Abstract
Anti-human immunodeficiency virus type 1 (HIV-1) antibodies whose binding to gp120 is enhanced by CD4 binding (CD4i antibodies) are generally considered nonneutralizing for primary HIV-1 isolates. However, a novel CD4i-specific Fab fragment, X5, has recently been found to neutralize a wide range of primary isolates. To investigate the precise nature of the extraordinary neutralizing ability of Fab X5, we evaluated the abilities of different forms (immunoglobulin G [IgG], Fab, and single-chain Fv) of X5 and other CD4i monoclonal antibodies to neutralize a range of primary HIV-1 isolates. Our results show that, for a number of isolates, the size of the neutralizing agent is inversely correlated with its ability to neutralize. Thus, the poor ability of CD4i-specific antibodies to neutralize primary isolates is due, at least in part, to steric factors that limit antibody access to the gp120 epitopes. Studies of temperature-regulated neutralization or fusion-arrested intermediates suggest that the steric effects are important in limiting the binding of IgG to the viral envelope glycoproteins after HIV-1 has engaged CD4 on the target cell membrane. The results identify hurdles in using CD4i epitopes as targets for antibody-mediated neutralization in vaccine design but also indicate that the CD4i regions could be efficiently targeted by small molecule entry inhibitors.
Human immunodeficiency virus type 1 (HIV-1) entry into host cells is initiated by the binding of the gp120 subunit of the viral envelope glycoprotein (Env) complex to the host cell receptor (CD4) (8, 20). This interaction induces conformational changes in gp120 resulting in the exposure of a conserved high-affinity binding site for the coreceptor (the chemokine receptors CCR5 or CXCR4) (46, 47, 54, 56, 59). A second obligatory binding step between the gp120-CD4 complex and the coreceptor is then thought to induce additional conformational changes that ultimately result in the fusion of viral and host cell membranes (9, 18).
Neutralizing antibodies are believed to act, at least in part, by binding to the exposed Env surface and obstructing the initial interaction between a trimeric array of gp120 molecules on the virion surface and receptor molecules on the target cell (36, 37, 57). In response, HIV-1 has evolved a number of strategies to evade recognition by neutralizing antibodies, particularly those directed to the conserved CD4 and coreceptor binding sites of Env. The extent of protection of these sites from antibody recognition is limited by the necessity to preserve the accessibility for receptor interaction. In the case of the CD4bs this has led to the following structural features: (i) it is partially obscured from antibody recognition by the V1/V2 loop and associated carbohydrate structures; (ii) the flanking residues are variable and modified by glycosylation; (iii) it is recessed to an extent that limits direct access by an antibody variable region; (iv) clusters of residues within the CD4bs that do not directly interact with CD4 are subject to variation among virus strains; (v) many gp120 residues interact with CD4 via main-chain atoms, allowing for variability in the corresponding amino acid side chains (26); and (vi) there is considerable conformational flexibility within the CD4-unbound state of gp120, and antibody binding therefore requires relatively large entropic decreases, thus “conformationally masking” the conserved CD4bs (23, 33).
The coreceptor binding site on gp120 is thought to be composed of a highly conserved element on the β19 strand and parts of the V3 loop (41, 42, 61). These elements are masked by the V1/V2 variable loops in the CD4-unbound state and largely unavailable for antibody binding (55, 59). Upon CD4 binding, conformational changes are induced; these changes include displacement of the V1/V2 stem-loop structure and consequent exposure of the coreceptor binding site (31, 47, 60). Binding studies with variable loop-deleted mutants suggest that CD4 induces additional rearrangement or stabilization of the gp120 bridging sheet near the β19 strand to form the final coreceptor binding surface (59, 61). Since the binding to CD4 occurs at the virus-cell interface, the exposed coreceptor binding site is optimally positioned for interaction with the coreceptor.
A highly conserved discontinuous structure on gp120 associated with the coreceptor binding site is recognized by monoclonal antibodies (MAbs) that bind better to gp120 upon ligation with CD4. These so-called CD4-induced (CD4i) antibodies, such as 17b and 48d (54, 60), recognize a cluster of gp120 epitopes that are centered on the β19 strand and partially overlap the coreceptor binding site (41, 42, 55, 59). Although such CD4i MAbs can neutralize some T-cell line-adapted HIV-1 strains, they are generally poorly neutralizing for primary isolates (40). However, we recently reported the isolation of an antibody Fab fragment, X5, from a phage display library, that is directed to a CD4i epitope and does neutralize a wide variety of primary isolates (32). Here we investigated the differences between Fab X5 and other CD4i MAbs at a molecular level. We provide evidence that size is the determining factor for the inability of CD4i MAbs to neutralize many primary HIV-1 isolates.
MATERIALS AND METHODS
Reagents.
The following materials were obtained from the National Institute of Health AIDS Research and Reference Reagent Program (ARRRP): molecular clones of HIV-1 89.6, HxB2, JR-FL, JR-CSF, and ADA; sCD4 (amino acids 1 to 370; contributed by N. Schülke); and recombinant gp120JR-FL and CD4-immunoglobulin G2 (IgG2; kindly provided by Paul Maddon and William Olson [Progenics, Tarrytown, N.Y.]).
Construction and purification of IgG1 X5.
Fab X5 was isolated from a phage display library constructed from the bone marrow of an HIV-1-seropositive donor as described previously (32). The DNA fragments encoding the heavy chain (Fd fragment) and light chain of X5 were transferred from the pComb3H phagemid vector (1) to the pDR12 mammalian expression vector (4) and transfected into Chinese hamster ovary (CHO) cells by using FuGENE6 transfection reagent (Roche, Indianapolis, Ind.). Stable transfected clones were selected under methionine sulfoximine (MSX; Sigma, St. Louis, Mo.) amplification. The clone with the highest production of IgG1 X5, as determined by enzyme-linked immunosorbent assay (ELISA) with cell culture supernatant, was chosen for scale-up in the CellCube System (Corning, Inc., Corning, N.Y.) and purified by affinity chromatography with protein A (Pharmacia, Uppsala, Sweden). Purified IgG1 X5 was concentrated and dialyzed against phosphate-buffered saline (PBS). Purity was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and the concentration was measured by determining the A280 value.
Production and purification of antibody fragments.
Antibody Fab fragments were produced by papain digest as described previously (24). Single chain Fv (scFv) X5 was engineered into the pComb3X vector by using antibody specific primers (1), produced in Escherichia coli and purified by nickel chelate chromatography (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Purified protein was dialyzed with PBS and stored at −80°C until use. Purity was determined by SDS-polyacrylamide gel electrophoresis, and the concentration was measured by determining the A280 value. The 17b single-chain Fv plasmid was a gift from Wayne Marasco (Dana-Farber Cancer Institute, Boston, Mass.). In short, the 17b scFv was produced by PCR amplification of the heavy- and light-chain variable-region fragments from the 17b hybridoma cell line and cloned into the prokaryotic expression vector, pHEN (16). From this construct, the 17b heavy chain, the (G4S)5 linker, the light-chain variable sequences and a His6 epitope tag were PCR amplified and subcloned into the inducible expression vector pMT (17) downstream of the Drosophila metallothionein promoter and in frame with the tissue plasminogen activator leader sequence. A stable S2 Drosophila cell line was established by cotransfection with a hygromycin resistance plasmid, pCo-hygro, and selection with hygromycin (300 μg/ml). Production of the 17b single chain was induced by the addition of 750 μM CuSO4 to the cells at a density of 107cells/ml in serum-free insect medium containing 0.1% pluronic (BASF, Mount Olive, N.J.). Purification of the single chain was performed in a single step by direct passage over a nickel chelating column (Pharmacia). The 33-kDa protein was eluted by 50 mM EDTA, dialyzed, and quantified by A280 and SDS gel analysis.
Pseudovirion production.
Plasmids containing the env genes of HIV-1 strains HxB2 (35), JR-CSF (65), ADA (53), ADAΔV1V2 (21), and SOS-JRFL (2) were constructed as described previously. Similarly, the env genes of HIV-1 strains JR-FL, HxB2, and 89.6 were cloned into pSVIIIenv (14) and those of strains SF162, 92HT594, JR-CSF, JR-FL, and ADA were cloned into pSV7d (35). Additionally the env gene of amphotropic murine leukemia virus (A-MLV) was cloned into pSV7d (38). Recombinant pseudovirions were produced as described previously (7, 28, 38, 62, 65).
Neutralization assay.
Neutralization was measured in various luciferase reporter gene assays as described previously (2, 28, 38, 62, 65).
(i) Standard neutralization assay A.
A pseudovirus inoculum, previously determined to yield ∼1,000,000 relative light units, was incubated with a serial dilution of MAb for 1 h at 37°C and added to 2 × 104 U87.CD4.CCR5 or U87.CD4.CXCR4 cells (ARRRP [H. Deng and D. Littman]). After 24 h of incubation, fresh medium was added, followed by incubation for 3 days (37°C, 5% CO2). Luciferase activity was measured by using the luciferase assay system (Promega, Madison, Wis.) according to the manufacturer's instructions.
(ii) Standard neutralization assay B.
A pseudovirus inoculum was incubated with a serial dilution of MAb for 18 h at 37°C and added to U87.CD4 expressing both CCR5 and CXCR4 cells (ARRRP [H. Deng and D. Littman]). After 72 h of incubation (37°C, 5% CO2), luciferase activity was measured by using the luciferase assay system (Promega) according to the manufacturer's instructions.
(iii) SOS neutralization assays.
Standard and postattachment neutralization assays using mutant SOS pseudovirions have been described previously (2). Briefly, for standard neutralization, a pseudovirus inoculum normalized for p24 content by ELISA was incubated with a serial dilution of MAb for 1 h at 37°C and added to 2 × 104 U87.CD4.CCR5 cells for 2 h. Unbound virus was removed by changing the medium, and the culture was incubated for an additional 1 h. Alternatively, to measure postattachment neutralization, a pseudovirus inoculum was incubated with 2 × 104 U87.CD4.CCR5 cells for 2 h to form an SOS-arrested intermediate. After replacement of the medium, a serial dilution of MAb was added for 1 h. Cells were subsequently treated with 5 mM dithiothreitol for 10 min to activate the fusion reaction, and the medium was replaced. After an additional 3-day incubation (37°C, 5% CO2), luciferase activity was measured as described above.
(iv) Temperature-regulated neutralization assay.
A pseudovirus inoculum, previously determined to yield ∼10,000 reverse transcriptase (RT) units, was incubated with a serial dilution of MAb for 1 h at 37°C and added to 6 × 103 Cf2Th cells expressing CD4 and CCR5 (22). Alternatively, the virus inoculum was incubated for 4 to 5 h at 4°C with the cells prior to washing. Serial dilutions of antibodies were added, and the temperature was raised to 37°C. After a 24-h incubation at 37°C (5% CO2), fresh medium was added, and the cells were incubated for an additional 3 days. Luciferase activity was measured as described above.
ELISA.
Microtiter plates (Costar, Corning, N.Y.) were coated overnight at 4°C with 5 μg of anti-gp120 antibody D7324 (Aalto BioReagents, Ltd., Dublin, Ireland)/ml in PBS (pH 7.5). Plates were blocked with 3% bovine serum albumin for 1 h at room temperature and then washed with PBS-0.05% Tween 20 (PBST). Lysed pseudovirions (1% Empigen; Sigma) were added, diluted in PBST-1% bovine serum albumin, and incubated for 4 h at room temperature, in the presence or absence of sCD4 (2 μg/ml). Next, serial dilutions of MAb were incubated with the captured proteins, and bound MAb was detected with horseradish peroxidase-labeled goat anti-human IgG F(ab′)2 (Pierce, Rockford, Ill.) and tetramethylbenzidine substrate (Bio-Rad). The color reaction was stopped after 20 min by the addition of 2 M H2SO4, and the absorbance at 450 nm was measured.
Modeling.
The positions of four-domain CD4 structures were generated by superimposing the four-domain CD4 structures (pdb accession numbers 1WIO, 1WIP, and 1WIQ, residues 1 to 363) (58) onto the trimeric model of the gp120-D1D2 complex (27) by using the main-chain atoms of CD4 D1D2 (residues 1 to 178). Root-mean-square (RMS) deviations for the superpositions are ∼1.0 Å for all superpositions. A model having an additional 35° rotation of D3D4 toward the target cell membrane was constructed by using the interactive graphics program “O” (19), by altering the backbone angle between CD4 residues 176 and 177 in 1WIP from ψ, φ values of −96 and 108° for residue 176 to values of −96° and 162°, respectively, and from ψ, φ values of −72° and 154° for residue 177 to values of −153 and 154°, respectively. The nine-amino-acid linker between the last crystallographically ordered residue and the transmembrane-spanning region contains two prolines. Its length was estimated by using calculations of polymer dimensions (48), where an Ala9 polymer has an average N-terminal and C-terminal distance of 16 Å and a Pro9 polymer a distance of 21 Å. All superpositions were carried out with the program LSQKAB, which is in the CCP4 suite of programs (6).
RESULTS
Antibody Fab and scFv fragments of CD4i MAbs are more effective in neutralization of HIV-1JR-CSF than whole IgG1 antibodies.
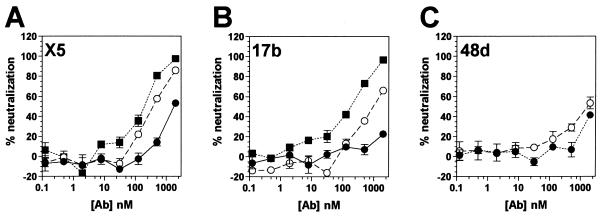
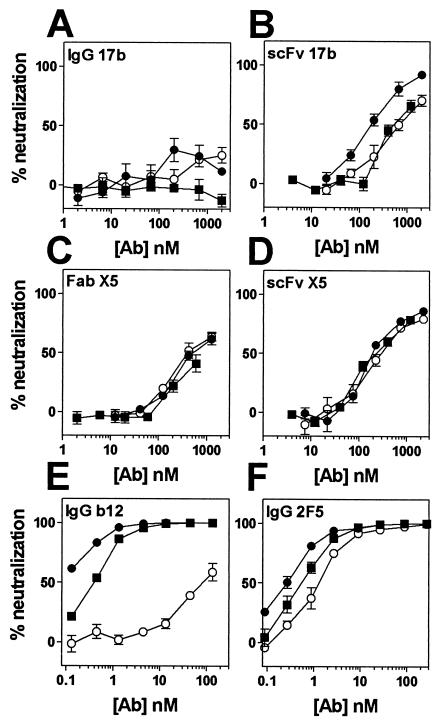
We recently reported the isolation of a novel CD4i Fab fragment, designated X5, that differed from previously isolated CD4i antibodies, such as 17b or 48d, in that it potently neutralized an array of primary HIV-1 isolates (32). Whole-antibody molecules are generally more effective in neutralization than their corresponding Fab fragments (57). We therefore converted Fab X5 to a whole IgG1 molecule, expressed it in CHO cells and purified it by affinity chromatography. IgG X5 was tested against CCR5-dependent (R5) HIV-1JR-CSF in a neutralization assay. Surprisingly, the whole antibody neutralized ∼5-fold less effectively than the Fab fragment (Fig. 1A and Table 1) .
FIG. 1.
Neutralization of HIV-1JR-CSF by CD4i antibodies and antibody fragments. Neutralization titers of whole antibodies (•), Fab fragments (○), and scFv fragments (▪) to HIV-1JR-CSF were determined in a pseudotyped luciferase-based neutralization assay with U87.CD4.CCR5 cells. Datum points are the means of triplicates ± the standard error of the mean (SEM).
TABLE 1.
Neutralization of HIV-1 by CD4i antibodies and antibody fragments (standard neutralization assay A)
| MAb | Format | Neutralization (IC50 [nM])a of HIV-1 isolate
|
||||
|---|---|---|---|---|---|---|
| JR-CSF R5 | JRFL R5 | ADA R5 | 89.6 X4 | HxB2 X4 | ||
| X5 | IgG | 1,799 | 1,950 | >2,000 (14) | 347 | 8.1 |
| Fab | 458 | 955 | >2,000 (33) | 759 | 74 | |
| scFv | 224 | 115 | 447 | 191 | 16 | |
| 17b | IgG | >2,000 (22) | >2,000 (20) | >2,000 (32) | 189 | 21 |
| Fab | 912 | 288 | 1,778 | 242 | 43 | |
| scFv | 158 | 331 | 447 | 122 | 61 | |
| 48d | IgG | >2,000 (41) | NT | 1,380 | 457 | 8.7 |
| Fab | 1,698 | NT | >2,000 (32) | >2,000 (36) | 39 | |
Neutralization was assessed on U87.CD4 cells expressing either the CCR5 (R5) or the CXCR4 (X4) coreceptor. The neutralization of the dual-tropic isolate 89.6 was studied on U87.CD4 cells expressing CXCR4. Where 50% neutralization was not achieved, the percent inhibition at the highest antibody concentration is given in parentheses. IC50, concentration of antibody causing a 50% reduction in viral infection; NT, not tested.
To assess whether the observed phenomenon was a general characteristic of CD4i MAbs, we tested whole antibody and antibody Fab fragments of other CD4i MAbs in neutralization against HIV-1JR-CSF. The Fab fragments of MAbs 17b and 48d were made by papain digestion. Both Fab fragments were more effective in neutralization compared to their corresponding whole antibody molecules, although the difference appeared less significant for 48d (Fig. 1B and C and Table 1). In addition, single-chain Fv (scFv) variants of X5 and 17b were constructed and tested for neutralization. The smaller scFvs were even more effective than the corresponding Fab fragments (Fig. 1A and B).
Lack of HIV-1JR-CSF neutralization by IgG is not due to a loss of structural integrity of the IgG molecule.
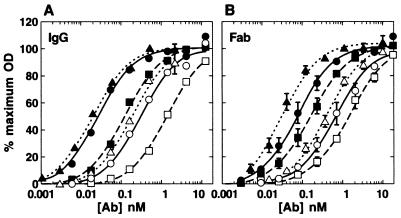
To assess whether the inability of the whole antibody molecules to neutralize HIV-1JR-CSF was caused by diminished recognition of gp120JR-CSF, we compared the binding of IgG with Fab fragments to monomeric gp120JR-CSF in ELISA in the presence or absence of sCD4. In contrast to the neutralization results, the IgG molecules of X5 and 17b had an approximately two- to threefold-higher apparent affinity, probably due to the bivalency of the whole IgG molecule (Fig. 2). Fab X5 obtained by papain digestion of IgG X5 had a similar apparent affinity to E. coli-expressed Fab X5 (data not shown). No differences in apparent binding affinities were observed between Fab and IgG 48d. All antibodies displayed an ∼10-fold increase in apparent affinity for gp120 in the presence of sCD4. Although affinity to monomeric gp120 does not correlate with neutralization, this finding does demonstrate that the lack of neutralizing activity was not due to a loss of structural integrity of the whole antibody molecule.
FIG. 2.
Binding curves of CD4i antibodies and antibody fragments to monomeric gp120JR-CSF as determined by ELISA. Titers of whole antibodies (A) and Fab fragments (B) to gp120JR-CSF were determined in the presence (solid symbols) or absence (open symbols) of saturating amounts of sCD4. Symbols: circles, 17b; squares, 48d; triangles, X5. Datum points are the means of at least two separate experiments ± the standard deviation.
Antibody fragment size-dependent neutralization is a function of viral isolate.
To determine whether the observed phenomenon was virus isolate specific, we assessed a panel of HIV-1 isolates for sensitivity to CD4i antibody (fragment)-mediated neutralization (Table 1). HIV-1JR-FL and HIV-1ADA, both R5 primary isolates, showed a broadly similar tendency to be better neutralized by smaller antibody fragments, with the exceptions that the 17b Fab and scFv fragments were equally effective against HIV-1JR-FL and that HIV-1ADA was somewhat better neutralized by the 48d whole IgG than by the Fab fragment. HIV-1ADA was overall somewhat less sensitive to CD4i MAb neutralization than was HIV-1JR-CSF. The relatively neutralization-resistant isolate HIV-1YU2 (29, 52) was weakly neutralized by scFv CD4i MAbs, but infectivity was enhanced by CD4i whole IgG1 molecules (data not shown).
In agreement with other studies (54, 60), the T-cell line-adapted, CXCR4-dependent (X4) HIV-1 strain HxB2 was relatively sensitive to all three tested CD4i MAbs as whole IgG molecules. In this case, the Fab fragments were generally somewhat less effective than whole IgG. The scFv fragment of X5 was more effective than the Fab fragment, whereas for 17b the two fragments were approximately equally effective (Table 1).
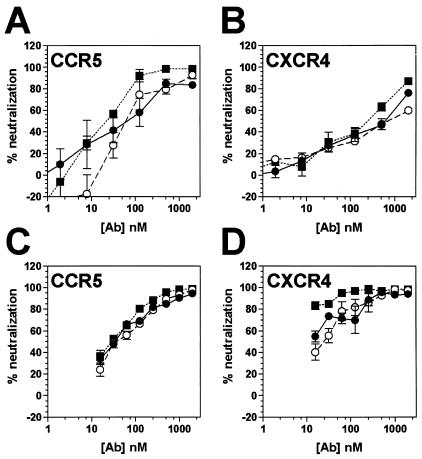
Two dualtropic (R5X4) primary isolates were evaluated, HIV-189.6 and a dualtropic variant of HIV-1JR-CSF, designated HIV-1JR-CSF/IR (P. Poignard et al., unpublished data). When HIV-189.6 neutralization by X5 was evaluated on CXCR4-expressing U87 cells, whole IgG and antibody fragments displayed similar potency (Fig. 3B and Table 1). For HIV-1JR-CSF/IR evaluated on CXCR4-expressing U87 cells, X5 scFv was clearly more effective than the Fab fragment and the whole IgG1 molecule (Fig. 3D). Next, we evaluated X5-mediated neutralization of HIV-189.6 and HIV-1JR-CSF/IR on CCR5-expressing U87 cells. For HIV-189.6, neutralization was achieved at somewhat lower antibody concentrations on CCR5-expressing U87 cells than on CXCR4-expressing cells, with whole IgG and Fab neutralization somewhat less effective than with scFv (Fig. 3A). When HIV-1JR-CSF/IR neutralization by X5 was evaluated on CCR5-expressing U87 cells, whole IgG and antibody fragments displayed similar potency that was comparable to whole IgG and Fab neutralization on CXCR4-expressing U87 cells (Fig. 3C). Thus, the precise pattern of neutralization potency displayed by the CD4i MAb X5 and antibody fragments is dependent upon isolate and coreceptor usage.
FIG. 3.
Neutralization of dualtropic HIV-1 by MAb X5 whole IgG and antibody fragments. Titers of whole antibody (•), Fab fragment (○) and scFv fragment (▪) to HIV-189.6 (A and B) and HIV-1JR-CSF/IR (C and D) were determined in a pseudotyped luciferase-based neutralization assay with U87.CD4 cells expressing either CCR5 (A and C) or CXCR4 (B and D). Datum points are the means of triplicates ± the SEM.
In a separate study, with a similar reporter gene neutralization assay in a different laboratory, a panel of HIV-1 isolates was assessed for sensitivity to CD4i antibody (fragment)-mediated neutralization (Table 2). In these experiments, whole IgGs of CD4bs-specific MAbs were included for reference. In addition, pseudovirions expressing the A-MLV env gene were included as negative control. As for the experiments described in Table 1, the R5 primary isolates HIV-1JR-FL, HIV-1JR-CSF, and HIV-1ADA, were sensitive to neutralization by both X5 and 17b scFv fragments and relatively resistant to Fab X5, IgG X5, and IgG 17b. Another R5 primary isolate, HIV-1SF162, was sensitive to CD4i IgG and antibody fragments, but this isolate was also sensitive to IgG b6, a generally weakly neutralizing anti-CD4bs MAb. Primary dualtropic R5X4 isolate HIV-192HT594 was neutralized by MAb X5 and Fab and scFv antibody fragments and by 17b scFv fragment but was resistant to 17b whole IgG (Table 2).
TABLE 2.
Neutralization of HIV-1 by CD4i antibodies and antibody fragments (standard neutralization assay B)a
| MAb | Format | Neutralization (IC50 [nM]) of HIV-1 isolate
|
||||||
|---|---|---|---|---|---|---|---|---|
| SF162 R5 | 92HT594 R5X4 | JR-CSF R5 | JR-FL R5 | ADA R5 | HxB2 X4 | A-MLVb | ||
| X5 | IgG | 0.22 | 67 | >333 (16) | >333 (15) | >333 (0) | 1.8 | >333 |
| Fab | 2.8 | 283 | >1,000 (25) | >1,000 (13) | >1,000 (15) | 35 | >1,000 | |
| scFv | 1.7 | 129 | 633 | 1,077 | 637 | 1.2 | >2,000 | |
| 17b | IgG | 3.1 | >333 (21) | >333 (0) | >333 (0) | >333 (0) | 15 | >333 |
| scFv | 3.9 | 117 | 369 | 573 | 294 | 45 | >1,160 | |
| b12 | IgG | 0.46 | 2.1 | 7.3 | 0.76 | 18 | 0.07 | >333 |
| b6 | IgG | 0.53 | >333 (42) | >333 (0) | >333 (0) | >333 (0) | 0.38 | >333 |
Neutralization was assessed on U87.CD4 cells expressing both CCR5 and CXCR4 coreceptors. Coreceptor tropism of the viral isolates is indicated (R5, X4, or R5X4). Where 50% neutralization was not achieved, the percent inhibition at the highest antibody concentration is given in parentheses. IC50, concentration of antibody causing a 50% reduction in viral infection.
As a negative control, pseudovirions expressing the A-MLV env gene were included.
CD4i antibody fragments neutralize HIV-1 subsequent to CD4 binding.
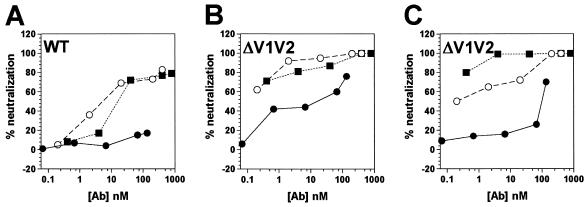
The CD4-induced conformational changes in gp120 include displacement of the V1/V2 variable loops, thus exposing the coreceptor binding site and the CD4i epitopes (31, 47, 60). To investigate the role of the V1/V2 variable loops in CD4i antibody- and antibody fragment-mediated neutralization, we compared the neutralizing activities of different 17b antibody forms against wild-type ADA (WT) (Fig. 4A) and V1V2-deleted ADA (ΔV1V2) (Fig. 4B). Removal of the V1/V2 loops resulted in increased sensitivity to neutralization by MAb 17b, as has been previously reported (5, 21). However, 17b scFv and Fab fragments neutralized the ΔV1V2 virus more efficiently than the whole antibody, suggesting that some steric constraints on MAb 17b neutralization operate even in the absence of the V1/V2 loops.
FIG. 4.
Pre- and postattachment neutralization of HIV-1ADA and HIV-1ADAΔV1V2 by MAb 17b whole antibody and antibody fragments. Neutralization titers of whole antibody (•), Fab fragment (○), and scFv fragment (▪) to HIV-1ADA (A) and HIV-1ADAΔV1V2 (B and C) were determined in a pseudotyped luciferase-based neutralization assay with Cf2Th cells expressing CD4 and CCR5. In these settings pre- and postattachment neutralizations were measured simultaneously (A and B) by preincubating virus and antibody fragments at 37°C prior to addition to cells. (C) Postattachment neutralization was measured exclusively by preincubating virus and cells at 4°C, washing the cells, and adding antibody fragments before the temperature was increased to 37°C.
Membrane fusion mediated by the HIV-1 envelope glycoproteins is a temperature-dependent process (12, 13). To investigate the sequence of events for MAb 17b-mediated neutralization, we tested the ΔV1V2 virus in a temperature-dependent neutralization assay. Virus and target cells were preincubated at 4°C and washed, and then 17b scFv, Fab, or whole IgG was added. The fusion reaction was resumed when the temperature was raised to 37°C. Under these conditions, compared to the standard neutralization assay (Fig. 4B), the neutralizing efficiency of the scFv was only minimally changed or slightly improved, the efficiency of the Fab fragment was slightly reduced, and the whole IgG molecule was markedly less efficient (Fig. 4C). This suggests that the antibody fragments can neutralize virus after it has attached to the target cell, whereas the whole antibody molecule is much less effective in this context. In other words, it seems likely that neutralization is most efficiently mediated by CD4i antibody binding to its epitope after CD4 engagement and that this is sterically restricted for the whole antibody but not for the fragments.
Finally, we used an activatable fusion intermediate system to look further at the role of CD4i antibodies and fragments. Introduction of an intermolecular disulfide bond between gp120 and gp41 subunits has been shown to stabilize their association (designated SOS Env) (3). Incorporation of SOS Env into pseudovirions produces particles that bind to target cells but do not fuse (2). The addition of a reducing agent activates the fusion reaction. Initially, the neutralization of reduced SOS pseudovirions of HIV-1JR-FL (SOSJR-FL) was compared to wild-type HIV-1JR-FL (WTJR-FL) pseudovirions under standard assay conditions. With the exception of whole IgG 17b, all CD4i antibody fragments could neutralize both SOS and WT virus to a similar extent (Fig. 5A to D). In addition, whole IgG b12 and IgG 2F5, which were included as control antibodies, could efficiently neutralize both viruses under standard conditions (Fig. 5E and F). We next used SOSJR-FL to examine the ability of CD4i antibody fragments to neutralize under postattachment conditions. SOSJR-FL pseudovirions were incubated with cells, unbound virus was washed away, and antibody fragments were added to cells with adsorbed SOSJR-FL pseudovirions. All CD4i antibody fragments could still efficiently neutralize SOSJR-FL under these conditions, in contrast to whole IgG 17b. Whereas neutralization of SOSJR-FL by whole IgG b12 was much weaker under postattachment conditions, neutralization by whole IgG 2F5 was comparable, a finding consistent with the concept that b12 neutralizes by inhibiting virus attachment to cells and that 2F5 neutralizes in a postattachment event (2, 37). This provides further evidence that smaller antibody fragments, but not whole antibody, can gain access to virions attached to target cells.
FIG. 5.
Standard and postattachment neutralization of SOSJR-FL and WTJR-FL by CD4i whole antibodies and antibody fragments. Antibodies 17b (IgG [A] and scFv [B]), X5 (Fab [C] and scFv [D]), b12 (IgG [E]), and 2F5 (IgG [F]) were tested in pseudotyped luciferase-based neutralization assays, under standard assay conditions against WTJR-FL (▪) and SOSJR-FL, and in a postattachment format against SOSJR-FL (○). Datum points are the means of triplicates ± the SEM.
A previous study suggested that the CCR5 binding site is engaged in the SOS-arrested intermediate (2). However, we show here that, on the SOS-arrested intermediate, the CD4i antibody fragments have access to their epitope, which overlaps the coreceptor binding site. One possible explanation for this apparent paradox is that not all CCR5 sites are engaged in the bound SOS intermediate, at least until dithiothreitol is added, or that the association of virus with CCR5 can be displaced by high concentrations of CD4i antibodies.
DISCUSSION
Both CD4 and coreceptor binding sites on gp120 form potential targets for antibody-mediated intervention. The primary isolate-neutralizing antibodies identified thus far that are specific for gp120 are directed to epitopes that appear to be present on the oligomeric Env complex before contact with CD4. They are believed to act, at least in part, by binding to the virion surface and sterically obstructing the interaction between virus and target cell (37, 49, 57). For these antibodies, the larger whole antibody molecules are more effective than the corresponding Fab fragments at neutralization due to greater steric obstruction but probably also because of increased avidity (bivalency) (57). Antibodies to the CD4i epitopes generally do not display primary isolate neutralizing activity at relevant concentrations.
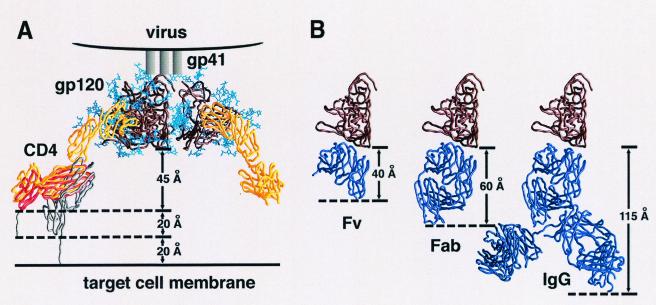
The major finding of our study is that, for some HIV-1 isolates, the size of CD4i-specific (antibody-derived) neutralizing agents is inversely correlated with neutralization efficiency. Thus, antibody fragments are more effective than whole antibody molecules in neutralization. Further, scFv (25 kDa) are generally more effective than Fab (50 kDa) fragments. The temperature-regulated (Fig. 4) and SOS-arrested (Fig. 5) neutralization data presented here are consistent with a model in which CD4i scFvs or Fabs, but not IgGs, can neutralize after attachment of the virus to CD4 (43). Because the difference in size is expected to have limited effects on the diffusion rates of the CD4i antibody fragments, these results strongly suggest that the restriction against CD4i antibody neutralization is steric, not temporal. Therefore, the likeliest explanation for the observed sensitivity of neutralization to antibody size is that, after CD4 binding to the virus, the available space between the virus and the target cell surface is not enough to accommodate a whole antibody molecule but is sufficient for antibody fragments. This is consistent with the estimated size of the antibody fragments and predictions of the distance between the CD4i epitope on the CD4-bound envelope glycoprotein trimer and the target cell membrane (Fig. 6). Recent findings demonstrating the inaccessibility of the 17b epitope at the fusing cell interface are in agreement with this hypothesis (10).
FIG. 6.
Steric restrictions on CD4i antibody neutralization after CD4 attachment. (A) HIV-1 viral spike attached to cell surface CD4. The gp120 trimer (brown) (27) is shown with the threefold axis oriented perpendicular to the target cell surface. Cα worm representations of gp120 and four-domain CD4 are shown in brown and yellow, respectively. N-linked carbohydrate on gp120 is shown in blue. The flexibility between the second and third extracellular CD4 domains allows the observed angle between the membrane-distal (D1D2) and membrane-proximal (D3D4) domains to vary by up to 10° (distinct crystal structure conformations are shown in red and yellow) (58). The distance between the membrane-proximal portion of gp120 and the bottom of the crystallographically ordered CD4 is ∼45 Å. Nine amino acids (shown in gray) occur between the last ordered crystallographic residue (position 363) and the transmembrane domain (starting at position 373), potentially increasing the gp120-target cell membrane distance by 20 Å. There is considerable uncertainty in the degree of flexibility between the second and third extracellular domains of CD4. If the interdomain rotation is increased by 35° (gray), the gp120-target cell membrane distance would increase by an additional 20 Å, resulting in a total gp120-transmembrane distance of up to 85 Å. (B) Dimensions of Fv, Fab, and IgG portions of 17b interacting with gp120. Both gp120 (brown) and antibody fragments (purple) are shown in the Cα worm representation. The gp120 molecule is depicted in precisely the same orientation as the front left protomer in panel A, with the 17b portion oriented by superimposing the X-ray structure (pdb accession number 1G9 M [25]) of the gp120 ternary complex with D1D2 and 17b Fab complex. The length of the 17b Fv along the trimer threefold axis is ∼40 Å, and the length of the Fab is ∼60 Å, with the dimensions measured for the distance between the target membrane-proximal portions of gp120 and 17b. For IgGs, considerable flexibility is found in the hinge region connecting Fab and Fc portions of the IgG. Superimposing the crystal structure of a full-length human antibody (45) onto 17b gives dimensions of 115 Å (shown here) and 140 Å (not shown) for the two alternative Fab superpositions. The figures were generated with GRASP (34).
The higher sensitivity to CD4i MAbs observed for some isolates is most likely a reflection of the exposure of the CD4i epitope on the oligomer prior to CD4 binding, as suggested previously (37). In addition, our data suggests that when the CD4i epitope is exposed, any advantage of Fab or scFv fragments disappears (Tables 1 and 2). It is, however, noteworthy that exposure of the CD4i epitope, through deletion of the V1/V2 stem-loop structure, not only facilitates virus neutralization by the whole IgG molecule but also increases the potency of the Fab and scFv fragments (Fig. 4). Comparison of the relative neutralization potencies of 17b antibody and antibody fragments under different assay conditions (Fig. 4A and C) implies that the V1/V2 variable loops continue to play a role in obstructing antibody binding even after CD4 attachment. Thus, our results suggest that CD4i MAb-mediated neutralization is biphasic, with (i) a “preattachment” phase that may not be antibody fragment size dependent but is dependent on the accessibility of the CD4i epitope on the resting oligomer, primarily governed by the V1/V2 variable loops, and (ii) a “postattachment” phase that is antibody fragment size dependent due to steric restrictions imposed by the cellular membrane and the V1/V2 variable loops. The fact that we observe more effective neutralization by X5 of HIV-1JR-CSF/IR on CCR5+ cells than on CXCR4+ cells (Fig. 3D) and the fact that scFv X5, but not scFv 17b, was more effective than the corresponding Fab fragments in neutralization of HIV-1HxB2 (Table 1), however, indicates that there are additional variables influencing these mechanisms. One could speculate that the precise orientation of the epitope may influence the susceptibility of the antibody fragments to steric constraints by the surrounding protein and carbohydrate structures at the virus-cell interface.
Attempts to improve Env immunogens include strategies to better expose the coreceptor binding site and the overlapping CD4i epitopes (11, 15, 30, 44). It is argued that increasing CD4i epitope exposure may elicit more-potent CD4i MAbs or higher serum antibody titers. Our data, however, suggest that the inability of CD4i MAbs to neutralize primary isolates is not due to lack of potency per se but to spatial constraints. Since non-syncytium-inducing, R5 HIV-1 variants establish primary infection in humans (39, 50, 51, 63, 64), the inability of CD4i-specific whole antibodies to neutralize the majority of R5 viruses used in the present study raises concerns as to whether the CD4i epitopes would be useful targets for antibody neutralization in vaccine design.
In conclusion, we show that CD4i-specific MAbs do not neutralize some primary isolates due to steric constraints. We propose that HIV-1 can exclude whole antibody molecules from the CD4i epitopes due to the close physical proximity of the cellular membrane and obstruction by the V1/V2 variable loops. The constraints were especially apparent for the primary R5 isolates tested. This raises questions about the utility of CD4i epitopes as targets for antibody-mediated neutralization in vaccine design. Understanding these viral defenses may suggest new strategies to circumvent them. The fact that the smaller antibody fragments were able to neutralize does, however, suggest that the CD4i epitopes could be used as targets for small molecule entry inhibitors.
Acknowledgments
This work was supported by NIH grants to D.R.B. (AI33292), P.P. (AI45357), J.B. (AI49566), and J.S. (AI24755, AI41851, AI31783 and AI39420); by a Center for AIDS Research grant (AI42848); by an unrestricted research grant from the Bristol-Myers Squibb Foundation; by a gift from the late William F. McCarty-Cooper; and by funds from the International AIDS Vaccine Initiative to D.R.B. and J.S. through the Neutralizing Antibody Consortium.
REFERENCES
- 1.Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled HIV-1 pseudovirions: implications for viral entry, therapy, and vaccine development. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 5.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 8.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 9.Dimitrov, D. S. 1996. Fusin: a place for HIV-1 and T4 cells to meet. Nat. Med. 2:640-641. [DOI] [PubMed] [Google Scholar]
- 10.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouts, T. R., R. Tuskan, K. Godfrey, M. Reitz, D. Hone, G. K. Lewis, and A. L. DeVico. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J. Virol. 74:11427-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey, S., M. Marsh, S. Gunther, A. Pelchen-Matthews, P. Stephens, S. Ortlepp, and T. Stegmann. 1995. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J. Virol. 69:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, Y. K., T. K. Hart, Z. L. Jonak, and P. J. Bugelski. 1993. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J. Virol. 67:3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoogenboom, H. R., A. D. Griffiths, K. S. Johnson, D. J. Chiswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivey-Hoyle, M., J. S. Culp, M. A. Chaikin, B. D. Hellmig, T. J. Matthews, R. W. Sweet, and M. Rosenberg. 1991. Envelope glycoproteins from biologically diverse isolates of immunodeficiency viruses have widely different affinities for CD4. Proc. Natl. Acad. Sci. USA 88:512-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 21.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 24.Kwong, P. D., R. Wyatt, E. Desjardins, J. Robinson, J. S. Culp, B. D. Hellmig, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115-4123. [DOI] [PubMed] [Google Scholar]
- 25.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 26.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, J. P., Q. J. Sattentau, H. Yoshiyama, M. Thali, M. Charles, N. Sullivan, S. W. Poon, M. S. Fung, F. Traincard, M. Pinkus, et al. 1993. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J. Virol. 67:6136-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 35.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parren, P. W., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed]
- 37.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 41.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 42.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 43.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed]
- 45.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 46.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schimmel, P. R., and P. J. Flory. 1967. Conformational energy and configurational statistics of poly-l-proline. Proc. Natl. Acad. Sci. USA 58:52-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schønning, K., O. Lund, O. S. Lund, and J. E. Hansen. 1999. Stoichiometry of monoclonal antibody neutralization of T-cell line-adapted human immunodeficiency virus type 1. J. Virol. 73:8364-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuitemaker, H., N. A. Kootstra, R. E. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatatos, L., A. Werner, and C. Cheng-Mayer. 1994. Differential regulation of cellular tropism and sensitivity to soluble CD4 neutralization by the envelope gp120 of human immunodeficiency virus type 1. J. Virol. 68:4973-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 56.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ugolini, S., I. Mondor, P. W. Parren, D. R. Burton, S. A. Tilley, P. J. Klasse, and Q. J. Sattentau. 1997. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T-cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, H., P. D. Kwong, and W. A. Hendrickson. 1997. Dimeric association and segmental variability in the structure of human CD4. Nature 387:527-530. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 60.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 62.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 65.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]