Abstract
TSC22D1, which encodes transforming growth factor β-stimulated clone 22 (TSC-22), is thought to be a tumor suppressor because its expression is lost in many glioblastoma, salivary gland, and prostate cancers. TSC-22 is the founding member of the TSC-22/DIP/Bun family of leucine zipper transcription factors; its functions have not been investigated in a multicellular environment. Genetic studies in the model organism Drosophila melanogaster often provide fundamental insights into mechanisms disrupted in carcinogenesis, because of the strong evolutionary conservation of molecular mechanisms between flies and humans. Whereas humans and mice have four TSC-22 domain genes with numerous isoforms, Drosophila has only one TSC-22 domain gene, bunched (bun), which encodes both large and small protein isoforms. Surprisingly, Drosophila Bun proteins promote cellular growth and proliferation in ovarian follicle cells. Loss of both large isoforms has the strongest phenotypes, including increased apoptosis. Cultured S2 cells depleted for large Bun isoforms show increased apoptosis and less frequent cell division, with decreased cell size. Altogether, these data indicate that Drosophila TSC-22/DIP/Bun proteins are necessary for cellular growth, proliferation, and survival both in culture and in an epithelial context. Previous work demonstrated that bun prevents recruitment of epithelial cells to a migratory fate and, thus, maintains epithelial organization. We speculate that reduced TSC22D1 expression generally reduces cellular fitness and only contributes to carcinogenesis in specific tissue environments.
Keywords: bunched, follicle cells, oogenesis
Cancer remains the second leading cause of death in the U.S. (1), despite successful therapeutics against many types of tumors. Mechanisms that permit tumors to evade chemotherapy must be identified for further progress. Correlations between gene expression and tumor progression suggest candidate tumor suppressors; their mechanisms of action often are tested in cultured tumor cells. However, cultured cells lack the complex biology of tumors in vivo. Recently, genes that promote tumor progression and invasion have been identified by using the model genetic organism Drosophila melanogaster (reviewed in ref. 2). The strong conservation of genes and molecular mechanisms between flies and humans makes this an ideal organism to evaluate candidate tumor-suppressor functions in vivo.
Human TSC22D1 is a candidate suppressor for several cancers (3–7). In graded tumors, the lowest levels of TSC22D1 antigen is detected in the most aggressive astrocytomas (8). In prostate biopsies, it is absent from carcinoma cells but still present in adjacent normal epithelium (5). TSC22D1 encodes two protein isoforms that share conserved domains with TSC-22/DIP/Bun (TDB) transcription factors from flies to humans. Functional tests have focused on overexpression of the smaller isoform, TSC-22 (transforming-growth-factor-β-stimulated clone-22) (9), in cultured cells. The prevailing hypothesis is that loss of TSC22D1 expression allows tumor cells to evade apoptotic or antigrowth signals. For example, forced expression of TSC-22 reverses resistance to genotoxic agents in salivary-gland tumor cell lines (3, 4, 10–13). To date, there is no report of TSC22D1 function in normal epithelia.
bunched (bun) is the only TDB family gene in the Drosophila genome (14). Like other TDB proteins, Bun proteins share strong sequence similarity with TSC-22 in the DNA-binding and protein–protein-interaction domains (9, 15–20). Like mammalian TDB genes, bun encodes large and small isoforms, but little is known about the relative functions of the two types of isoforms.
bun is required for development, oogenesis and viability (18, 19, 21, 22). We have studied bun function in the follicular epithelium that overlies each maturing oocyte. Here, bun prevents epithelial cells from being recruited to a migratory fate (23); the boundary between epithelial and migrating cells is regulated through EGF and BMP regulation of bun expression (24). Expression of vertebrate TSC22D1 is similarly up-regulated by receptor tyrosine kinase signaling and down-regulated by BMP signaling during initiation of feather bud outgrowth in chicken skin (25). Thus, the function of TDB genes as targets of EGF and BMP signaling may be conserved between flies and vertebrates.
The follicular epithelium is an outstanding model to investigate mechanisms for tissue growth and invasive behavior (2, 26). Follicle cells (FCs) proliferate as the epithelium matures during oogenesis stages 1–6 (27). FCs enlarge by endoreplication, a modified cell cycle that replicates DNA without cell division (27, 28) and migrate to new positions before terminal differentiation (reviewed by refs. 29–31).
Here, we test for a bun function in proliferation. Surprisingly, large Bun isoforms promote proliferation, growth, and survival of Drosophila cells in epithelia and cultured cells.
Results
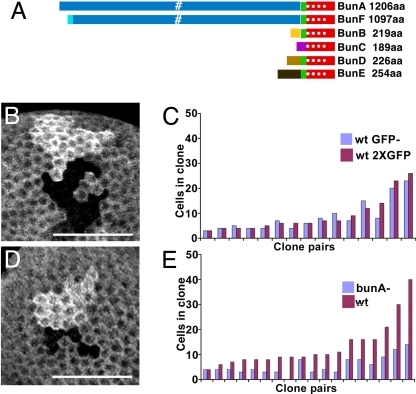
The hypothesis that human TSC22D1 inhibits cell proliferation (4, 10, 13, 32–35) makes the simple prediction that loss of the fly homolog would lead to increased cell number. bun expresses six protein isoforms that have the same DNA-binding and leucine zipper domains but can be divided into two size classes [Fig. 1A and supporting information (SI) Fig. S1]. The four small isoforms, B–E, range from 189 to 254 aa; each has a unique N-terminal domain. The two large isoforms, A (1,206 aa) and F (1,097 aa), share most of a large N-terminal domain and share small sequence motifs with mammalian large TDB proteins (36). Previous studies used transposon insertion alleles that disrupt multiple transcripts (e.g. refs. 19 and 23). We obtained lethal loss-of-function alleles that eliminate the DNA-binding and dimerization domains of only the large isoforms (SI Text and ref. 36). These alleles provided a unique opportunity to investigate the function of TDB large isoforms in the follicular epithelium, where we know the most about bun functions (23, 24). Expression of the large A isoform alone rescues null and heteroallelic mutations, suggesting that the small isoforms may be dispensable (36). Initial characterization of FCs lacking large Bun isoforms suggested that absence of large isoforms has qualitatively similar, but more severe, phenotypes during late oogenesis compared with insertional mutations (data not shown).
Fig. 1.
bunA is required for normal follicle cell proliferation. (A) Schematic of Bun protein isoforms. Red, shared domain with the DNA binding and leucine zipper domains (white dots). Domains from unique exons are depicted in different colors. BunA and BunF share a large exon that contains two motifs (white) found in all large TDB proteins. (B) Wild-type sister clones marked by absence of GFP (GFP−, black region) or intense fluorescence from two copies of the GFP gene (2XGFP, bright region). (D) Sister clones of a homozygous bunA (bun[A-Q988X]) mutant clone marked by GFP− and a homozygous wild-type clone marked by 2XGFP. (Scale bars, 40 μm.) (C and E) Cell numbers for sister clones, GFP− (blue) and 2XGFP (red) sisters are paired in graphs. Cell number is not significantly different for wild-type sisters. (C, P = 0.4). Each bunA mutant clone (blue) has fewer cells than its wild-type sister (red) (E, P = 0.00007).
We tested for bun regulation of proliferation using clonal analysis (Fig. 1). Paired sister clones, one composed of homozygous bun mutant cells and one of wild-type cells, were generated by FLP/FRT-mediated mitotic recombination (37). Control experiments generated sister pairs of a wild-type GFP− clone and a wild-type GFP+ clone; each averaged nine cells per clone, a ratio of 1 (Fig. 1 B and C). If bun were a negative regulator of proliferation, then each homozygous bun clone would have more cells than its wild-type sister. For the large isoform-specific allele bun[A-Q988X], mutant and wild-type sister clones averaged 6 and 13 cells per clone, respectively, a ratio of 0.45 (Fig. 1 D and E). A lethal transposon insertion with very strong FC phenotypes, bun[6903] (Fig. S1) (23), gave similar results, means of 11 mutant and 20 wild-type cells per clone, a ratio of 0.55 (n = 7, P = 0.01; data not shown). Thus, bun, particularly the large Bun isoforms, promotes FC proliferation.
Premature exit from mitotic cell cycles can lead to fewer cells. FCs proliferate until stage 6, when Notch signaling halts mitosis and initiates endoreplication cycles (27, 28). Because bun blocks Notch signaling during late oogenesis (23), we tested for altered expression of two Notch target genes, cut and emc (38, 39), in proliferating bunA mutant FCs (Fig. S2). Expression was unaffected, indicating that large Bun isoforms do not influence cell number through regulation of these Notch targets.
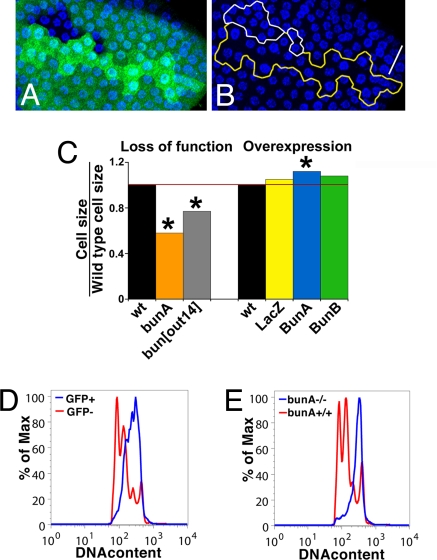
Studies of bun function in the adult eye indicate that bun influences photoreceptor size (36), so we examined FC size. Immediately after proliferation, there was no difference in mean cell size for mutant FCs (n = 16) compared with sister clone wild-type FCs (data not shown). Because Drosophila cells enlarge chiefly by endoreplication (40), we also examined cell size after endoreplication. At stage 10, bunA mutant FCs were significantly smaller, 200 ± 80 pixels per cell for mutant versus 350 ± 90 for wild type (Fig. 2 A –C and Table S1). We confirmed that bunA mutant FCs undergo all three endoreplication cycles using FACS analysis for DNA content (Fig. 2 D and E). Thus, large Bun isoforms are required for normal growth of FCs.
Fig. 2.
BunA promotes follicle cell growth. (A and B) A sister pair of bun[A-Q578X] mutant clone (GFP−) and wild-type clone (2XGFP) at stage 10. GFP, green; nuclei, blue. (Scale bar, 100 μm.) (A) Merged image. (B) Nuclei only. Nuclei in the bunA mutant clone (outlined in white) are more densely packed than nuclei in the wild-type sister clone (outlined in yellow). (C) Bar graphs showing mean sizes of follicle cells mutant for or overexpressing bun. *, P < 0.01. Cell sizes are normalized to the respective wild-type (wt) control for each treatment, which is set to be 1.0. (D and E) Overlay of DNA content profiles between control wild-type FCs, GFP+ (blue) and GFP− (red) (D) or between bunA mutant (GFP+, blue) and wild-type (GFP−, red) FCs (E). Wild-type profile includes cells from all oogenesis stages; the 4N, 8N, and 16N populations are from first, second, and third rounds of endoreplication, respectively. Most bunA mutant FCs were in the 16N population, indicating that three rounds of endoreplication occurred between clone induction and analysis.
This requirement in FC growth and proliferation was unexpected. We speculated that large isoforms might regulate growth differently from insertional mutations that additionally disrupt function of small isoforms. To test this, we examined stage-10 FCs mutant for bun[out14], a semiviable allele that disrupts the bunB transcription unit with strong effects on FC morphogenesis (Figs. S1 and S3). Both the large BunA isoform and the small BunB isoform are expressed in FCs (24). Mutant FCs again were significantly smaller than sister clone wild-type FCs, 3.6 ± 1.5 versus 4.7 ± 2.4 arbitrary units (au) respectively (Table S1 and Fig. S3A). We next tested the effects of overexpression, using clonal expression of Gal4 to generate GFP+ cells overexpressing BunA, BunB, or lacZ (Fig. 2 C and Fig. S3 B–E and Table S1). BunB-overexpressing FCs were not significantly different from nearby wild-type FCs. However, BunA-overexpressing FCs were significantly larger than their neighbors, 7.6 ± 2.3 versus 6.8 ± 2.1 au, respectively. We conclude that large Bun isoforms are both necessary and sufficient for normal FC growth.
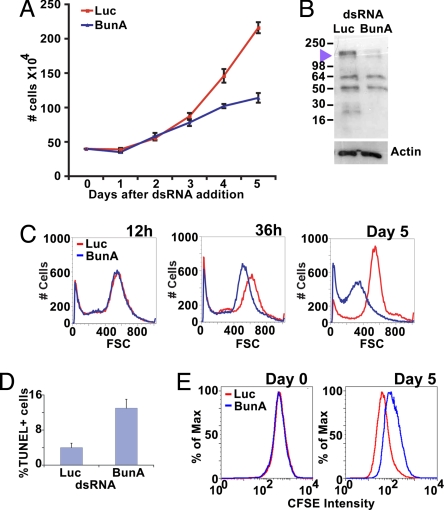
We speculated that TDB proteins might function differently in cultured cells than in intact epithelia. Thus, we tested the requirement for large Bun isoforms in a nonadherent Drosophila cell line, using double-stranded RNA-mediated interference to deplete BunA and BunF in S2 cells. Double-stranded RNAs (dsRNA) were synthesized from two different sequences unique to the BunA and BunF mRNAs. Control cultures were treated with dsRNA synthesized from the firefly luciferase gene, which is not present in the Drosophila genome. A large polypeptide, detected with antibody against the shared sequences of Bun isoforms, was significantly reduced after 4 days of treatment with either BunA/F dsRNA compared with controls (Fig. 3B).
Fig. 3.
bunA promotes S2 cell growth, division, and survival. Ds RNA-mediated interference was used to deplete BunA and BunF (labeled as BunA) from S2 cells. (A) Growth curves of BunA/F-depleted cells (blue) and controls (red). Luc, luciferase dsRNA control. (B) Western blot showing depletion of BunA/F 4 days after dsRNA treatment. (C) Size distribution of BunA/F-depleted cells (blue) and control cells (red) at 12 h, 36 h, and day 5 after addition of dsRNA, as determined by forward scatter (FSC). (D) Percentages of TUNEL-positive nuclei between BunA/F-depleted and control cells on day 4, P = 0.01. (E) Example of carboxyfluorescein diacetate succinimidyl ester (CFSE) fluorescent intensity in BunA/F-depleted cells (blue) and control cells (red) at day 0 after dye treatment and day 5, when control cells had significantly less dye (P = 0.00006). n = 3 for all experiments.
Consistent with in vivo results, large isoform-depleted cultures proliferated more slowly than control-treated cultures (Fig. 3A). Control cell number increased exponentially between days 3 and 5 after dsRNA treatment. In contrast, numbers of BunA/F-depleted cells increased only slightly during this period. This effect was observed with both dsRNAs, indicating that it is specific to BunA/F. Proliferation rates became markedly different on day 4, so we assayed for apoptotic cells at this time. Significantly, more TUNEL-positive cells were observed in BunA/F-depleted cultures than in controls (Fig. 3D and Fig. S4), indicating that large Bun isoforms promote cell survival in culture.
To determine whether cellular growth was altered by BunA/F depletion, we examined cell size distribution using forward light scatter in flow cytometry. At 12 h, there was no difference in the profile of cell sizes between BunA/F-depleted cultures and controls. A significant shift to smaller sizes for BunA/F-depleted cells was detected at 18 h (Figs. S4C and S5) and became pronounced by 36 h (Fig. 3C), the time at which BunA/F-depletion was detectable by Western blotting (data not shown). More BunA/F-depleted cells were smaller with each additional day, whereas control cells were unchanged. The smallest, apoptotic fraction of BunA/F-depleted cells did not increase above that of the control until day 3 (Fig. S4C). Thus, decreased cellular growth preceded the increase in apoptosis.
Normal cells balance growth with cell division by coordinating cell cycle progression with the rate of protein synthesis (reviewed in refs. 41 and 42). Reduced cell size could arise from decoupling the cell cycle from cellular growth in either of two ways: an increased rate of cell division without a proportionate increase in growth or a decreased rate of cellular growth without a proportionate decrease in proliferation (43). However, there was no consistent difference in the proportion of cells in each phase of the cell cycle between BunA/F-depleted cells and controls (Fig. S4D). If all cell cycle stages are lengthened proportionately, the cell cycle profile appears normal (e.g., ref. 44). To assess the overall cell division rate, we labeled cell contents with the fluorophore carboxyfluorescein diacetate succinimidyl ester (CFSE) (45). CFSE-labeled cytoplasmic contents are divided between daughter cells at each division, so that relative rates of cell division are detected by flow cytometry as decreases in fluorescence over time (Fig. 3E). BunA/F-depleted and control cells had the same fluorescence intensity on day 0, but fluorescence of control cells was substantially lower than BunA/F-depleted cells on day 5. Thus, BunA/F-depleted cells divide less often, indicating that smaller size is not a secondary consequence of accelerated division.
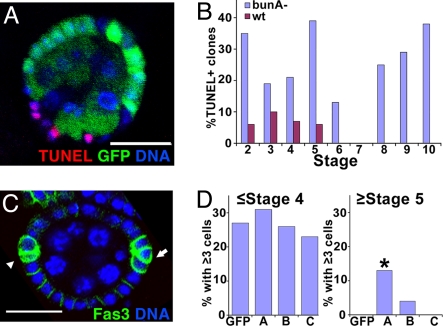
Slow proliferation of BunA/F-depleted S2 cells resulted from a combination of slower cell division and increased cell death, so we assayed for apoptotic cells in mutant FCs (Fig. 4). With good nutrition, no cell death is observed in epithelial follicle cells after stage 1 (reviewed in ref. 46), but approximately half the polar cells undergo apoptosis during stages 1–4 (47). Consistent with this, wild-type sister clones had TUNEL+ FCs only during stages 1–5. In contrast, mutant clones had TUNEL+ FCs throughout oogenesis (Fig. 4B). Furthermore, 26% of bunA mutant clones had TUNEL+ FCs (Fig. 4A) compared with only 4% of wild-type sister clones (data not shown). Control sister clones were indistinguishable (data not shown). Thus, increased apoptosis likely contributes to reduced proliferation of mutant FCs.
Fig. 4.
bunA promotes follicle cell survival. (A and B) Apoptotic cells are detected more frequently in bunA loss-of-function clones than in wild-type clones. (A) Example of a bun[A-Q988X] mutant clone with TUNEL+ nuclei (red). bunA mutant cells lack GFP (green); nuclei are blue. (B) Many more bunA mutant clones have TUNEL+ nuclei than do wild-type clones. Bar graphs indicate the percentage of clones with TUNEL+ nuclei for bunA clones (blue) and wild-type sisters (red), by stage of oogenesis; n = 113 for each. (C and D) Overexpression of bunA influences polar cell number. (C) Example of egg chamber from a female with upd-Gal4-driven expression of UAS-bunA. Polar cells have high Fas3 levels (green); arrowhead, group of two; arrow, group of three; nuclei (blue). (D) Effect of expression of bun gene products on polar cell number. Control polar cell clusters all had two cells after stage 4 (n = 64). Survival of excess polar cells after stage 4 was seen in ≈10% of the clusters expressing bunA (n = 31; *, P = 0.01); bunB rarely had excess polar cells after stage 4. (Scale bars, 20 μm.)
As a final test, we overexpressed individual Bun isoforms specifically in polar cells. Excess polar cells undergo apoptosis, leaving only two in each cluster by stage 5 (47). We overexpressed BunA, BunB, or BunC using upd-Gal4 (48). At stage 5 or later, 13% of polar cell clusters with BunA overexpression had at least three cells (Fig. 4D), whereas all control clusters had two cells. Expression of the small-isoform BunC did not alter polar cell number, and the small-isoform BunB had a minimal effect (Fig. 4D). Before stage 5, expression of BunA did not increase the mean number of cells per cluster, and ectopic mitosis was not observed in 69 clusters stained for phosphohistone H3 (data not shown). Prolonged survival was not due to a change in cell fate, because only Fas3+ cells were counted in this experiment; moreover, BunA promotes loss of Fas3 from epithelial FCs (Fig. S6). Thus, increased BunA levels promote polar cell survival, and large Bun isoforms are necessary to promote epithelial FC survival. Tests for genetic interactions with the central regulators of Drosophila apoptosis also indicate that endogenous bun antagonizes apoptosis during eye development (Fig. S7).
Discussion
These data demonstrate that large Bun isoforms promote cellular growth in culture and cell autonomously in vivo. This is a rare phenotype in S2 cells; a dsRNA interference screen found that only ≈4% of Drosophila genes had a cell size, cell cycle, or cell death phenotype (49). Reduced proliferation of large isoform-depleted S2 cells is a secondary consequence of reduced cellular growth and increased apoptosis. Similarly, FCs lacking large Bun isoforms proliferate poorly, in part from increased apoptosis. Large isoforms are required for proliferation in the wing primordium and for full size of adult photoreceptors (36). In adult primordia, slowly growing cells die when they compete with adjacent cells for survival factors (e.g., in ref. 50); it is unknown whether such competition occurs in FCs.
Large Bun isoforms are important to reach normal adult size (19), but genetic studies suggest they are not central to the insulin receptor or TOR signaling pathways (36). FCs lacking the large BunA/F isoforms maintained normal size while proliferating but not during endoreplication. Unlike other genes necessary for final FC size (27, 28, 51), BunA/F do not regulate entry into endocycles via Notch target genes. Importantly, FCs lacking BunA/F can complete all three endocycles by stage 10 of oogenesis. Oogenesis slows substantially under poor nutrient conditions (52), a developmental flexibility that might permit slowly growing FCs to reach normal size. The growth pathway regulated by bun remains to be identified.
This work does not distinguish whether only the large Bun isoforms are necessary for FC growth or whether small isoforms are also needed. Insertion mutations in the bunB transcription unit, bun[6903] and bun[out14], are associated with reduced cellular proliferation and growth, but do not directly test the requirement for bunB because they may alter splicing of other isoforms. In FCs, overexpression of BunA gave increased FC size, but overexpression of BunB had no significant effect. Notably, BunA overexpression rescues size and viability of putative bun null larvae, suggesting that loss of all small isoforms has no effect on larval cell growth (36). Overexpression of BunA prolonged polar cell survival, whereas the effect of BunB was minimal. In contrast, ectopic BunB, but not BunA, could alter expression of downstream genes during late oogenesis (24). These differences in phenotypes may result from differential expression of different isoforms, or from mechanistic differences in the functions of large and small isoforms. Detailed studies of specific phenotypes are needed to conclusively distinguish the functions of large and small isoforms.
Mammalian TDB genes, called TSC22D1, TSC22D2, TSC22D3, and TSC22D4, encode both large and small isoforms, similar to bun (16, 53). All protein isoforms encoded by these genes share the same TSC-22 DNA-binding domain and a leucine zipper motif that permits both homo- and heterodimerization (16). We expected that bun would have a function similar to TSC22D1, the most similar mammalian gene by polypeptide sequence and exon/intron structure (14). Despite this similarity, we find that ectopic BunB does not accelerate apoptosis of polar cells and that cells overexpressing BunB continue to proliferate. This was a surprise, because increased levels of TSC-22 can promote cell death in tumor cell lines (10, 32, 33) and promotes cell cycle arrest induced by TGF-β or PPAR-γ (35), the opposite of the proliferation-promoting functions of large bun isoforms in flies. This apparent paradox may indicate that a different mammalian TDB protein shares the BunB function, such as Gilz, encoded by TSC22D3 (54). Gilz, promotes lymphocyte survival (17) and has functional similarities with BunB (56).
Studies of TSC22D1 focused on only the small protein isoform, TSC-22; the large isoform has not been investigated. In a genetic study of murine TSC22D1, we find similarities to bun growth phenotypes (M. Guitard, C. E. Dohrmann, T. Soma, L. L. Dobens, J. Brissette, and L.A.R., unpublished work), suggesting that this gene also shares in vivo functions with bun. Although TSC22D1 is suspected to be a tumor-suppressor gene for some tissues (7, 8, 13, 32, 55), it is overexpressed in renal cell carcinoma (56). Reduced TSC22D1 expression may promote carcinogenesis only when accompanied by mutations that suppress apoptosis, such as p53 mutations, or in tissues where expression of another TDB gene can rescue growth. Alternatively, reduced expression might promote chemotherapeutic resistance by slowing growth and proliferation (57, 58).
An emerging concept in tumor biology is that mutations can both promote carcinogenesis in one context and decrease cellular fitness in others (59). In flies, bun has context-dependent functions that might antagonize tumor progression. In most FCs, bun is dispensable for epithelial morphology; however, bun mutant FCs are sensitized to migration-inducing signals (23) and display position-dependent loss of monolayer organization (data not shown). Perhaps TSC22D1 similarly prevents neoplasia in the presence of potentially oncogenic signals. Consistent with this model, overexpression of TSC-22 inhibits anchorage-independent growth in salivary-gland tumor cells (11). The functions of bun in the follicular epithelium provide a framework for evaluating the importance of TSC22D1 in tumor progression.
Materials and Methods
Expanded methods are in SI Text.
Fly Culture and Genetics.
Flies were cultured on glucose-cornmeal medium, at 25°C unless indicated. Strains and bunA alleles are in SI Text; all others are in Flybase.
UAS–BunC Transgenic Flies.
BunC cDNA (clone GH13775; Open Systems) was subcloned into pUAST; transgenic flies made by the Cutaneous Biology Research Center Transgenic Fly Core.
Clonal Analyses.
Loss of function clones induced with FLP/FRT mediated mitotic recombination (37): 2- to 4-day-old females were treated for from 15 min to 1 h at 37°C and, depending on the stage to be analyzed, reared another 2–4 days before dissection. Clones with multilayered organization were not scored. Statistical analyses used the paired t test. GFP+ mutant clones were made by MARCM (60). For overexpression, we used Flip-out Gal4 (24, 61), and statistical analyses used the unpaired t test. Numerical results are expressed as mean ± SD.
Immunostaining and Cell Size Measurement.
Primary antibodies from mouse: α-GFP (1:200; Invitrogen), anti-Cut (1:20; Developmental Studies Hybridoma Bank) (DSHB) and anti-Notch (1:30, C17.9C6; DSHB). From rabbit: anti-Emc (1:500; from Y. Jan, University of California, San Francisco), α-GFP (1:200; Invitrogen). Anti-Bun#63 is in SI Text. Secondary antibodies were from goat and labeled with Alexa Fluor 488 or Alexa Fluor 568 (1:200; Invitrogen). DNA was stained with ToPro3 (1:5,000; Invitrogen). Ovaries were dissociated and mounted in Vectashield (Vector Laboratories) for confocal imaging.
Stage 6/7: Individual cells were visualized by Alexa Fluor 546-phalloidin (Invitrogen) and the area measured from confocal images by using NIH Image software. Stage 10: the area of each clone was measured in pixels by using Adobe Photoshop, and divided by the number of cells. Individual bun[out14] cells were measured from clones produced for ref. 23. Clones were selected to avoid regions of curvature or cell migration.
Ds RNA Interference.
DsRNA was made with the dsRNA Synthesis kit (Ambion). Genomic DNA from y w flies was prepared with a DNAeasy kit (Qiagen). Regions from the unique exons for BunA and BunF were amplified by PCR: nucleotides 1,000–1,437 and 1,698–2,517 of the bunA cDNA sequence. Primers are in SI Text. Data shown is from dsRNA against nucleotides 1,000–1,437. Ds-RNA treatment of S2 cells followed ref. 62.
TUNEL Labeling.
S2 cells were plated on Lab-Tek chamber slides (Nunc) for 2 h. S2 cells or ovaries were fixed, permeabilized, blocked with immunostaining buffer containing 3%BSA, and then incubated for 1 hr at 37°C in reaction solution (In Situ Cell Death Detection kit; Roche).
FACS Analysis.
FACS was performed on a FACSCalibur cytometer (Becton Dickinson). For cell cycle analysis we used propidium iodide staining (63). For CFSE, we used a protocol for T cells (46). FACS of FCs followed refs. 64 and 65.
Acknowledgments.
We thank U. Makhija and P. Gomez-del Arco for technical assistance; M. Guitard and K. White for discussions; D. Harrison (University of Kentucky, Lexington) and Bloomington Stock Center (Bloomington, IN) for flies; Y. Jan and Developmental Studies Hybridoma Bank (Iowa City, IA) for antibodies. X.W. was supported by a Massachusetts General Hospital Medical Discovery Fund Fellowship. This work was also supported by National Institutes of Health Grants R01GM53203 (to N.J.D.) and 2R01-GM60501 (to L.A.R.), an American Cancer Society grant (to L.A.R.), and a grant from Shiseido Corp., Ltd. of Japan to the Cutaneous Biology Research Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800945105/DCSupplemental.
References
- 1.Heron M, Smith B. Deaths: Leading causes for 2003. Natl Vital Stat Rep. 2007;55:1–92. [PubMed] [Google Scholar]
- 2.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 3.Kawamata H, et al. Induction of TSC-22 by treatment with a new anti-cancer drug, vesnarinone, in a human salivary gland cancer cell. Br J Cancer. 1998;77:71–78. doi: 10.1038/bjc.1998.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omotehara F, et al. In vivo enhancement of chemosensitivity of human salivary gland cancer cells by overexpression of TGF-beta stimulated clone-22. Oncol Rep. 2000;7:737–740. doi: 10.3892/or.7.4.737. [DOI] [PubMed] [Google Scholar]
- 5.Rentsch CA, et al. Differential expression of TGFbeta-stimulated clone 22 in normal prostate and prostate cancer. Int J Cancer. 2005 doi: 10.1002/ijc.21449. [DOI] [PubMed] [Google Scholar]
- 6.Shostak KO, et al. Downregulation of putative tumor suppressor gene TSC-22 in human brain tumors. J Surg Oncol. 2003;82:57–64. doi: 10.1002/jso.10180. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, et al. Identification of TSC-22 as a potential tumor suppressor that is upregulated by Flt3–D835V but not Flt3-ITD. Leukemia. 2007;21:2246–2257. doi: 10.1038/sj.leu.2404883. [DOI] [PubMed] [Google Scholar]
- 8.Shostak KO, et al. Patterns of expression of TSC-22 protein in astrocytic gliomas. Exp Oncol. 2005;27:314–318. [PubMed] [Google Scholar]
- 9.Shibanuma M, Kuroki T, Nose K. Isolation of a gene encoding a putative leucine zipper structure that is induced by transforming growth factor β1 and other growth factors. J Biol Chem. 1992;267:10219–10224. [PubMed] [Google Scholar]
- 10.Hino S, et al. Cytoplasmic TSC-22 (transforming growth factor-beta-stimulated clone-22) markedly enhances the radiation sensitivity of salivary gland cancer cells. Biochem Biophys Res Commun. 2002;292:957–963. doi: 10.1006/bbrc.2002.6776. [DOI] [PubMed] [Google Scholar]
- 11.Hino S, et al. Leucine zipper structure of TSC-22 (TGF-beta stimulated clone-22) markedly inhibits the anchorage-independent growth of salivary gland cancer cells. Oncol Rep. 2002;9:371–374. [PubMed] [Google Scholar]
- 12.Kawamata H, Fujimori T, Imai Y. TSC-22 (TGF-beta stimulated clone-22): a novel molecular target for differentiation-inducing therapy in salivary gland cancer. Curr Cancer Drug Targets. 2004;4:521–529. doi: 10.2174/1568009043332844. [DOI] [PubMed] [Google Scholar]
- 13.Nakashiro K, et al. Down-regulation of TSC-22 (transforming growth factor beta-stimulated clone 22) markedly enhances the growth of a human salivary gland cancer cell line in vitro and in vivo. Cancer Res. 1998;58:549–555. [PubMed] [Google Scholar]
- 14.FlyBase. The Drosophila genetic database. Nucleic Acids Res. 1994;22:3456–3458. doi: 10.1093/nar/22.17.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sillard R, et al. A novel 77-residue peptide from porcine brain contains a leucine-zipper motif and is recognized by an antiserum to delta-sleep-inducing peptide. Eur J Biochem. 1993;216:429–436. doi: 10.1111/j.1432-1033.1993.tb18160.x. [DOI] [PubMed] [Google Scholar]
- 16.Kester HA, Blanchetot C, den Hertog J, van der Saag PT, van der Burg B. Transforming growth factor-β-stimulated clone-22 is a member of a family of leucine zipper proteins that can homo- and heterodimerize and has transcriptional repressor activity. J Biol Chem. 1999;274:27439–27447. doi: 10.1074/jbc.274.39.27439. [DOI] [PubMed] [Google Scholar]
- 17.D'Adamio F, et al. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- 18.Dobens L, et al. The Drosophila bunched gene is a homologue of the growth factor stimulated mammalian TSC-22 sequence and is required during oogenesis. Mech Dev. 1997;65:197–208. doi: 10.1016/s0925-4773(97)00080-4. [DOI] [PubMed] [Google Scholar]
- 19.Treisman JE, Lai Z-C, Rubin GM. shortsighted acts in the decapentaplegic pathway in Drosophila eye development and has homology to a mouse TGF-β-responsive gene. Development. 1995;121:2835–2845. doi: 10.1242/dev.121.9.2835. [DOI] [PubMed] [Google Scholar]
- 20.Ohta S, Shimekaka Y, Nagata K. Molecular cloning and characterization of a transcription factor for the C-type natriuretic peptide gene promoter. Eur J Biochem. 1996;242:460–466. doi: 10.1111/j.1432-1033.1996.460rr.x. [DOI] [PubMed] [Google Scholar]
- 21.Perrimon N, Lanjun A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kania A, et al. P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics. 1995;139:1663–1678. doi: 10.1093/genetics/139.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobens L, Jaeger A, Peterson J, Raftery L. bunched sets a boundary of Notch signaling to pattern anterior eggshell structures during Drosophila oogenesis. Dev Biol. 2005;287:425–437. doi: 10.1016/j.ydbio.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Dobens LL, Peterson J, Treisman J, Raftery LA. Drosophila bunched integrates opposing DPP and EGF signals to set the operculum boundary. Development. 2000;127:745–754. doi: 10.1242/dev.127.4.745. [DOI] [PubMed] [Google Scholar]
- 25.Dohrmann CE, Noramly S, Raftery LA, Morgan BA. Opposing effects on TSC-22 expression by BMP and receptor tyrosine kinase signals in the developing feather tract. Dev Dyn. 2002;223:85–95. doi: 10.1002/dvdy.1236. [DOI] [PubMed] [Google Scholar]
- 26.Montell DJ. The social lives of migrating cells in Drosophila. Curr Opin Genet Dev. 2006;16:374–383. doi: 10.1016/j.gde.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Schier H, St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng WM, Althauser C, Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–4746. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- 29.Dobens L, Raftery L. Integration of epithelial patterning and morphogenesis in the Drosophila ovarian follicle cells. Dev Dyn. 2000;218:80–93. doi: 10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Horne-Badovinac S, Bilder D. Mass transit: Epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 31.Berg CA. The Drosophila shell game: Patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Ohta S, Yanagihara K, Nagata K. Mechanism of apoptotic cell death of human gastric carcinoma cells mediated by transforming growth factor β. Biochem J. 1997;324:777–782. doi: 10.1042/bj3240777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida D, et al. Over-expression of TSC-22 (TGF-beta stimulated clone-22) markedly enhances 5-fluorouracil-induced apoptosis in a human salivary gland cancer cell line. Lab Invest. 2000;80:955–963. doi: 10.1038/labinvest.3780098. [DOI] [PubMed] [Google Scholar]
- 34.Hino S, et al. Nuclear translocation of TSC-22 (TGF-beta-stimulated clone-22) concomitant with apoptosis: TSC-22 as a putative transcriptional regulator. Biochem Biophys Res Commun. 2000;278:659–664. doi: 10.1006/bbrc.2000.3840. [DOI] [PubMed] [Google Scholar]
- 35.Gupta R, et al. Peroxisome proliferator-activated receptor γ and transforming growth factor-β pathways inhibit intestinal epithelial growth by regulating levels of TSC-22. J Biol Chem. 2003;278:7431–7438. doi: 10.1074/jbc.M208076200. [DOI] [PubMed] [Google Scholar]
- 36.Gluderer S, et al. Bunched, the Drosophila homolog of the mammalian tumor suppressor TSC-22, promotes cellular growth. BMC Dev Biol. 2008;8:10. doi: 10.1186/1471-213X-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 38.Adam JC, Montell DJ. A role for extra macrochaetae downstream of Notch in follicle cell differentiation. Development. 2004;131:5971–5980. doi: 10.1242/dev.01442. [DOI] [PubMed] [Google Scholar]
- 39.Sun J, Deng WM. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development. 2005;132:4299–4308. doi: 10.1242/dev.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–27. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Tapon N, Moberg KH, Hariharan IK. The coupling of cell growth to the cell cycle. Curr Opin Cell Biol. 2001;13:731–737. doi: 10.1016/s0955-0674(00)00284-2. [DOI] [PubMed] [Google Scholar]
- 43.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 44.McCollum G, Keng PC, States JC, McCabe MJ., Jr Arsenite delays progression through each cell cycle phase and induces apoptosis after G2/M arrest in U937 myeloid leukemia cells. J Pharmacol Exp Ther. 2005;313:877–887. doi: 10.1124/jpet.104.080713. [DOI] [PubMed] [Google Scholar]
- 45.Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 46.McCall K. Eggs over easy: Cell death in the Drosophila ovary. Dev Biol. 2004;274:3–14. doi: 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Besse F, Pret AM. Apoptosis-mediated cell death within the ovarian polar cell lineage of Drosophila melanogaster. Development. 2003;130:1017–1027. doi: 10.1242/dev.00313. [DOI] [PubMed] [Google Scholar]
- 48.Xi R, McGregor JR, Harrison DA. A gradient of JAK pathway activity patterns the anterior–posterior axis of the follicular epithelium. Dev Cell. 2003;4:167–177. doi: 10.1016/s1534-5807(02)00412-4. [DOI] [PubMed] [Google Scholar]
- 49.Bjorklund M, et al. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 50.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 51.Maines JZ, Stevens LM, Tong X, Stein D. Drosophila dMyc is required for ovary cell growth and endoreplication. Development. 2004;131:775–786. doi: 10.1242/dev.00932. [DOI] [PubMed] [Google Scholar]
- 52.Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- 53.Fiol DF, Mak SK, Kultz D. Specific TSC22 domain transcripts are hypertonically induced and alternatively spliced to protect mouse kidney cells during osmotic stress. FEBS J. 2007;274:109–124. doi: 10.1111/j.1742-4658.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 54.Levine B, Jean-Francois M, Bernardi F, Gargiulo G, Dobens L. Notch signaling links interactions between the C/EBP homolog slow border cells and the GILZ homolog bunched during cell migration. Dev Biol. 2007;305:217–231. doi: 10.1016/j.ydbio.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Kester HA, van der Leede BM, van der Saag PT, van der Burg B. Novel progesterone target genes identified by an improved differential display technique suggest that progestin-induced growth inhibition of breast cancer cells coincides with enhancement of differentiation. J Biol Chem. 1997;272:16637–16643. doi: 10.1074/jbc.272.26.16637. [DOI] [PubMed] [Google Scholar]
- 56.Rae FK, Stephenson SA, Nicol DL, Clements JA. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int J Cancer. 2000;88:726–732. doi: 10.1002/1097-0215(20001201)88:5<726::aid-ijc7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 57.Dolle L, Depypere HT, Bracke ME. Anti-invasive/anti-metastasis strategies: New roads, new tools and new hopes. Curr Cancer Drug Targets. 2006;6:729–751. doi: 10.2174/156800906779010263. [DOI] [PubMed] [Google Scholar]
- 58.Hedley BD, Allan AL, Chambers AF. Tumor dormancy and the role of metastasis suppressor genes in regulating ectopic growth. Future Oncol. 2006;2:627–641. doi: 10.2217/14796694.2.5.627. [DOI] [PubMed] [Google Scholar]
- 59.Davis SR, Meltzer PS. Modeling synovial sarcoma: Timing is everything. Cancer Cell. 2007;11:305–307. doi: 10.1016/j.ccr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 60.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 61.Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 62.Stevaux O, et al. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryant Z, et al. Characterization of differentially expressed genes in purified Drosophila follicle cells: Toward a general strategy for cell type-specific developmental analysis. Proc Natl Acad Sci USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cayirlioglu P, Bonnette P, Cickson M, Duronio R. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development. 2001;128:5085–5098. doi: 10.1242/dev.128.24.5085. [DOI] [PubMed] [Google Scholar]