Abstract
When gene conversion is initiated by a double-strand break (DSB), any nonhomologous DNA that may be present at the ends must be removed before new DNA synthesis can be initiated. In Saccharomyces cerevisiae, removal of nonhomologous ends depends not only on the nucleotide excision repair endonuclease Rad1/Rad10 but also on Msh2 and Msh3, two proteins that are required to correct mismatched bp. These proteins have no effect when DSB ends are homologous to the donor, either in the kinetics of recombination or in the proportion of gene conversions associated with crossing-over. A second DSB repair pathway, single-strand annealing also requires Rad1/Rad10 and Msh2/Msh3, but reveals a difference in their roles. When the flanking homologous regions that anneal are 205 bp, the requirement for Msh2/Msh3 is as great as for Rad1/Rad10; but when the annealing partners are 1,170 bp, Msh2/Msh3 have little effect, while Rad1/Rad10 are still required. Mismatch repair proteins Msh6, Pms1, and Mlh1 are not required. We suggest Msh2 and Msh3 recognize not only heteroduplex loops and mismatched bp, but also branched DNA structures with a free 3′ tail.
In Saccharomyces cerevisiae, homologous recombination initiated by double-strand breaks (DSBs) can occur by at least two distinct pathways: gene conversion and single-strand annealing (SSA) (1–6). In both cases, the ends of the DSB are resected by a 5′ to 3′ exonuclease to produce long 3′ ended single-strand tails (7, 8). In gene conversion, these tails invade a homologous donor sequence and act as primers of new DNA synthesis. However, for this to occur, any nonhomologous bases at the 3′ end must be removed, so that the primer end may basepair with the donor template. Similarly, in SSA, complementary strands of homologous regions flanking a DSB can anneal, producing an intermediate that has two nonhomologous 3′ ended tails that must be removed before new DNA synthesis and ligation can occur. In both instances, removal of nonhomologous tails depends on the Rad1 and Rad10 proteins (9), which have been shown in vitro to cleave 3′ ended nonhomologous tails and which carry out a related function in nucleotide excision repair (NER) (10–12). Other NER proteins are not required (13).
In NER, Rad1/Rad10 are presumably recruited to the site of DNA damage by other proteins of the NER repair complex. We were interested in whether other proteins were required to attract Rad1/Rad10 to the sites of strand invasion or strand annealing. One set of proteins that recognize DNA distortions are the mismatch repair proteins, most notably Msh2, which has been shown to bind to mismatched bp (14), heteroduplex loops (14), and Holliday junctions (15). Msh2 has been shown to form heterodimers with Msh6 (16, 17) and studies of the specificity of mismatch repair have led to the conclusion that Msh2/Msh6 primarily recognize and correct single bp mismatches, while Msh2 and Msh3 act to correct heteroduplex DNA containing small loops formed by frameshift mutations (16, 18). All of these mismatch repair events require the participation of the Pms1/Mlh1 heterodimer (19, 20). Evidence that mismatch repair proteins might be involved in the removal of 3′ ended single-strand DNA tails came from the observations by Saparbaev et al. (21) that Msh2 and Msh3 were required in spontaneous recombination events that also depend on Rad1 and Rad10. It was not known whether these events were initiated by single-strand or double-strand DNA lesions, nor at which molecular step Msh2 and Msh3 acted, but Saparbaev et al. suggested that these proteins might be involved in the resolution of Holliday junctions.
We report here experiments showing that Msh2 and Msh3, but not other mismatch repair proteins, are required to remove nonhomologous DNA ends during both the initiation of gene conversion and the resolution of SSA intermediates that are initiated by a DSB. When the ends of recombining DNA are perfectly matched to their donor templates, Msh2 and Msh3 are not required, nor do they appear to affect the resolution of Holliday junctions. Additional experiments support our conclusion that Msh2 and Msh3 act to facilitate Rad1/Rad10-dependent removal of nonhomologous DNA ends.
MATERIALS AND METHODS
Strains and Growth Conditions.
All strains were isogenic derivatives of either JKM111 (22) or tNR85 (23), both of which contain mutations of the HO cleavage site at MATa, so that only the introduced HO cutting site is cleaved. The two strains are not isogenic; however we have found that HO-induced recombination of the same substrate (plasmid pJF5; ref. 1) yielded a nearly identical efficiency and kinetics of DSB repair and the proportion of gene conversion events accompanied by crossing-over was the same (data not shown). Deletions of the RAD1, MSH2, and MSH3 were made by gene replacement (24) using the following constructs: rad1::LEU2 in plasmid pL962, from R. Keil; msh2::LEU2 in pRHB113 (25) and msh3::LEU2 (26). G. Marsischky and R. Kolodner constructed and provided strains RKY3111 (msh6::hisG) and RKY3112 (msh3::hisG) in tNR85. The GAL::HO gene was integrated at the ADE3 locus (27) in JKM146 and was carried on plasmid pFH800 (28) in the tNR85 derivatives. Induction of HO endonuclease was accomplished by adding galactose (2% final concentration) to cells grown in lactate-containing medium at 30°C, or by plating them on yeast extract/peptone-galactose.
Measurement of DSB Repair Efficiency.
To measure repair and retention of pFP122, pFP140, and pFP120, cells were plated on yeast extract/peptone/dextrose and yeast extract/peptone-galactose plates, and the resulting colonies replicated to plates lacking uracil, to determine plasmid retention. The percent plasmid retention, a measure of successful gene conversion, was calculated from the fraction of colonies retaining the plasmid on galactose medium divided by the same fraction on dextrose medium. Cell viability and deletion formation in the SSA assay were measured by plating cells on yeast extract/peptone/dextrose and replica plating cells to synthetic complete medium lacking tryptophan (29) to score for the retention of pFH800.
Recombination Substrates.
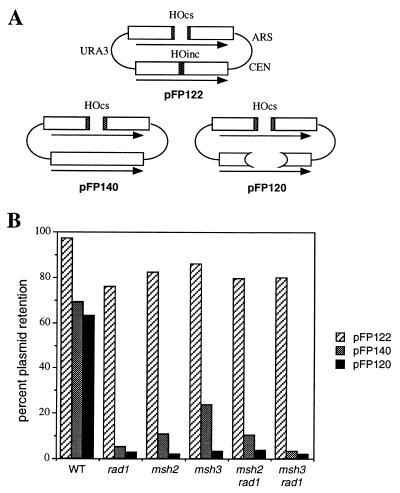
Plasmids pFP120, pFP122, and pFP140 were similar to the centromeric URA3-marked plasmids containing inverted copies of the Escherichia coli LacZ sequence used previously (30), except that they did not carry LEU2. Their structure is shown in Fig. 1A. In pFP122, the recipient LacZ sequence contains a 40-bp MATa cleavage site. The donor locus carries a mutant cut site differing by only a single bp substitution at position Z6 that prevents HO cleavage (31). Therefore, the DSB generates a cut plasmid whose ends are almost exactly identical to the uncleaved donor sequence that is used to repair the break by gene conversion. In pFP140, the recipient LacZ sequence carries a 60-bp MATa cleavage site, so that the DSB creates two ends with 30 bp of nonhomology that must be removed before gene conversion can begin. pFP120 contains a 40-bp cleavage site in the recipient LacZ sequence, and the donor lacZ copy is deleted for the 878-bp ClaI–SacI fragment; consequently, the HO cleavage site of the recipient is now surrounded by two large sequences (308 and 610 bp) without any homologous counterpart in the donor.
Figure 1.
Effect of MSH and RAD genes on HO endonuclease-induced gene conversion repair of a DSB. (A) Structure of the pFP122, pFP140, and pFP120 plasmids. (B) DSB repair efficiency in wild-type, rad1, msh2, msh3, msh2 rad1, and msh3 rad1 strains.
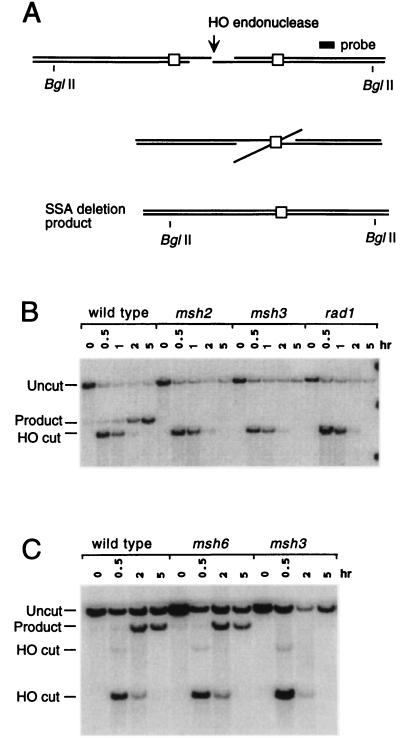
The SSA chromosomal substrates were derived from a triplication of URA3 sequences similar to tNS62 (8) except that the distal ura3–52 gene was completely deleted and replaced by a THR4 gene. The resulting structure on chromosome V has a 205-bp region of the 5′ end of the HindIII URA3 fragment duplicated on either side of an HO cleavage site. In Fig. 3 both duplicated regions were shown by sequencing to be identical to the URA3 sequence derived from strain +D4 (32), except that they both possess the 5-bp deletion found in strain FL100 (32) and a T to C substitution at nucleotide 57 of the +D4 sequence. In the strains shown in Fig. 4, the 205-bp segment to the left of the cleavage site is identical to that from FL100 and hence has seven single bp substitutions or frameshifts relative to the 205-bp region to the right of the cut site. In Fig. 5 similar substrates with 205, 415, or 1,170 bp of identical sequences were examined. Additional experiments were carried out using a LEU2-marked centromeric plasmid, pNSU208, similar to pJF6 (9), but carrying only 240 bp of directly repeated LacZ sequences.
Figure 3.
Requirement for Msh2, Msh3, and Rad1 in SSA. (A) SSA between identical 205-bp URA3 gene segments flanking an HO-induced DSB. Extensive 5′ to 3′ exonuclease digestion produces 3′ ended DNA tails in which complementary regions can anneal. Cleavage of the 3′-ended tails allows new DNA synthesis to fill in gaps and ligate the deletion. Diagnostic BglII fragments are indicated. (B) Southern blot of the time course of deletion formation in wild-type cells. BglII-digested DNA reveals HO cleavage and the formation of the deletion in the wild-type strain (tNS1379). Deletion formation is significantly reduced in msh2 (tNS1406), msh3 (tNS1390), and rad1 (tNS1429) strains. SSA was analyzed by probing with a HindIII–BamHI DNA fragment centromere proximal to the URA3 gene (8). (C) Southern blot of the time course of deletion formation on plasmid pNSU208 carrying 240 bp of directly repeated LacZ sequences in wild-type cells (tNS865), compared with msh3 (tRKY3112) and msh6 (tRKY3111) strains. DNA was digested with EcoRI. All strains were derivatives of tNR85 (22) and carried GAL::HO on a TRP1-marked plasmid, pFH800 (27).
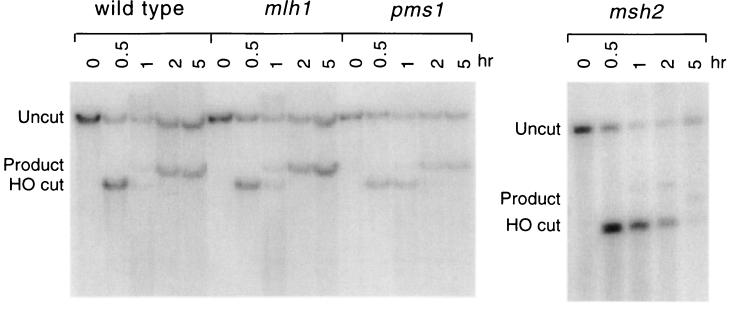
Figure 4.
Effect of PMS1 and MLH1 on SSA. An experiment similar to that of Fig. 3 was performed except that the 205-bp repeats flanking the DSB differed by seven heterologies (see Materials and Methods). Strains included wild type (tNS1357), mlh1 (tNS1396), pms1 (tNS1394), and msh2 (tNS1359).
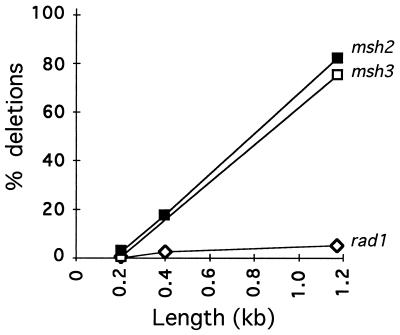
Figure 5.
Effect of length of the annealed region on requirements for RAD and MSH genes during SSA. Experiments identical to those shown in Fig. 3 were carried out with SSA substrates with different lengths of homology flanking the HO-induced DSB. Strains were identical to those used in Fig. 3, whose data are plotted here for the experiment with a length of 205 bp. The lengths of the nonhomologous tails to be excised were the same in all cases. Data for rad1, msh2, and msh3 were normalized to the wild-type value at each point.
DNA Analysis.
DNA was extracted at intervals after HO induction as described (8) and appropriate restriction endonuclease-digested DNA was analyzed on Southern blots. Gene conversions of plasmid pFP122 both with and without crossing-over were analyzed by Southern blots probed with LacZ sequences (30). SSA was analyzed by probing with a HindIII–BamHI DNA fragment centromere proximal to the URA3 gene (8). The efficiency of SSA was measured by dividing the intensity of the product band after 5 hr of induction by the intensity of the parental band prior to induction. Values were normalized to LEU2 or HIS3 sequences to control for the amount of total DNA per sample.
RESULTS
MSH2 and MSH3 Are Required to Remove Nonhomologous Ends During Gene Conversion.
The site-specific HO endonuclease can be expressed under the control of a galactose-inducible promoter (33), to initiate DSB-mediated recombination in plasmid or chromosomal substrates containing an HO recognition site (31). We have examined gene conversion in a centromeric plasmid where a 60-bp HO endonuclease recognition site was introduced into one of a pair of inverted repeated LacZ sequences (pFP140; Fig. 1A). The terminal 30 bp of the cut ends are not homologous to the intact donor. In wild-type strains, 69% of the cells complete recombination and retain the repaired plasmid. DSB repair is reduced 12-fold in the absence of Rad1 (Fig. 1B). These results are similar to those reported in similar plasmids with a 117-bp HO recognition site (9).
Efficient DSB repair also depends on Msh2 and Msh3. Completion of HO induced gene conversion was reduced 6-fold in msh2 or 3-fold in a msh3 deletion (Fig. 1B). A rad1 msh2 and a rad1 msh3 double mutant was reduced 7- to 10-fold. Similar results were found with plasmid pFP120 (Fig. 1A), with nonhomologous ends of 308 bp on one side and 610 bp on the other. msh2, msh3, and rad1 reduced gene conversion by 33-, 20-, and 23-fold, respectively, relative to a wild-type control (Fig. 1B).
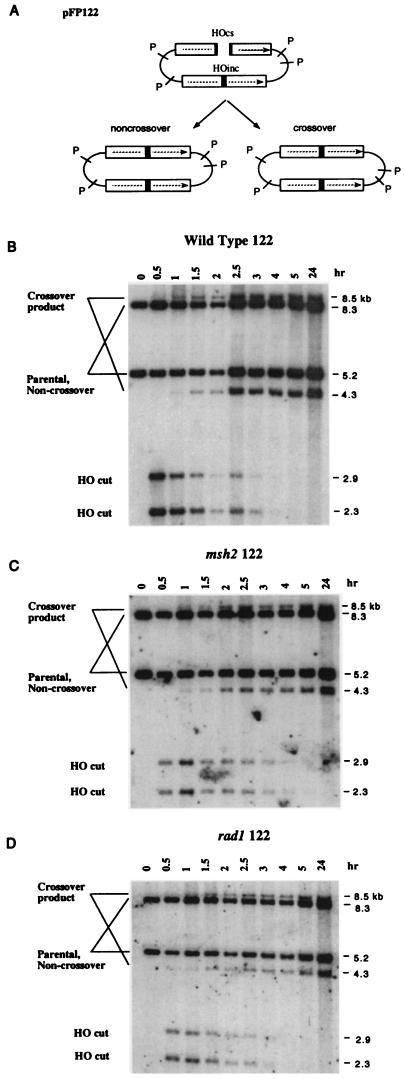
In contrast, msh2, msh3, and rad1 had no effect when the donor also contained a (mutant) HO cut site and the HO-cleaved ends of the recipient were therefore homologous to the donor, except for a 1 bp difference. A similar plasmid (pFP122; Fig. 1A) was constructed, where HO-cut ends are almost perfectly homologous to the donor DNA, by placing a 40-bp HO cut site in the recipient copy of LacZ and a mutant 40-bp HO cutting site, differing by a single bp substitution, into the donor. Neither Rad1, Msh2, nor Msh3 was required for efficient recombination (Fig. 1B). We conclude that Msh2 and Msh3 are required for the removal of nonhomologous tails in conjunction with the Rad1/Rad10 endonuclease and are not required when the DNA ends match their donor template. We have only used plasmid-borne substrates to analyze the requirement for MSH2 and MSH3 on gene conversion. However, using chromosomal substrates we have also demonstrated that RAD1 is required when the ends are nonhomologous (data not shown) and we presume that MSH2/MSH3 would also be required for chromosomal events.
Lack of Effect of MSH2 and RAD1 on Later Steps in Gene Conversion.
Since rad1 and msh2 had little or no effect on viability when the HO-cut ends were homologous to the donor, we more closely examined these strains by Southern blots of DNA extracted at intervals after HO induction, to discern whether these mutations had an effect on the kinetics of recombination or on the proportion of gene conversion events associated with crossing-over. Southern blots for pFP122 in wild-type, msh2, and rad1 strains (Fig. 2) show the kinetics of appearance and disappearance of HO-cleaved DNA fragments and the time of appearance of the two PstI restriction fragments representing gene conversions associated with crossing-over. PhosphorImager analysis of these Southern blots showed that the proportion of gene conversions associated with crossing-over was ≈23.5% in the wild-type strain. This proportion was not significantly affected by msh2 (19%) or rad1 (20.5%). Moreover, there was no change in the kinetics of appearance of the crossover products in the msh2 or rad1 strains. Thus, Msh2 and Rad1 are required at the same step: to remove nonhomologous single-stranded DNA from the end of recombining DNA. Neither msh2 nor rad1 affects any of the other steps that should be common to all gene conversion events (e.g., DNA pairing, strand invasion per se, branch migration, or Holliday junction resolution).
Figure 2.
Kinetics of HO-induced crossing-over. (A) When pFP122 undergoes HO cleavage, gene conversions both with and without crossing-over arise. The crossover products can be distinguished on Southern blots, probed with LacZ sequences, by the sizes of PstI-restriction fragments homologous to LacZ (1, 10). HO-induced re-combination in wild-type (B), msh2 (C), and rad1 (D) strains exhibit the appearance of two crossover products one hr after HO cleavage of the 5.2-kb PstI LacZ fragment. One of the two gene conversion products comigrates with the 5.2-kb parental fragment. However, in these strains carrying an integrated ADE3::HO gene, HO cleaves essentially all of the plasmid targets (data not shown), so that virtually all of the 5.2-kb band remaining at the end of the experiment represents gene conversions without crossing-over and not the original parental fragment. The sum of the intensities of the 4.3- and 8.5-kb crossover bands divided by the sum of the crossover bands and the 5.2- and 8.3-kb noncrossover bands, measured 24 hr after HO induction, indicates the proportion of gene conversions associated with crossing-over.
Role of Mismatch Repair Genes in SSA.
When a DSB is flanked by homologous regions, repair can occur by nonreciprocal recombination leading to a deletion. Most of these events arise by SSA (Fig. 3A) but could also occur by “one-ended” recombination events, in which one DNA end could invade the other flanking homologous region before it became single-stranded (1, 9). In contrast to efficient deletion formation between 205-bp URA3 sequences in wild-type cells (Fig. 3B), deletions were greatly reduced in a rad1 derivative (Fig. 3B). The effect of msh2 and msh3 deletion mutations was very similar to rad1 (Fig. 3B). Cell viability (those that repaired the DSB) went from 77% in the wild type to 3.7% in msh2 and 5.4% in msh3, very similar to a rad1Δ strain (1.0%).
Similar experiments were carried out with a deletion of MSH6, whose gene product has been shown to form heterodimers with Msh2, but which has been implicated principally in the repair of single bp substitutions rather than small loops in heteroduplex DNA (16–18). In these experiments (Fig. 3C), SSA was monitored on a centromeric plasmid containing 240 bp of LacZ homology, in direct orientation, flanking the DSB, similar to plasmids described (1, 9). As shown in Fig. 3C, msh6 does not impair deletion formation by SSA, while msh3 does. A similar result has been obtained using chromosomal substrates identical to those shown in Fig. 3A (data not shown). This result argues that Msh2/Msh3 recognizes nonhomologous tails during recombination in a fashion similar to their recognition of insertion/deletion heterologies, whereas Msh2/Msh6, which mainly recognizes bp substitutions, plays no significant role in SSA.
Mismatch repair of bp substitutions and frameshifts requires not only Msh2, Msh3, and Msh6, but also the Mlh1 and Pms1 proteins (19, 20). However, while both MSH2 and MSH3 are required to remove nonhomologous DNA tails, PMS1 and MLH1 are not (Fig. 4). We first showed this for pms1 when the 205-bp repeats were perfectly homologous (data not shown). It was then of interest to know if SSA would be differently affected if the two repeats were divergent, since previous studies had shown that homologous recombination was improved by mutations in msh2, msh3, and pms1 (25, 26, 34). We therefore examined SSA in substrates that carried seven heterologies in the 205-bp chromosomal regions flanking the DSB. SSA was reduced in the wild-type strain to ≈25% of the value for fully homologous sequences (Fig. 3B vs. Fig. 4). Neither pms1 nor mlh1 affected the efficiency of deletion formation suggesting that (i) they are not required to excise nonhomologous tails and (ii) they are not responsible for the discouragement of annealing by 3% heterology. In contrast, the absence of msh2 caused a dramatic decrease in deletion formation (Fig. 4). These results argue that MSH2 and MSH3 play a role in the removal of nonhomologous tails from the annealed intermediate that is distinct from their roles in mismatch correction or heteroduplex DNA formation. Whatever improvement msh2 might have caused in increasing recombination between diverged substrates was overwhelmed by its very strong negative effect on the completion of SSA.
Distinguishing the Roles of RAD1 and MSH2/MSH3 During SSA.
Msh2 and Msh3 might act in several different ways in removing nonhomologous DNA tails. Msh2/Msh3 may recognize and bind to branched intermediates and present them to Rad1/Rad10 for cleavage. They might also stabilize initially unstable strand annealing intermediates, allowing Rad1/Rad10 more time to act. Finally they might form a complex with Rad1/Rad10 to increase its endonuclease activity. Further experiments have shown that the requirement for Msh2 and Msh3 in SSA depends on the length of the regions that anneal. Three SSA substrates were constructed on chromosome V in which the lengths of the single-stranded DNA tails that must be removed remained constant, but the length of the annealed region increased from 205 bp to 415 bp or to 1,170 bp (Fig. 5). The absence of RAD1 had the same severe effect on deletion formation, regardless of the length of the flanking homologous regions. In contrast, while the requirement for MSH2 and MSH3 was as great as for RAD1 with 205-bp flanking regions, the absence of the mismatch repair proteins was less profound with longer flanking regions. With 1,170-bp flanking homology, msh2 and msh3 reduced SSA only 25% relative to wild type. The significance of this difference will be discussed below.
DISCUSSION
We propose that the Msh2/Msh3 heterodimer recognizes and binds to a branched DNA structure such as that formed by strand invasion or strand annealing. Msh2/Msh3 either (i) stabilizes this structure so that Rad1/Rad10 can trim off the nonhomologous tail, and/or (ii) directly participates in recruiting Rad1/Rad10 to the sites of recombination, in the way that other NER proteins may recruit Rad1/Rad10 to the sites of DNA photodamage.
Interestingly, the requirements for Msh2/Msh3 are different during the initiation of gene conversion and the removal of 3′ ended tails during SSA. In SSA, Msh2 and Msh3 are less important as the length of the flanking homologous regions increases from 205 bp to 1 kb. In contrast, Msh2 and Msh3 are as necessary as Rad1 during gene conversion, even though the lengths of the homologous regions shared between the donor and recipient are ≈2 kb on either side of the DSB. We believe this difference reflects the nature of the intermediate structures on which Rad1/Rad10 acts in SSA and in gene conversion. In gene conversion, the initial encounter between the invading strand and the target duplex DNA is a relatively unstable paranemic joint that cannot be converted into a more stable plectonemic configuration until the 3′ ended tail is excised (35). We suggest that such unstable intermediates require the presence of Msh2/Msh3 to facilitate Rad1/Rad10 cleavage. In contrast, annealing between two single-strand tails can intertwine more readily to form plectonemic joints, though initial encounters could also be via paranemic associations. It appears that the stability of these interactions significantly increases between 0.2–1 kb of homology and that Msh2/Msh3 are especially critical when the homologous regions are short. These results argue that Msh2/Msh3 recognize branched structures and stabilize them, thus enabling Rad1/Rad10 to cleave off the tails.
These data provide the first direct evidence of the specific steps in recombination that require Msh2 and Msh3. Previously Saparbaev et al. (21) had reported that MSH2 and MSH3 are required in a RAD1–RAD10-dependent spontaneous recombination, either in deletion formation (which we presume occurred by SSA) or in a RAD52-independent pathway for spontaneous recombination involving unequal sister-chromatid crossing-over. They suggested that Msh2 and Msh3 proteins might aid the resolution of Holliday junctions. However, our data demonstrate that DSB-initiated gene conversions are not impeded by a msh2 mutant, so long as the invading strands have homologous ends. These conversions occur both with and without crossing-over (and presumably involve Holliday junctions, ref. 36). Why then should the events studied by Saparbaev et al. (21) be rad1- and msh2-sensitive, since the two sister chromatids are identical and should have perfect homology? First, if the lesions initiating these events (either DSBs or single-strand nicks) occurred in the interval between the repeated sequences, then unequal sister recombination between the misaligned repeats would also require removal of a nonhomologous tail and thus require RAD1 and MSH2. Second, it must be noted that previous studies have shown that recombination in a rad52 strain between the same his4 alleles used by Saparbaev et al. does not actually occur by reciprocal exchange; instead it occurs by a nonreciprocal event in which only one of the two participating chromatids is recovered (37). The actual mechanism, which might involve the invasion of only one DNA end (e.g., ref. 38), remains unknown. It is of course possible that Msh2 and Msh3 play still other roles in spontaneous recombination, different from those we observe for DSB-mediated recombination. However, RAD52-dependent spontaneous recombination between homologous substrates that occur by a true reciprocal crossing-over are not affected by msh2 or msh3 (26, 34).
Our results imply that Msh2 and Msh3 recognize a branched DNA structure with a free 3′ end. This structure is different from all previously reported interactions of Msh2 and Msh3 with DNA, where these proteins bound to mismatches or heteroduplex containing loops in continuous duplex strands of DNA, in vitro (14) and apparently in vivo (39). A recent report also shows that Msh2 can, by itself, bind to another branched structure, the Holliday junction (15); however, our data do not support suggestions (15, 25) that the absence of Msh2 would affect the resolution of this structure and hence the proportion of gene conversions that are resolved with crossing-over. Still, Msh2 might affect the extent of branch migration (40) in a way that our assays do not detect. The key point of our study is that MSH2 and MSH3, but not MSH6 nor PMS1 nor MLH1, play a role in removing nonhomology from the ends of recombining DNA.
Thus, the removal of nonhomologous tails to initiate new DNA synthesis during recombination brings together an endonuclease from the NER pathway with heterology-recognizing proteins from the mismatch repair pathway, to carry out a novel role in recombination.
Acknowledgments
G. Marsischky and Richard Kolodner generously constructed and provided msh6 and msh3 strains. We thank R. Keil, R. Lahue, and G. Crouse for plasmids. Susan Lovett, Bernard Lopez, and members of the Haber lab made helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM20056. M.C. was supported by U.S. Public Health Service Training Grant in Genetics GM 01722 and National Institutes of Health Minority Predoctoral Fellowship GM18050. F.P. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: DSB, double-strand break; SSA, single-strand annealing; NER, nucleotide excision repair.
References
- 1.Fishman-Lobell J, Haber J E. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickoloff J A, Chen E Y, Heffron F. Proc Natl Acad Sci USA. 1986;83:7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray A, Machin N, Stahl F W. Proc Natl Acad Sci USA. 1989;86:6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liefshitz B, Parkett A, R, Maya R, Kupiec M. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng Y S, Whelden J, Gunn L, Nickoloff J A. Curr Genet. 1996;29:335–343. doi: 10.1007/BF02208614. [DOI] [PubMed] [Google Scholar]
- 6.Haber J. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 7.White C I, Haber J E. EMBO J. 1990;9:663–674. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara N, Haber J E. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishman-Lobell J, Haber J E. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 10.Tomkinson A E, Bardwell A J, Bardwell L, Tappe N J, Friedberg E C. Nature (London) 1993;362:860–862. doi: 10.1038/362860a0. [DOI] [PubMed] [Google Scholar]
- 11.Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 12.Sung P, Reynolds P, Prakash L, Prakash S. J Biol Chem. 1993;268:26391–26399. [PubMed] [Google Scholar]
- 13.Ivanov E L, Haber J E. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alani E, Chi N W, Kolodner R D. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 15.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R E, Kovvali G K, Prakash L, Prakash S. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 17.Alani E. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsischky G T, Filosi N, Kane M F, Kolodner R D. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 19.Prolla T A, Christie D M, Liskay R M. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer B, Kramer W, Williamson M S, Fogel S. Mol Cell Biol. 1989;9:4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saparbaev M, Prakash L, Prakash S. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin N, Haber J E. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 25.Hunter N, Chambers S R, Louis E J, Borts R H. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 26.Selva E M, New L, Crouse G F, Lahue R S. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 28.Nickoloff J A, Singer J D, Hoekstra M F, Heffron F. J Mol Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink G R, Lawrence C W. Methods in Yeast Genetics: Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1971. [Google Scholar]
- 30.Rudin N, Sugarman E, Haber J E. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiffenbach B, Rogers D T, Haber J E, Zoller M, Russell D W, Smith M. Proc Natl Acad Sci USA. 1983;80:3401–3405. doi: 10.1073/pnas.80.11.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose M, Grisafi P, Botstein D. Gene. 1984;29:113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- 33.Jensen R, Herskowitz I. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bianchi M, DasGupta G, Radding C M. Cell. 1983;34:931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- 36.Schwacha A, Kleckner N. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 37.Haber J E, Hearn M. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malkova A, Ivanov E, Haber J E. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkpatrick D R, Petes T D. Nature (London) 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 40.Alani E, Reenan R A, Kolodner R D. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]