reported by Inamura et al. (2007) (approximately −1.8-fold) using qRT-PCR. However, if either 5S (P < 0.005) or total RNA (P < 0.0004) were used as normalizers, the fold change for let-7a down-regulation was roughly twice (−2.2 to −2.8-fold) that measured after normalization with the most stable reference miRNAs. Most alarmingly, normalization to miR-30d reported twofold higher, rather than lower, let-7a expression in tumors with statistical confidence (P = 0.008). This misleading result is a consequence of the systematically lower expression of miR-30d in LuCa.
FIGURE 3.
Differential expression of hsa-let-7a in human lung cancer: impact of normalizer stability on qRT-PCR expression analyses from both flash-frozen and FFPE tumor and normal adjacent tumor tissues. (A) Hsa-let-7a differential expression in 12 flash-frozen lung cancer NAT pair as normalized to (1) geometric mean (GeoMean) of miR-191 and miR-24, (2) miR-191, (3) miR-103, (4) 5S rRNA, (5) total RNA, and (6) miR-30d. The dotted line indicates the respective average differential expression value. Negative values indicate reduced let-7a expression in lung cancer samples and positive values indicate increased let-7a expression in lung cancer samples. (B) Hsa-let-7a differential expression in 16 FFPE lung cancer NAT pairs as normalized to (1) geometric mean (GeoMean) of miR-17–5p and miR-24, (2) miR-103, (3) miR-191, (4) miR-16, (5) miR-25, and (6) total RNA. The dotted line indicates the respective average differential expression value. (*) Indicates an outlier. Negative values indicate reduced let-7a expression in lung cancer samples and positive values indicate increased let-7a expression in lung cancer samples.
Next, a subset of the most stable miRNAs was evaluated in the FFPE LuCa sample set (Fig. 3B). In this case, normalization to miR-103 resulted in 1.5-fold lower expression for let-7a in tumor (P = 0.01). Similarly, normalization to the most stable pair, miR-17–5p and miR-24, revealed 1.5-fold lower expression in tumors (P = 0.02). In contrast, if let-7a were normalized to less stable targets, such as miR-25, miR-16, or even total RNA, expression was interpreted as either lower (miR-25) or higher (miR-16, or total RNA) in tumors, with none of the outcomes reaching statistical significance. Another important finding was that normalization to total RNA failed to correct for large variations in expression as observed with some samples. For example, three samples (see starred samples in Fig. 3B) produced unexpectedly high Ct values in either the tumor or the NAT RNA. Normalization to total RNA cannot correct for inadvertent errors in total RNA quantification, or the effects of inhibitors that reduce the efficiency of the RT and/or PCR steps that can produce such anomalies. In contrast, endogenous reference miRNAs are coamplified in the same sample, and thus can help correct data outliers that are sometimes observed when working with real-world clinical specimens.
As a result, normalization to the most stable pair of miRNAs for the frozen and FFPE sample panels, respectively, both supported the statistically significant conclusion of down-regulation of let-7a in lung tumors, consistent with literature reports (Takamizawa et al. 2004; Johnson et al. 2005; Yanaihara et al. 2006; Inamura et al. 2007). Moreover, the quantification of differential expression was highly reproducible between the independent frozen (−1.4-fold) and FFPE (−1.5-fold) sample sets (Fig. 3).
Normalization with the most stable miRNAs identifies novel oncomirs in LuCa
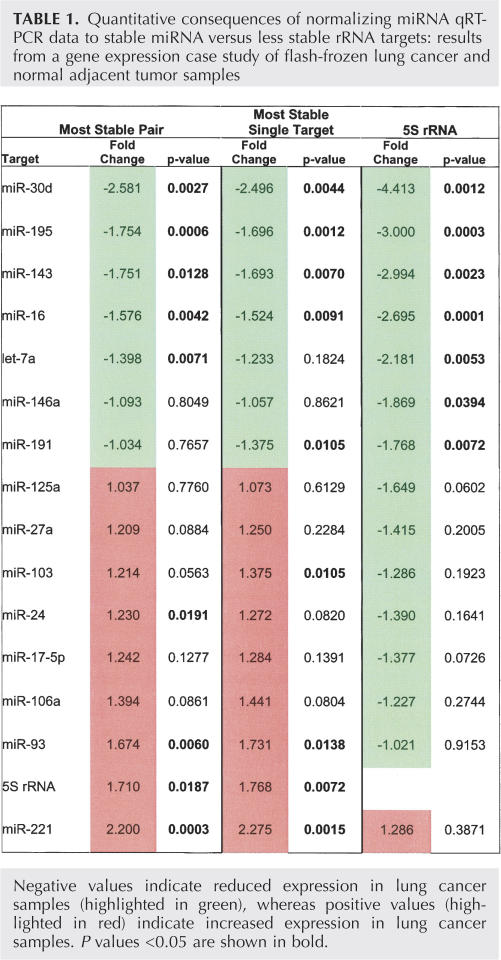
Having determined the most invariant miRNA normalizers in LuCa tissue, we next measured the expression differences associated with each of the RNAs that were evaluated in the frozen sample collection. The 16 RNA targets were individually normalized to: (1) the most stable pair or targets, (2) the most stable single target, or (3) 5S rRNA. The fold change and P-value was determined in each case (Table 1). Normalization to stable RNA targets identified ∼30% more targets that were differentially expressed at statistical confidence (P < 0.05), and, overall, a much more balanced group of up- and down-regulated targets, as would be expected from this random sampling of miRNA targets. Moreover, two miRNAs, miR-30d and miR-221, were determined to be >2-fold differentially expressed in the LuCa samples, a result supported by a separate analysis of the FFPE LuCa sample panel (data not shown). We note miR-30d has been previously reported to be reduced in LuCa (Volinia et al. 2006); however, miR-221, which was 2.2-fold up-regulated in our study, has not been described as differentially expressed in LuCa. Importantly, miR-221 would not have been identified as differentially expressed if 5S had been used as a normalizer (P = 0.39).
TABLE 1.
Quantitative consequences of normalizing miRNA qRT-PCR data to stable miRNA versus less stable rRNA targets: results from a gene expression case study of flash-frozen lung cancer and normal adjacent tumor samples
DISCUSSION
The goal of normalization is to minimize data variation that can mask or exaggerate biologically meaningful changes, thereby increasing the accuracy of expression measurements. The choice of a normalization strategy, however, is anything but trivial. In microarray studies, normalization methods usually invoke sophisticated, population-based approaches that exploit abundance measures of hundreds, if not thousands, of genes. This luxury is absent in qRT-PCR, where assays are typically designed to target a few to dozens of targets, and limitations in optical bandwidth constrain the number of multiplexed targets that can be quantified in a single tube to five or less. Other considerations, such as cost and the availability of sample material, may also restrict the number of normalizers that can be evaluated.
Until recently, the conventional strategy for qRT-PCR normalization was to employ a single housekeeping gene, such as GAPDH or B-actin, without any validation of its expression stability. This approach is ill-advised as such genes can vary by 10-fold or more across different samples (Warrington et al. 2000). Vandesompele et al. (2002) first proposed an empirical approach to selecting suitable normalizers for mRNA qRT-PCR, advocating that one to three reference genes should be validated for each sample type and corresponding sample preparation method. Yet, to our knowledge, no study has been published that has systematically evaluated normalization targets in miRNA qRT-PCR assays, despite the surge of interest in miRNA identification and quantification.
To date, only a handful of miRNA qRT-PCR studies have described some form of normalization, using RNA targets such as 5S (Takamizawa et al. 2004; Pineles et al. 2007), U6 (Choong et al. 2007; Corney et al. 2007; Shell et al. 2007), 18S (Iorio et al. 2007), or miR-16 or let-7a (Mattie et al. 2006). Rigorous experimental justification for the selection of these targets in these publications has been lacking. Importantly, our data suggests that total RNA is inferior to miRNAs such as miR-191, miR-103, and miR-17–5p in more than one biological context. In fact, 5S or U6 were the two least stable RNA species in a panel of 12 RNA targets evaluated across 13 discrete normal human tissues (Fig. 2). The fact that all of the miRNAs that were tested were superior to these two commonly used normalizers may in part reflect the effectiveness of the miRNA microarray selection criteria (Fig. 1).
Interestingly, the most stable miRNAs identified in the normal tissue survey were also among the most stable RNA targets in experiments with other sample sets. For example, miR-191, miR-25, and miR-17–5p were three of the four most stable RNAs in the NAT group from a panel of five distinct cancer tissues, consistent with their stability in the normal tissue survey. In tumor samples, miR-103 was the most invariant RNA, whereas the combination of miR-191 and miR-25 was the most stable pair.
To extend these findings, we next evaluated RNA stability in 12 pairs of flash-frozen LuCa/NAT tissues, and, separately, in 16 pairs of FFPE LuCa/NAT tissues. The rank order of RNA stabilities for those seven RNAs in common to both sample sets were extremely consistent, with six out of seven miRNAs ordering the same from most to least stable. This conservation in rank order between the two unique sample sets was noteworthy, since each of the reference RNAs demonstrated uniform expression in the independent frozen LuCa set, and were clustered within a relatively small range of expression stabilities. Consistent with previous data, miR-191 and miR-103 were the most stable single reference targets in both sample sets. Thus, the miRNA stability ranking is robust to the sample treatment and processing for human lung tissue.
The practical consequences of miRNA normalization were then evaluated using let-7a as a case study, given the evidence from multiple groups that let-7a is down-regulated in lung tumors (Takamizawa et al. 2004; Johnson et al. 2005; Yanaihara et al. 2006; Inamura et al. 2007). The results revealed that the reduced expression of let-7a in LuCa was determined with quantitative consistency (<10% difference in fold change) and statistical confidence (P < 0.05) across both the frozen and FFPE sample sets when the most stable pair of reference miRNAs were used to normalize the data in each case. Additionally, normalization to less stable reference species caused both quantitative (i.e., magnitude of fold change) and qualitative (i.e., direction of fold change) errors in let-7a differential expression. Worse yet, inappropriate normalization can support statistical confidence in the wrong conclusion (see miR-30d panel in Fig. 3A, which suggests that let-7a expression is up-regulated in LuCa).
There are several important implications from these findings. First, large differences in expression between normal and disease groups may be tolerant to poor normalization, but small differences certainly will not. It has been suggested that the capability of miRNAs to regulate multiple targets within the same pathway can amplify their biological effects (Calin and Croce 2006b). As a result, even relatively small changes in miRNA expression may be biologically significant. For this reason, rigorous normalization of miRNA data may be even more critical than that of other RNA functional classes. In fact, stringent normalization may be required in some cases to enable a detailed understanding of miRNA biology, and it may be critical for the potential development of reliable diagnostic assays. Second, a single miRNA normalizer may be sufficient in some experimental situations, but more than one may be required to produce accurate data that can be interpreted with confidence. Last, the consistency of the results between frozen and FFPE LuCa sample sets suggests that normalization of miRNA qRT-PCR assays using reference miRNA can be extremely robust. This is a particularly remarkable result given that the two tissue sources were procured independently of one another, and, further, that FFPE samples present significant sources of variation in RNA expression profiling compared to flash-frozen samples (Xi et al. 2007). For example, FFPE samples are subjected to harsh chemical fixation and high temperature during the embedding process—procedures that both damage RNA species and increase the variability in RNA representation following extraction and amplification. The observed conservation in miRNA stability across frozen and FFPE sample groups is a testament to their utility in normalizing qRT-PCR data using very different tissue preparation methods. It has been speculated that miRNA, owing to their small size, can survive this harsh tissue processing with less variability than mRNA (Xi et al. 2007).
Finally, we recognize that the effort, cost, and sample requirements necessary for the experimental selection of miRNA normalizers is not always possible. In such cases, the data presented here suggests that miR-191, miR-103, and/or miR-17–5p, are likely more reasonable choices than the default use of U6, 5S, or even a more or less randomly selected miRNA such as miR-16. However, it is clear that empirical validation is the optimal strategy for ensuring accurate miRNA quantification by qRT-PCR.
MATERIALS AND METHODS
Total RNA samples
Samples sets included: (1) 13 individual normal flash-frozen human tissue RNAs (Fig. 1), (2) five flash-frozen human tumor/NAT RNA pairs (Fig. 1; Supplemental Table 3), (3) 12 flash-frozen human lung tumor/NAT RNA pairs (Supplemental Table 5), and (4) 16 FFPE human lung tumor/NAT RNA pairs. Samples derived from human patients were acquired from commercial suppliers by Asuragen's Tissue Procurement Group in compliance with the regulations as outlined in 45 CFR 46 and other regulatory guidance (Supplemental Table 6).
Accurate and sensitive miRNA quantification is only possible when methods are used that efficiently isolate small RNA species. For this reason, FirstChoice Total RNA samples (Ambion), certified to contain small RNAs (miRNA, siRNA, and snRNA) as well as large RNAs (rRNA, mRNA, and tRNA), were used to generate microRNA expression profiling and real-time RT-PCR data unless otherwise noted. Total RNA from the 12 pair of flash-frozen lung tissues was extracted using mirVana miRNA Isolation Kit (Ambion) following the manufacturer's protocol to ensure the recovery of small RNA. Total RNA from 16 pair of FFPE lung tissues was isolated using RecoverAll Total Nucleic Acid Isolation Kit (Ambion) as described by Doleshal et al. (in press).
RNA concentrations were verified by measuring absorbance (A260) on the NanoDrop Spectrophotometer ND-1000 (NanoDrop) and total RNA profiles were assessed on the Agilent 2100 bioanalyzer (Agilent Technologies) with equal mass loadings of 100 ng per sample onto the RNA 6000 Nano LabChip kit. The Eukaryote Total RNA Nano assay on the 2100 bioanalyzer expert software reported 28S/18S ratios in the typical range of 1.1–1.8 for RNA isolated from flash-frozen tissues. RNA profiles for FFPE RNA samples were not assessed on the bioanalyzer.
Selection of candidate targets for normalization
miRNA array expression profiling data sets were prepared and generated in-house using mirVana miRNA Bioarrays V1 (Ambion) as previously described (Shingara et al. 2005). Within each array data set, miRNA targets were further standardized by a modified z-score ranking as described in Supplemental Table 1. Following the modified z-score filtering, each normalization candidate was transformed to a quantity as outlined by the authors of geNorm and NormFinder (Vandesompele et al. 2002; Andersen et al. 2004), creating a list of targets ranked accordingly relative to a stability value (Supplemental Table 1). In addition, 5S and U6 TaqMan assays were included based on their historical use for Northern blot normalization. Both assays for 5S and U6 had comparable reproducibility and performance to the miRNA TaqMan assays.
Real-time RT-PCR
qRT-PCR was performed in duplicate, including minus reverse transcription (RT) controls to assess genomic DNA and nontemplate controls that ensured a lack of signal in the assay background. The RT reaction consisted of 1.0 μL 10× RT Buffer (Ambion), 1.0 μL dNTPs 2.5 mM each (Ambion), 0.5 μL 5× RT Primer (Applied Biosystems), 0.1 μL RNase Inhibitor Protein 40 U/μL (Ambion), 0.1 μL wt-MMLV-RT 100 U/ul (Ambion), and 500 pg total RNA in a final volume of 10 μL. Reactions were incubated on a 384-well GeneAmp PCR System 9700 at 16°C for 30 min, 42°C for 30 min, then 85°C for 5 min. Following the RT step, 2 μL of the RT product was transferred into a 15 μL PCR consisting of 1.5 μL 10× PCR Buffer (Invitrogen), 1.5 μL 50 mM Magnesium Chloride (Invitrogen), 1.5 μL dNTP's 2.5 mM each (Ambion), 0.3 μL 20× TaqMan Assay (Applied Biosystems), 0.3 μL 50× ROX Standard (Ambion), and 0.1 μL Platinum Taq 5 U/μL (Invitrogen). PCR cycling began with template denaturation and hot start Taq activation at 95°C for 1 min, then 40 cycles of 95°C for 5 sec, and 60°C for 30 sec performed on a 7900HT Fast Real-Time PCR System with data collected during each cycle at the 60°C extension step with 7900HT SDS v2.3 (Applied Biosystems). Threshold and baselines were manually determined with thresholds typically set between 0.05 to 0.1 paired with a baseline starting at 1 – 3 Cts and finishing at 15 – 17 Cts.
qRT-PCR data analysis
The qRT-PCR results were imported into Microsoft Excel and the average value of duplicate Ct values converted to quantities for geNorm and NormFinder analysis (Vandesompele et al. 2002; Andersen et al. 2004). The assessment of let-7a expression comparing different targets as normalizers in LuCa was determined by the ddCt comparative threshold (ΔΔCt) method. P-values were determined by a two-tailed paired Student's t test from the ΔCt values of tumor and NAT or Ct values in the case of normalization to total RNA (Fig. 3A,B). Normality of the data was assessed by Lilliefor's hypothesis test.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work is supported in part by grant R44CA118785 from the National Cancer Institute (NCI). We thank Ania Szafranska for generous access to FFPE LuCa tumor/NAT samples, Tim Davison for helpful statistical analysis discussions, and Bernie Andruss for his critical review of manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.939908.
REFERENCES
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andersen, C.L., Jensen, J.L., Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bas, A., Forsberg, G., Hammarström, S., Hammarström, M.L. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand. J. Immunol. 2004;6:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006a;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Croce, C.M. MicroRNA–cancer connection: The beginning of a new tale. Cancer Res. 2006b;15:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Choong, M.L., Yang, H.H., McNiece, I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp. Hematol. 2007;4:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Corney, D.C., Flesken-Nikitin, A., Godwin, A.K., Wang, W., Nikitin, A.Y. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;18:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Doleshal, M., Magotra, A.A., Choudhury, B., Cannon, B.D., Labourier, E., Szafranska, A.E. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J. Mol. Diag. 2008 doi: 10.2353/jmoldx.2008.070153. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher, A., Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Inamura, K., Togashi, Y., Nomura, K., Ninomiya, H., Hiramatsu, M., Satoh, Y., Okumura, S., Nakagawa, K., Ishikawa, Y. Let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58:392–396. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Iorio, M.V., Visone, R., Di Leva, G., Donati, V., Petrocca, F., Casalini, P., Taccioli, C., Volinia, S., Liu, C.G., Alder, H., et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;18:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Johnson, S.M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K.L., Brown, D., Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., Kosik, K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Ridzon, D., Wong, L., Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie, M.D., Benz, C.C., Bowers, J., Sensinger, K., Wong, L., Scott, G.K., Fedele, V., Ginzinger, D., Getts, R., Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W., Tichopad, A., Prgomet, C., Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;6:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Pineles, B.L., Romero, R., Montenegro, D., Tarca, A.L., Han, Y.M., Kim, Y.M., Draghici, S., Espinoza, J., Kusanovic, J.P., Mittal, P., et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 2007;3:261.e1–e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Shell, S., Park, S.M., Radjabi, A.R., Schickel, R., Kistner, E.O., Jewell, D.A., Feig, C., Lengyel, E., Peter, M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingara, J., Keiger, K., Shelton, J., Laosinchai-Wolf, W., Powers, P., Conrad, R., Brown, D., Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., Perou, C.M., Karaca, M., Perreard, L., Quackenbush, J.F., Bernard, P.S. Statistical modeling for selecting housekeeper genes. Genome Biol. 2004;5:R59. doi: 10.1186/gb-2004-5-8-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska, A.E., Davison, T.S., John, J., Cannon, T., Sipos, B., Maghnouj, A., Labourier, E., Hahn, S.A. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:1–11. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;11:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., Parker, J., Perou, C.M., Hammond, S.M. Custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tricarico, C., Pinzani, P., Bianchi, S., Paglierani, M., Distante, V., Pazzagli, M., Bustin, S.A., Orlando, C. Quantitative real-time reverse transcription polymerase chain reaction: Normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2007-3-8-research0034. RESEARCH0034.1; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia, S., Calin, G.A., Liu, C.G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington, J.A., Nair, A., Mahadevappa, M., Tsyganskaya, M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics. 2000;3:143–147. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- Wu, W., Sun, M., Zou, G.M., Chen, J. MicroRNA and cancer. Current status and prospective. Int. J. Cancer. 2006;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- Xi, Y., Nakajima, G., Gavin, E., Morris, C.G., Kudo, K., Hayashi, K., Ju, J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., Stephens, R.M., Okamoto, A., Yokota, J., Tanaka, T., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]