Abstract
Mammalian microRNAs are emerging as key regulators of the development and function of the immune system. Here, we report a strong but transient induction of miR-155 in mouse bone marrow after injection of bacterial lipopolysaccharide (LPS) correlated with granulocyte/monocyte (GM) expansion. Demonstrating the sufficiency of miR-155 to drive GM expansion, enforced expression in mouse bone marrow cells caused GM proliferation in a manner reminiscent of LPS treatment. However, the miR-155–induced GM populations displayed pathological features characteristic of myeloid neoplasia. Of possible relevance to human disease, miR-155 was found to be overexpressed in the bone marrow of patients with certain subtypes of acute myeloid leukemia (AML). Furthermore, miR-155 repressed a subset of genes implicated in hematopoietic development and disease. These data implicate miR-155 as a contributor to physiological GM expansion during inflammation and to certain pathological features associated with AML, emphasizing the importance of proper miR-155 regulation in developing myeloid cells during times of inflammatory stress.
Mammalian hematopoiesis involves the generation of blood cells from a common hematopoietic stem cell (HSC) through many intermediate stages, each of which can give rise to various types of malignancies upon their dysregulation. However, the molecular mechanisms that govern this process are incompletely understood. In particular, the quantitative decisions regarding how many cells take which pathway of maturation remain obscure. Most studies of this process have focused on cytokines and transcription factors, which can control cellular proliferation and differentiation decisions (1). MicroRNAs (miRNAs) are a novel class of small, regulatory RNA molecules that play evolutionarily conserved roles in cellular development and function, and mediate target gene repression through 3′ untranslated region (UTR) interactions (2–4). Recently, a growing body of evidence has implicated specific miRNAs in the modulation of mammalian hematopoiesis during both physiological and pathological conditions (5, 6).
Among miRNAs expressed by hematopoietic cells, miR-155 has emerged as having significant impact on the biology of lymphocytes (7–9). MiR-155 is up-regulated to high levels in response to B or T cell receptor engagement (10, 11) and plays a B cell–intrinsic role in germinal center formation and subsequent antibody production in vivo after antigen challenge (8, 9). Furthermore, miR-155–deficient T cells exhibit a Th2 bias, likely through repression of c-Maf. Beyond its apparent contribution to the humoral immune response, the need to properly regulate miR-155 levels is suggested by its dramatically elevated expression in several types of human B cell lymphomas (11–13), and its reported sufficiency in triggering B cell lymphoma when overexpressed in a B cell–restricted manner in mice (7).
In contrast to the emerging picture of miR-155 functions in lymphocytes, the role or consequences of miR-155 expression in hematopoietic cells of myeloid origin have been largely uncharacterized. Our group recently reported high levels of miR-155 expression in cells of the innate immune system, such as monocytes and macrophages, after exposure to inflammatory stimuli (14, 15). As in lymphocytes, miR-155 expression is tightly regulated in myeloid cells, suggesting a specialized function during times of inflammatory stress. Interestingly, the inflammatory process is known to have a significant impact on hematopoiesis by enhancing production of granulocyte/monocyte (GM) populations to replenish those that become depleted while combating infection (16, 17). This developmental reprogramming is driven in part by cytokines and growth factors produced during inflammation, and by direct recognition of pathogen-associated molecular patterns, such as LPS by mammalian Toll-like receptors expressed on hematopoietic stem and progenitor cell populations (18). Because miR-155 is part of the Toll-like receptor–induced gene program, we have examined its potential role in regulating GM expansion in the bone marrow during inflammation.
In this study, we demonstrate that miR-155 expression is greatly increased in mouse bone marrow cells after LPS injection and is sufficient to drive GM expansion when constitutively expressed in mouse HSCs in vivo. However, sustained expression of miR-155 also leads to several features characteristic of pathological myeloid proliferations, correlating with its overexpression in samples from human acute myeloid leukemia (AML) patients. Finally, miR-155 directly repressed a broad range of target mRNAs implicated in myeloid hyperplasia and/or hematopoiesis. These data suggest an important physiological role for miR-155 in GM expansion during times of inflammation, yet underscore the importance of its proper regulation for maintaining the balance between an efficient immune response and the potential for inducing malignant disease.
RESULTS
LPS induces bone marrow expression of miR-155 before myeloid expansion in vivo
Although miR-155 is expressed at low levels in mice under steady-state conditions, we examined whether its expression is elevated in the bone marrow compartment after the onset of inflammation in vivo, as seen in cultured macrophages (14). Mice were injected i.p. with a sublethal dose of LPS (50 μg) or PBS control, and their bone marrow cells were analyzed for miR-155 expression. Strong induction of miR-155 levels were observed after 24 h of LPS treatment, which returned to control levels by 72 h (Fig. 1 A). Up-regulated miR-155 expression was also detected upon direct LPS or GM-CSF stimulation of isolated bone marrow cells from WT or Rag1−/− mice, demonstrating that cells other than mature B and T lymphocytes contribute to this response (Fig. 1 B). Furthermore, both populations enriched in mature cells (Mac-1, B220, and Ter-119 positive), and those enriched in immature cells (Mac-1, B220, and Ter-119 low to negative) responded to LPS by up-regulating miR-155, with the immature population having a distinctly stronger response than the mature (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20072108/DC1).
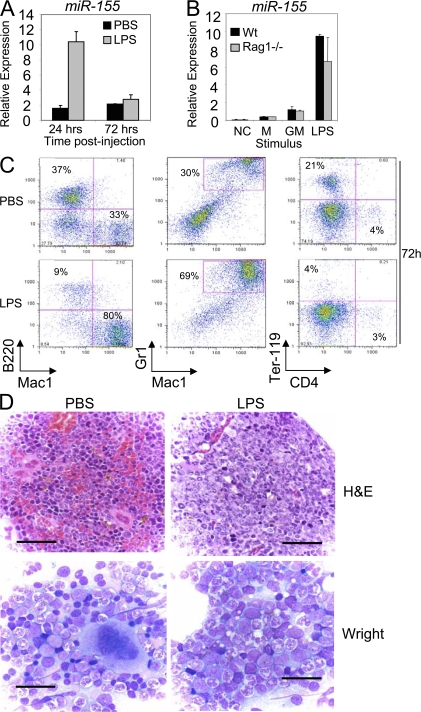
Figure 1.
LPS treatment induces bone marrow expression of miR-155 before GM expansion. (A) WT mice (n = 3 per group) were injected i.p. with 50 μg LPS dissolved in PBS or PBS alone. RNA was collected from total bone marrow cells, and miR-155 expression was assayed by quantitative PCR (mean ± SD). (B) Bone marrow was flushed out of the femurs and tibias of WT and Rag1−/− mice (n = 3 per group), stimulated with 100 ng/ml LPS, 100 ng/ml GM-CSF (GM), or medium (M) for 24 h, and RNA was then assayed for miR-155 expression levels (mean ± SD). NC, no template control. (C) BM cells collected from mice in A at the 72-h time point were stained with antibodies against Mac1, Gr1, B220, Ter-119, or CD4 and analyzed by FACS. (D) Wright-stained bone marrow smears (bottom; bar, 25 μM) or hematoxylin and eosin–stained bone marrow sections (top; bar, 60 μM) from WT mice injected with LPS or PBS for 72 h.
In addition to miR-155 expression, we also monitored bone marrow cell dynamics in response to LPS in vivo. Although there was little change in bone marrow GM (Mac1+ Gr1+), B cell (B220+), and erythroid precursor (Ter-119+) populations by 24 h after LPS treatment (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20072108/DC1), substantial expansion of GM cells and reductions in B cells and erythroid precursors were evident by 72 h (Fig. 1 C), consistent with a previous study (17). Histological analyses also showed myeloid preponderance and hyperplasia, with relative erythroid hypoplasia, after 72 h of LPS treatment (Fig. 1 D). Collectively, these data indicate that LPS-induced miR-155 expression in the bone marrow precedes GM cell expansion.
Enforced expression of miR-155 in HSCs causes a myeloproliferative disorder in the bone marrow
We next investigated whether miR-155 is sufficient to mediate GM expansion in the mouse bone marrow in vivo. Retroviral-mediated gene transfer was used to force expression of GFP and miR-155 in HSCs (Fig. 2 A), followed by engraftment of these cells into lethally irradiated C57BL6 mouse recipients. By 2 mo after reconstitution, mice were killed and coexpression of miR-155 and GFP was detected in various hematopoietic tissues, including the bone marrow (Fig. 2 B), thymus, spleen, and lymph nodes (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20072108/DC1). Control mice only expressed GFP but not miR-155.
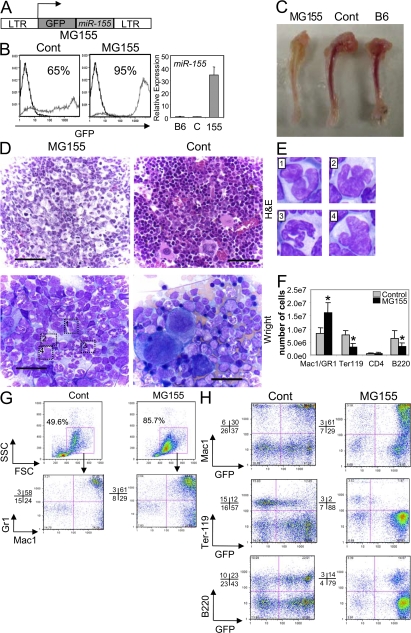
Figure 2.
Expression of miR-155 in HSCs causes a myeloproliferative disorder in the bone marrow. (A) Depiction of the retroviral construct used to enable both miR-155 and GFP expression in HSCs. (B) Bone marrow cells of mice reconstituted with MG155- or control vector–transduced HSCs were analyzed for their expression of GFP by FACS, where the percentage of GFP+ cells is indicated. Black line, C57BL6 control; gray line, MG155 or control vector. Cells from the same compartments were analyzed for miR-155 expression using quantitative PCR (mean ± SD). (C) Tibias were removed from mice reconstituted with MG155 or control vector HSCs for 2 mo, or untreated C57BL6 (B6) mice, and photographs were taken. (D) Hematoxylin and eosin–stained bone marrow sections from miR-155–expressing or control mice (top; bar, 60 μM). Wright-stained bone marrow smears from mice reconstituted with MG155 or control vector HSCs (bottom; bar, 25 μm). (E) Examples of dysplastic myeloid cells observed in miR-155–expressing bone marrow are enlarged. (F) Number of specified cell types found in the bone marrow (one femur plus one tibia) of mice reconstituted with MG155 or control vector HSCs (mean ± SD). (G) GFP-gated bone marrow cells from mice reconstituted with MG155 or control vector HSCs were analyzed for FSC and SSC counts and expression of Mac1 and Gr1. (H) Bone marrow cells from mice reconstituted with MG155 or control vector HSCs were analyzed for expression of Mac1, Ter-119, or B220 on both GFP+ and GFP− cells by FACS. Data represent at least six independent animals in each group, and p-values (*) of <0.05 were considered significant after a Student's two-tailed t test.
Gross analyses of femurs and tibias from mice expressing miR-155 revealed a white-tan bone marrow coloration unlike the vibrant red seen in controls (Fig. 2 C). Upon microscopic inspection of hematoxylin and eosin–stained bone marrow sections and Wright-stained bone marrow smears (Fig. 2 D), miR-155–expressing bone marrow was dominated by GM cells at a variety of either normal or abnormal developmental stages based upon their morphology. Indeed, many of the cells that appeared to be granulocytic precursors showed irregular segmentation of their nuclei and lacked condensation of nuclear chromatin (Fig. 2 E). Conversely, miR-155 expression also led to a reduction in erythrocytes, megakaryocytes, and lymphocytes in the bone marrow (Fig. 2 D).
Flow cytometry identified approximately twice as many Mac1+ GR1+ GM cells, very few Ter-119+ erythroid precursors, and a reduction in B220+ B cells in the bone marrow of mice expressing miR-155 versus the control vector (Fig. 2 F). When gated on GFP+ cells (expressing miR-155), there was a dramatic increase in large granular cells, as defined by having high forward scatter (FSC) and side scatter (SSC), respectively (Fig. 2 G). Back-gating confirmed that these cells were Mac1+, with a majority also positive for Gr1. Furthermore, the cells responsible for the overall GM, B, and erythroid precursor differences were largely GFP+ (Fig. 2 H). These observations reveal profound myeloid proliferation with dysplastic changes in the bone marrow of mice expressing miR-155 compared with controls.
MiR-155 expression in HSCs causes splenomegaly and extramedullary hematopoiesis
Splenomegaly was observed in miR-155–expressing compared with control mice (Fig. 3 A). Hematoxylin and eosin staining of splenic sections from miR-155–expressing mice revealed expanded interfollicular regions containing various hematopoietic elements, as well as constricted and disrupted B cell follicles compared with control spleens (Fig. 3 B). Upon analyses of Wright-stained splenic touch preparations, we observed a large number of erythroid precursors, megakaryocytes, and some developing GM cells in the spleens of miR-155–expressing mice, whereas very few of these cell types were observed in control spleens (Fig. 3 B).
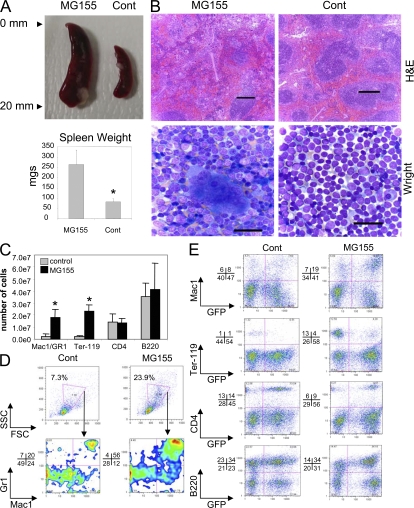
Figure 3.
Expression of miR-155 in HSCs triggers extramedullary hematopoiesis in the spleen. (A) Spleens were removed from mice reconstituted with MG155 or control vector HSCs for 2 mo, and photographs were taken (top). Spleen weight was also determined in the two groups (bottom; mean ± SD). (B) Hematoxylin and eosin–stained sections (top; bar, 200 μm) or Wright-stained touch preps (bottom; bar, 25 μm) from mice reconstituted with MG155 or control vector HSCs. (C) Number of specified cell types found in the spleens of mice reconstituted with MG155 or control vector HSCs (mean ± SD). (D) GFP-gated spleen cells from mice reconstituted with MG155 or control vector HSCs were analyzed for FSC and SSC counts and expression of Mac1 and Gr1. (E) Splenocytes from mice reconstituted with MG155 or control vector HSCs were analyzed for expression of Mac1, Ter-119, CD4, or B220 on GFP+ and GFP− cells by FACS. Data represent at least six independent animals in each group, and p-values (*) of <0.05 were considered significant using a Student's two-tailed t test.
FACS analyses corroborated these observations. We saw elevated numbers of Mac1+ Gr1+ myeloid cells and Ter-119+ erythroid cells, with little change in CD4+ T cells and B220+ B cells in miR-155–expressing compared with control spleens (Fig. 3 C). When gated on GFP+ cells (expressing miR-155), there were elevated numbers of large granular cells, as defined by having high FSC and SSC, respectively, with a majority coexpressing Mac1 and Gr1 (Fig. 3 D). Furthermore, miR-155–expressing splenocytes contained overall higher numbers of Mac1+ cells that expressed GFP compared with controls (Fig. 3 E). Conversely, the Ter-119+ cell population from miR-155–expressing spleens was largely negative for GFP, possibly arising from nontransduced HSCs. These results clearly demonstrate the presence of splenic extramedullary hematopoiesis in miR-155–expressing mice, likely compensating for the bone marrow defects.
Expression of miR-155 in HSCs perturbs peripheral blood cell populations
Consistent with the disrupted hematopoiesis observed in miR-155–expressing mice, their peripheral blood demonstrated several distinct abnormalities compared with controls. By 2 mo after reconstitution, FACS detected significantly elevated numbers of Mac1+ cells (Fig. 4 A), and Wright-stained blood smears revealed the presence of morphologically abnormal GM cells in miR-155–expressing mice (Fig. 4 B). Complete blood cell counts showed a significant reduction in red blood cell, hemoglobin, and platelet levels (Fig. 4 C), FACS found decreased B220+ B cells and CD4+ T lymphocytes (Fig. 4 C), and Wright staining identified several immature erythrocytes demonstrating polychromatophilia in miR-155–expressing animals (Fig. 4 D).
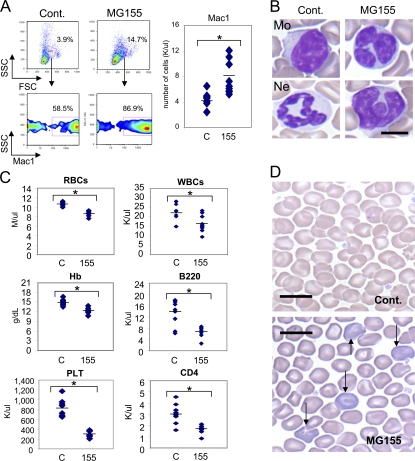
Figure 4.
Expression of miR-155 in HSCs perturbs peripheral blood cell populations. (A) Peripheral blood was collected from mice reconstituted with MG155 or control vector HSCs for 2 mo and analyzed by FACS to determine FSC and SSC counts and expression of Mac1. The total number of Mac1 cells was also determined. (B) Photomicrographs of a normal Wright-stained monocyte (Mo) and neutrophil (Ne) from the blood of mice reconstituted with control vector HSCs, and two examples of the Wright-stained irregular myeloid cells found in MG155 HSC-reconstituted animals (bar, 5 μm). (C) RBC, hemoglobin (Hb), platelet, white blood cell, B220 B cell, and CD4 T cell levels in the blood of mice reconstituted with MG155 or control vector HSCs. (D) Microscopic photographs of Wright-stained peripheral blood RBCs from mice reconstituted with MG155 or control vector HSCs (bar, 10 μm). Data represent at least nine independent animals in each group, and p-values (*) of <0.05 were considered significant after a Student's two-tailed t test.
A subset of human AML patients overexpress miR-155
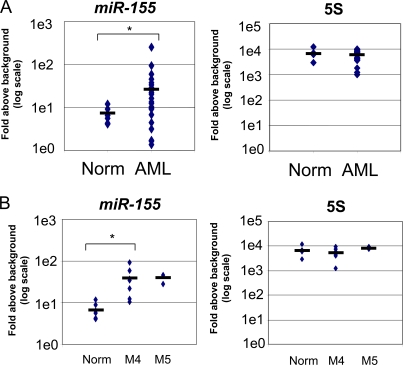
Several of the pathological features observed in miR-155–expressing mice are associated with human myeloid malignancies, including AML. Therefore, bone marrow samples from 24 AML patients and 6 healthy donors were assayed for miR-155 and 5S expression levels by quantitative PCR. On average, the AML samples significantly overexpressed miR-155 compared with healthy donors, with a level approximately four and a half times higher (Fig. 5 A). A few AML samples had miR-155 levels that were lower than the normal samples, whereas the overall AML sample distribution had a wide variance. In contrast, no significant difference in the average expression levels of 5S RNA was observed between the groups (Fig. 5 A). MiR-155 levels in different subtypes of AML were next ascertained using the World Health Organization (WHO) classification system. Patients with acute myelomonocytic leukemia and acute monocytic leukemia, corresponding to FAB-AML-M4 and FAB-AML-M5, respectively, exhibited significant overexpression of miR-155 compared with normal samples (Fig. 5 B). These observations demonstrate that miR-155 expression in the bone marrow is significantly elevated in a subset of patients suffering from AML.
Figure 5.
Overexpression of miR-155 in a subset of human AML patients. (A) RNA was collected from the bone marrow of 6 normal patients and 24 patients diagnosed with AML. MiR-155 (left) and 5S RNA (right) expression levels were assessed using quantitative PCR. (B) MiR-155 and 5S RNA expression data were compared between normal subjects and AML patients of the FAB subtypes M4 and M5. Group differences were considered statistically significant when the p-value (*) was <0.05.
MiR-155 can directly repress genes implicated in hematopoietic development and disease
MiRNAs exert their biological functions through the degradation and/or translational repression of target mRNAs. To identify miR-155 target genes that may be involved in the observed myeloproliferative phenotype, we first transduced RAW 264.7 myeloid cells with a miR-155–expressing retrovirus that increased mature miR-155 cellular levels within twofold of those observed after LPS stimulation (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20072108/DC1). A mRNA microarray analysis was next performed on RNA samples collected from miR-155–expressing and control cells to identify genes regulated by miR-155 (Fig. S5). Some 1,080 transcripts were down-regulated >1.2-fold with a p-value of <0.08, and 89 of the repressed mRNAs contained conserved (human and mouse) miR-155 binding sites with 7- or 8-mer seeds in their 3′ UTRs according to published lists of computationally predicted target genes found on the Targetscan 4.0 website (19, 20). Finally, genes with reported roles in myeloid hyperplasia and/or hematopoiesis were identified through literature searching. Using these criteria, our attention was drawn to 10 candidate targets: Bach1, Sla, Cutl1, Csf1r, Jarid2, Cebpβ, PU.1, Arntl, Hif1α, and Picalm (Fig. 6 A). To confirm the microarray results, quantitative PCR was performed using gene-specific primers. It showed that the mRNAs encoding these proteins were down-regulated ∼20–70% in RAW 264.7 cells expressing miR-155 versus empty vector control (Fig. 6 A). We also observed repression of Cebpb, PU.1, Cutl1, and Picalm protein levels in RAW 264.7 cells expressing miR-155 (Fig. 6 B), albeit to somewhat varying degrees between experiments (not depicted).
Figure 6.
Repression of specific target genes involved in myeloid hyperplasia and/or hematopoiesis by miR-155. (A) Messenger RNA from Raw 264.7 cells infected with MSCVpuro-155 or empty vector control was subjected to a microarray analysis, and results indicate expression changes mediated by miR-155. The intensities of red and green correlate with increased or decreased mRNA levels, respectively, and numerical repression values for each mRNA are listed. RNA from the same cell types were converted to cDNA and used to assay expression of these genes by quantitative PCR (mean ± SD). All values have been normalized to L32 mRNA levels, are displayed as percent expression of control, and are the average of three independent experiments. (B) Western blotting was performed to assay Cebpb, PU.1, Cutl1, Picalm, or αTubulin using extract from Raw 264.7 cells stably expressing miR-155 or empty vector, and data are representative of at least three independent experiments (left and middle). Expression of miR-155 in Raw 264.7 cells infected with MSCVpuro-155 or empty vector control was assayed by Northern blotting to ensure proper expression of mature miR-155 (right).
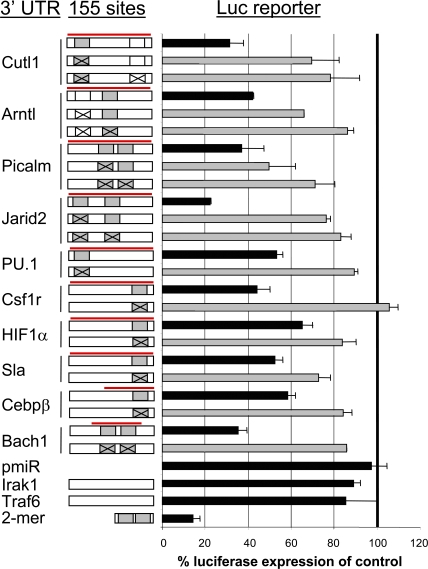
Next, we tested whether miR-155 could directly repress the identified mRNA targets through 3′ UTR interactions. Each full-length 3′ UTR, or in two cases (Bach1 and Cebpβ) the region of the UTR containing the miR-155 binding site(s), was cloned into a reporter vector downstream from luciferase. These vectors were then used to assess whether miR-155 could repress luciferase gene expression in 293T cells. Luciferase expression was repressed between 35 and 78% depending on the 3′ UTR tested (Fig. 7). There was even a rough correlation between the quantitative PCR results in RAW 264.7 cells and the luciferase repression in 293T cells. To demonstrate a direct interaction between miR-155 and the 3′ UTRs tested, we systematically mutated each conserved miR-155 7– or 8-mer seed region and found that a majority of the miR-155–mediated repression was abolished (Fig. 7). As a control, miR-155 repressed a reporter construct containing tandem miR-155 sites ∼80%. However, luciferase levels were relatively unaffected when the Traf6 or Irak1 3′ UTRs were tested, consistent with their lack of miR-155 binding sites (Fig. 7). These results provide strong evidence that miR-155 can directly regulate several genes with relevance to hematopoiesis and the myeloproliferative phenotype.
Figure 7.
MiR-155 repression of specific target genes occurs through direct 3′ UTR interactions. (A) The 3′ UTR regions from identified miR-155 target mRNAs containing miR-155 binding site(s) with conserved 7- or 8-mer seeds (gray boxes), nonconserved 7-mer seed (Arntl, white box), or conserved 6-mer seed (Cutl1, white box) were cloned downstream from luciferase (pmiReport vector). Mutations to these specific seed regions are marked with an X. The region of the 3′ UTR cloned is designated with a red line, and the cartoon schematics of the UTRs are not drawn to scale. These constructs were used for reporter assays in 293T cells by cotransfection with a control β-galactosidase expression plasmid and a miR-155 expression vector (FUW-155) or empty vector control (FUW). A positive control vector contained tandem miR-155 binding sites, whereas negative controls contained no 3′ UTR or the 3′ UTR from Irak1 or Traf6, which lack miR-155 sites. Data using WT 3′ UTRs are in black, and mutant UTRs are in gray. All luciferase values have been normalized to β-galactosidase and are represented as percent luciferase expression of control (mean ± SD). All data are a triplicate set representing at least three independent experiments.
DISCUSSION
As cells of the innate immune system combat infectious pathogens, their numbers are often depleted and must be replenished. This process is characterized by an increased production of GM populations, a response shown to involve GM-CSF– and LPS-mediated signaling events (17, 18, 21). Our present findings demonstrate that miR-155 is induced by GM-CSF and LPS in the bone marrow compartment and is sufficient to increase the relative and absolute numbers of GM cells when expressed in HSCs and throughout hematopoietic development. This expansion appears to be at the expense of B lymphocytes and erythroid precursors in the bone marrow, as also observed after LPS treatment. Thus, miR-155 may play a regulatory role in hematopoietic cell fates during times of inflammatory stress when factors like LPS or GM-CSF are present. Because stimulation of most Toll-like receptors induces miR-155 in macrophages (14), our results with LPS can probably be generalized to many conditions of microbial invasion.
It remains unclear whether miR-155 participates in hematopoiesis under steady-state conditions. A recent study detected miR-155 expression in human CD34+ cells, a population containing HSCs and early progenitors of lymphoid and myeloid lineages (5). However, defective myeloid populations in miR-155–deficient mice were not observed when analyzed under steady-state (non-inflammatory) conditions (8).
Whether miR-155 is only sufficient or in fact required for increased myelopoiesis during inflammation, our findings demonstrate that its unregulated expression triggers a myeloproliferative disorder, exhibiting many preleukemic aspects. Because frank myeloid leukemia was not observed in any of our mice analyzed within 2 mo of reconstitution with miR-155-expressing HSCs, such a transition may require additional mutations. However, the relevance of our observed phenotype in mice is substantiated by the elevated expression levels of miR-155 seen in human patients with AML. Interestingly, the two AML subgroups found to be overexpressing miR-155 are characterized as myelomonocytic (M4) and monocytic (M5), both thought to be derived from cells demonstrating aspects of GM cell differentiation, similar to the expanded GM cells in our miR-155–expressing mice. It is of note that miR-155 may also be elevated in other subtypes of AML where we did not have enough samples to make such a conclusion. If miR-155 does prove to be dysregulated in specific subtypes of AML, it might complement the recent finding that miR-181 expression positively correlates with M1 and M2 subtypes of AML, but not M4 or M5 (22). Furthermore, as the full spectrum of miRNAs that become dysregulated during AML is defined, these small RNAs may prove to have utility as diagnostic indicators of AML subtypes.
Despite the similarities mentioned above, certain aspects of the miR-155–induced myeloproliferative phenotype were not observed during the 3-d analysis period after LPS-mediated acute inflammation, such as GM dysplasia, peripheral blood leukopenia and polychromatophilic RBCs, and reduced megakaryocyte and platelet levels (not depicted). These observations may reflect a differential effect of sustained miR-155 expression in cell types that require strictly regulated levels of this miRNA. However, chronic inflammation (which may also sustain high miR-155 expression levels) might trigger some of these pathological events given enough time. For instance, after months of polymicrobial sepsis in mice, there is reported extramedullary hematopoiesis in the spleen and significantly increased numbers of morphologically heterogeneous GM cells in both the spleen and bone marrow compartments (23). The inflammatory response in the bone marrow involves profound myeloid proliferation and, through factors such as miR-155, may prove to create a microenvironment suitable for cancer formation and development if not resolved in a timely manner.
Unlike a previous report, which found that B cell–restricted transgenic expression of miR-155 triggers B cell lymphoma in mice (7), we did not observe a B cell malignancy in our model. This may be explained by differences in the systems used because our model allows for miR-155 expression beginning in adult HSCs, which precedes formation of pro–B cells during hematopoietic development (1). These observations suggest that miR-155 may trigger unique phenotypes when expressed at different stages or in distinct cell type(s) during hematopoiesis. There is also evidence that developing B cells and GMs may occupy an overlapping bone marrow niche. Based upon our current findings, miR-155 expression may allow for GM progenitors to dominate this compartment and inhibit B cell development, which has been proposed to occur during inflammation (17). This mechanism might also block events required for miR-155–dependent B cell transformation.
In an effort to explore the mechanistic basis for the myeloproliferative phenotype caused by HSC expression of miR-155, we identified several mRNA targets that were directly repressed by miR-155 according to 3′ UTR reporter assays. Of note, the reduced expression or altered function of some of these targets has been linked to AML, as in the case of PU.1 and Picalm (24, 25), or myeloproliferative conditions, as is true for Cutl1 and Csf1r (26, 27). Other identified targets have been implicated in control of various aspects of hematopoiesis involving many of the cell types that are perturbed in mice expressing miR-155 in HSCs. These include Cebpb (28), Bach1 (29), Arntl (30), Sla (31), Jarid2 (32), and Hif1α (33). Thus, miR-155 could mediate its overall biological affects, both physiological and pathological, through the combinatorial repression of a broad range of targets in a variety of cell types. Such a multi-target regulation has recently been described for T cell receptor signaling (34). Therefore, it is possible that complete rescue of this phenotype will not be achieved through replacing any one of the specific miR-155 target genes. However, the specific spatial and temporal contributions of individual targets to the myeloproliferative phenotype and AML in the context of miR-155 repression remain an area for future investigation.
There is emerging evidence that individual miRNAs are part of a more complex regulatory network involving other miRNAs and transcriptional regulators that cooperate to govern myelopoiesis. For example, Csf1r is important for monocyte development and has recently been reported to be regulated indirectly by miRNAs 17-5p-20a-106a (26). miRNAs 17-5p-20a-106a repress the transcriptional regulator AML1 required for Csf1r transcription. Therefore, both miRNAs 17-5p-20a-106a and miR-155 can influence Csf1r expression through different mechanisms. In the case of miR-155 targets PU.1 and Cebpb, they have been shown to transcriptionally regulate expression of myeloid-specific miR-223 (35). miR-223 is subsequently involved in unleashing CEBPα function, a central transcription factor in myelopoiesis, through the direct repression of its inhibitor, NFI-A (36). Such dynamic systems require appropriate miRNA expression levels and kinetics to carefully orchestrate hematopoietic development, as has been recently described during T cell development in the thymus (37). However, it is easy to see how this delicate process would be vulnerable to dysregulated miRNA expression leading to pathological outcomes. This concept is exemplified by the dysplastic features observed in many GM cells from our miR-155–expressing mice. It is possible that although initial miR-155 expression expands GM numbers, its timely down-regulation is necessary for these cells to complete their developmental programs. Such a model would be consistent with the transient expression of miR-155 that precedes expansion of morphologically normal GM populations in the bone marrow after LPS treatment.
Based upon our current study, miR-155 appears to play a role in promoting GM cell expansion during inflammatory responses while initiating pathological processes under forced expression. Due to the enhanced expression of miR-155 in a subset of AML patients, and its ability to repress several genes relevant to myeloid malignancies, therapeutic targeting of miR-155 with such agents as antagomirs may provide a beneficial option (38). Because miR-155 knockout mice display few detrimental phenotypes in the absence of infection (8), decreasing miR-155 function in human patients suffering from myeloid, lymphoid, or other malignancies correlated with enhanced miR-155 expression may provide more benefits than harm.
MATERIALS AND METHODS
Cell culture and reagents.
RAW 264.7 and 293T cells were both cultured in complete DMEM containing 10% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin with 5% C02. Mouse bone marrow–derived macrophages were made using M-CSF–containing media. LPS from Escherichia coli strain 055:B5 was purchased from Sigma-Aldrich, and recombinant mouse GM-CSF was purchased from eBioscience.
DNA constructs.
A miR-155 expression cassette containing the human miR-155 hairpin sequence and flanking regions was cloned from a B cell cDNA library into pcDNA3 as described previously (12). This cassette was subcloned into pMSCVpuro, FUW, or pMG. pMG155 is a modified MSCV vector whereby GFP was placed downstream from the 5′ LTR, and the miR-155 expression cassette was cloned downstream from the GFP stop codon (detailed cloning strategy available upon request). For reporter assays, the 3′UTRs of the respective mRNAs were cloned into pmiReport (Ambion) after amplification from a mouse macrophage cDNA library. Primer sequences are described in Table S1, which is available at http://www.jem.org/cgi/content/full/jem.20072108/DC1. The Bach1 3′ UTR region was amplified from a human B cell library. Site-directed mutagenesis was used to change specific nucleotides found within the miR-155 seed regions (Table S2). The 2-mer control insert consists of a tandem repeat of the complimentary sequence to the mature mouse miR-155 sequence. Cloning of the TRAF6 and IRAK1 3′ UTR into pmiReport was described previously (15).
Mice.
WT C57BL6 mice were purchased from The Jackson Laboratory, and Rag1−/− mice were bred in-house. All experiments involved female mice and were performed according to IACUC-approved protocols.
Retroviral infections, stable cell lines, and bone marrow reconstitution.
To generate VsVg-pseudotyped retroviruses containing the miR-155 expression cassette, 2 × 106 293T cells were transfected with pMSCVpuro–miR-155, pGag-Pol, and pVsVg using a standard calcium phosphate protocol. After 48 h, viral supernatant was harvested and used to infect 5 × 105 RAW 264.7 cells for 8 h in the presence of polybrene at 10 μg/ml. After 48 h, stably transduced cells were selected using puromycin at 7 μg/ml for 7–10 d, and miR-155 expression was assessed at the same time as experiments were performed by Northern blotting or quantitative PCR for all batches made.
To obtain HSC-enriched bone marrow cells, mice were injected i.p. with 5 μg 5-fluorouracil for 5 d before bone marrow harvest (39). Cells were collected from the bone marrow, and RBCs were removed using an RBC lysis solution (Invitrogen). Cells were cultured for 24 h in 20 ng/ml IL-3, 50 ng/ml IL-6, and 50 ng/ml SCF (all from eBioscience) containing complete RPMI (10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin, and 50 μM β-marcaptoethanol) before initial retroviral infection. To generate retroviruses for infecting HSC-enriched bone marrow cells, 293T cells were transfected with pMG155 and pCL-Eco. After 48 h, 8 μg/ml polybrene was added to culture supernatant–containing retroviruses, and this was used to spin-infect 106 HSC-enriched cells per donor for 1.5 h at 2,500 RPM and 30°C. This procedure was repeated three times once daily, followed by injection of 106 retrovirally infected HSC-enriched cells per lethally irradiated (1,100 rads from Cesium 137 source at 50 rads/minute) recipient. Recipients were maintained on Septra throughout the reconstitution period.
RNA quantification.
Northern blotting and quantitative PCR were used to assay miR-155 and other mRNAs as described previously (14). Gene-specific primer sequences used for quantitative PCR are shown in Table S3. For the microarray study, total RNA was collected from five RAW 264.7 stably infected clones expressing miR-155 or empty vector using the RNeasy Mini kit per the manufacturer's instructions (QIAGEN). The Affymetrix Mouse Genome 430 2.0 microarray analysis was then performed using pooled RNA from each group by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech according to detailed protocols (http://affymetrix.com/products/arrays/specific/mouse430_2.affx). Data were analyzed using Rosetta Resolver Software. Microarray data were deposited in the GEO database under accession number GSE10467.
Western blotting.
Western blotting was performed using standard protocols and the following antibody clones from Santa Cruz Biotechnology, Inc.: Cebpb (C-19), PU.1 (T-21), Cutl1 (M-222), Picalm (C-18), and αTubulin (AA12). Protein expression differences were determined using Scion Image software.
Flow cytometry and cell separation.
Fluorophor-conjugated monoclonal antibodies specific for either Mac1, Gr1, Ter-119, B220, and CD4 (all from eBioscience) were used in various combinations to stain RBC-depleted splenocytes, bone marrow, or peripheral blood mononuclear cells that were fixed after washing using paraformaldehyde (1% final). Stained cells were assayed using a BD FACSCalibur flow cytometer and further analyzed with FloJo software. Cell separation was performed using biotinylated monoclonal antibodies against Mac-1, Ter-119, and B220 (eBioscience), streptavidin-conjugated magnetic beads (Miltenyi Biotec), and MACS LS Separation Columns (Miltenyi Biotec).
Luciferase reporter assays.
8 × 104 293T cells were plated in DMEM media containing 5% FBS for 18 h, followed by transfection of relevant plasmids using lipofectamine (Invitrogen) per the manufacturer's instructions. Luciferase assays were performed 48 h later using a dual luciferase kit (Promega). A β-gal expression plasmid was cotransfected and β-gal levels were assayed and used to normalize the luciferase values.
Human AML sample collection and analysis.
Bone marrow biopsy samples collected from patients with AML were flash-frozen and stored at −80°C after the completion of diagnostic work in a tissue bank at University of California, Los Angeles. For this study, 24 samples were rapidly thawed and subjected to TRIzol purification of RNA. In addition, six RNA samples were isolated from healthy donors. AML cases were categorized according to the WHO “Classification of Tumors” using anonymous clinical reports. All work performed on these tissues was approved by the Institutional Review Board at UCLA.
Morphological assessment of hematolymphoid tissues.
For histological sectioning, organs were placed into 10% neutral-buffered formalin immediately after necropsy, fixed for 12–18 h, washed, and transferred to 70% ethanol before standard paraffin embedding, sectioning, and staining with hematoxylin and eosin. Bones were also decalcified. For cytological assessment, touch preparations of the cut surface of the spleen were performed. Peripheral blood smears were obtained from tail vein bleeds or from the heart at necropsy. Bone marrow smears were prepared from extracted bone marrow of reconstituted mice. All cytological preparations were air-dried and stained with Wright's stain. Both histological and cytological preparations were examined on an Olympus BX-51 microscope and photographed using a Spot Digital Camera and software. Complete blood cell counts were performed at UCLA's Department of Laboratory Animal Medicine.
Statistical tests.
All statistical analyses were performed using Microsoft Excel statistical software module. For patient samples, an F-test determined that the distributions of miR-155 expression in normal samples versus AML samples were heteroscedastic (P = 4.5 × 10−6 for F-test). Similarly, the distributions of miR-155 expression in normal versus AML-M4 was determined to be heteroscedastic (P = 1.8 × 10−5 for F-test). After this, a two-tailed t test was performed assuming heteroscedastic distributions for both comparisons. For all other data, a Student's two-tailed t test was used.
Online supplemental material.
Fig. S1 demonstrates that both immature and mature cell-enriched bone marrow populations up-regulate miR-155 in response to LPS. Fig. S2 provides FACS data showing the percentage of Mac1+, Gr1+, B220+, Ter-119+, or CD4+ cells in the bone marrow after 24 h of LPS versus PBS treatment. Fig. S3 presents the coexpression of miR-155 and GFP in several different immune organs from reconstituted mice. Fig. S4 compares miR-155 expression in Raw 264.7 cells and primary macrophages mediated by retroviral overexpression versus LPS stimulation. Fig. S5 outlines the scheme used to identify miR-155 targets with relevance to hematopoiesis. Table S1 provides the primer sequences used for cloning different 3′ UTRs into pmiReport. Table S2 provides sequence information regarding mutations introduced into the 3′ UTRs used for reporter assays. Table S3 provides the primer sequences used to assay miR-155 target mRNAs by quantitative PCR. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20072108/DC1.
Supplemental Material
Acknowledgments
We thank Jose Luis Riechmann, Vijaya Rao, Jaclyn Shingara, and David Brown for help with microarray work.
R.M. O'Connell was funded by the Irvington Institute Fellowship Program of the Cancer Research Institute. D.S. Rao was funded by the American Society of Hematology Basic Science Fellow Scholar Award. A.A. Chaudhuri was funded by the Howard Hughes Medical Institute's Medical Research Training Fellow Award. Part of this work was also funded by National Institutes of Health (NIH) grants. This work was supported by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology and by NIH Grants.
D. Baltimore is a scientific advisor to Regulus, a company devoted to microRNA therapeutics. All other authors have no conflicting financial interests.
Abbreviations used: AML, acute myeloid leukemia; FSC, forward scatter; GM, granulocyte/monocyte; HSC, hematopoietic stem cell; miRNA, microRNA; SSC, side scatter; UTR, untranslated region.
References
- 1.Rosenbauer, F., and D.G. Tenen. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V. 2004. The functions of animal microRNAs. Nature. 431:350–355. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, D.P., and C.Z. Chen. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5:396–400. [DOI] [PubMed] [Google Scholar]
- 4.He, L., and G.J. Hannon. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5:522–531. [DOI] [PubMed] [Google Scholar]
- 5.Georgantas, R.W. III, R. Hildreth, S. Morisot, J. Alder, C.G. Liu, S. Heimfeld, G.A. Calin, C.M. Croce, and C.I. Civin. 2007. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA. 104:2750–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kluiver, J., B.J. Kroesen, S. Poppema, and A. van den Berg. 2006. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 20:1931–1936. [DOI] [PubMed] [Google Scholar]
- 7.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C.M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 103:7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez, A., E. Vigorito, S. Clare, M.V. Warren, P. Couttet, D.R. Soond, S. van Dongen, R.J. Grocock, P.P. Das, E.A. Miska, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thai, T.H., D.P. Calado, S. Casola, K.M. Ansel, C. Xiao, Y. Xue, A. Murphy, D. Frendewey, D. Valenzuela, J.L. Kutok, et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608. [DOI] [PubMed] [Google Scholar]
- 10.Haasch, D., Y.W. Chen, R.M. Reilly, X.G. Chiou, S. Koterski, M.L. Smith, P. Kroeger, K. McWeeny, D.N. Halbert, K.W. Mollison, et al. 2002. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217:78–86. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg, A., B.J. Kroesen, K. Kooistra, D. de Jong, J. Briggs, T. Blokzijl, S. Jacobs, J. Kluiver, A. Diepstra, E. Maggio, and S. Poppema. 2003. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 37:20–28. [DOI] [PubMed] [Google Scholar]
- 12.Eis, P.S., W. Tam, L. Sun, A. Chadburn, Z. Li, M.F. Gomez, E. Lund, and J.E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 102:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluiver, J., S. Poppema, D. de Jong, T. Blokzijl, G. Harms, S. Jacobs, B.J. Kroesen, and A. van den Berg. 2005. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 207:243–249. [DOI] [PubMed] [Google Scholar]
- 14.O'Connell, R.M., K.D. Taganov, M.P. Boldin, G. Cheng, and D. Baltimore. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 104:1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taganov, K.D., M.P. Boldin, K.J. Chang, and D. Baltimore. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 103:12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shortman, K., and S.H. Naik. 2007. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 7:19–30. [DOI] [PubMed] [Google Scholar]
- 17.Ueda, Y., M. Kondo, and G. Kelsoe. 2005. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J. Exp. Med. 201:1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai, Y., K.P. Garrett, S. Ohta, U. Bahrun, T. Kouro, S. Akira, K. Takatsu, and P.W. Kincade. 2006. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 24:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimson, A., K.K. Farh, W.K. Johnston, P. Garrett-Engele, L.P. Lim, and D.P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis, B.P., I.H. Shih, M.W. Jones-Rhoades, D.P. Bartel, and C.B. Burge. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798. [DOI] [PubMed] [Google Scholar]
- 21.Zhan, Y., G.J. Lieschke, D. Grail, A.R. Dunn, and C. Cheers. 1998. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood. 91:863–869. [PubMed] [Google Scholar]
- 22.Debernardi, S., S. Skoulakis, G. Molloy, T. Chaplin, A. Dixon-McIver, and B.D. Young. 2007. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 21:912–916. [DOI] [PubMed] [Google Scholar]
- 23.Delano, M.J., P.O. Scumpia, J.S. Weinstein, D. Coco, S. Nagaraj, K.M. Kelly-Scumpia, K.A. O'Malley, J.L. Wynn, S. Antonenko, S.Z. Al-Quran, et al. 2007. MyD88-dependent expansion of an immature GR-1+ CD11b+ population induces T cell suppression and Th2 polarization in sepsis. J. Exp. Med. 204:1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metcalf, D., A. Dakic, S. Mifsud, L. Di Rago, L. Wu, and S. Nutt. 2006. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc. Natl. Acad. Sci. USA. 103:1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada, Y., Q. Jiang, M. Lemieux, L. Jeannotte, L. Su, and Y. Zhang. 2006. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat. Cell Biol. 8:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana, L., E. Pelosi, P. Greco, S. Racanicchi, U. Testa, F. Liuzzi, C.M. Croce, E. Brunetti, F. Grignani, and C. Peschle. 2007. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9:775–787. [DOI] [PubMed] [Google Scholar]
- 27.Sinclair, A.M., J.A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P.W. Tucker, E.J. Neufeld, and R.H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 98:3658–3667. [DOI] [PubMed] [Google Scholar]
- 28.Hirai, H., P. Zhang, T. Dayaram, C.J. Hetherington, S. Mizuno, J. Imanishi, K. Akashi, and D.G. Tenen. 2006. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7:732–739. [DOI] [PubMed] [Google Scholar]
- 29.Toki, T., F. Katsuoka, R. Kanezaki, G. Xu, H. Kurotaki, J. Sun, T. Kamio, S. Watanabe, S. Tandai, K. Terui, et al. 2005. Transgenic expression of BACH1 transcription factor results in megakaryocytic impairment. Blood. 105:3100–3108. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Y., Z. Yang, Z. Niu, J. Peng, Q. Li, W. Xiong, A.N. Langnas, M.Y. Ma, and Y. Zhao. 2006. MOP3, a component of the molecular clock, regulates the development of B cells. Immunology. 119:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dragone, L.L., M.D. Myers, C. White, T. Sosinowski, and A. Weiss. 2006. SRC-like adaptor protein regulates B cell development and function. J. Immunol. 176:335–345. [DOI] [PubMed] [Google Scholar]
- 32.Kitajima, K., M. Kojima, K. Nakajima, S. Kondo, T. Hara, A. Miyajima, and T. Takeuchi. 1999. Definitive but not primitive hematopoiesis is impaired in jumonji mutant mice. Blood. 93:87–95. [PubMed] [Google Scholar]
- 33.Yoon, D., Y.D. Pastore, V. Divoky, E. Liu, A.E. Mlodnicka, K. Rainey, P. Ponka, G.L. Semenza, A. Schumacher, and J.T. Prchal. 2006. Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development. J. Biol. Chem. 281:25703–25711. [DOI] [PubMed] [Google Scholar]
- 34.Li, Q.J., J. Chau, P.J. Ebert, G. Sylvester, H. Min, G. Liu, R. Braich, M. Manoharan, J. Soutschek, P. Skare, et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 129:147–161. [DOI] [PubMed] [Google Scholar]
- 35.Fukao, T., Y. Fukuda, K. Kiga, J. Sharif, K. Hino, Y. Enomoto, A. Kawamura, K. Nakamura, T. Takeuchi, and M. Tanabe. 2007. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 129:617–631. [DOI] [PubMed] [Google Scholar]
- 36.Fazi, F., A. Rosa, A. Fatica, V. Gelmetti, M.L. De Marchis, C. Nervi, and I. Bozzoni. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 123:819–831. [DOI] [PubMed] [Google Scholar]
- 37.Neilson, J.R., G.X. Zheng, C.B. Burge, and P.A. Sharp. 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krutzfeldt, J., N. Rajewsky, R. Braich, K.G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438:685–689. [DOI] [PubMed] [Google Scholar]
- 39.Yang, L., and D. Baltimore. 2005. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 102:4518–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.