Abstract
Duplication of genes encoding transcription factors plays an essential role in driving phenotypic variation. Because regulation can occur at multiple levels, it is often difficult to discern how each duplicated factor achieves its regulatory specificity. In these cases, a “systems approach” may distinguish the role of each factor by integrating complementary large-scale measurements of the regulatory network. To explore such an approach, we integrate growth phenotypes, promoter binding profiles, and gene expression patterns to model the DNA damage response network controlled by the Yeast-specific AP-1 (YAP) family of transcription factors. This analysis reveals that YAP regulatory specificity is achieved by at least three mechanisms: (i) divergence of DNA-binding sequences into two subfamilies; (ii) condition-specific combinatorial regulation by multiple Yap factors; and (iii) interactions of Yap 1, 4, and 6 with chromatin remodeling proteins. Additional microarray experiments establish that Yap 4 and 6 regulate gene expression through interactions with the histone deacetylase, Hda1. The data further highlight differences among Yap paralogs in terms of their regulatory mode of action (activation vs. repression). This study suggests how other large TF families might be disentangled in the future.
Keywords: ChIP–chip, evolution, systems biology
Gene duplication and subsequent divergence is the major driving force for the evolution of phenotypic complexity (1, 2). This duplication-and-divergence mode of evolution is particularly true for genes encoding transcription factor (TF) proteins. Approximately 55% of human TFs are part of gene families with multiple functional members (3); well known examples include the basic leucine zipper (bZIP), the C2H2 zinc finger, and the Hox gene families. Members of these TF families enable a variety of critical cellular functions. For instance, the Hox gene family plays a pivotal role in regulating morphogenesis in higher eukaryotes and arose by duplication and divergence of an ancestral homeobox cluster (4).
One of the best examples of gene duplication in Saccharomyces cerevisiae is the Yeast-specific AP-1 (Yap) family of transcription factors, which has a total of eight members (5). The Yap family belongs to the bZIP superfamily of TFs that is widely conserved from yeast to human. Specificity of gene regulation among paralogous Yaps has been attributed to two mechanisms: (i) slight but important differences in the DNA binding motifs targeted by different Yap TFs, as in the case of Yap1 versus Yap2 (6), and (ii) Variation in the regulatory domains present within each Yap protein that are modulated by specific upstream activation signals, i.e., Yap1 is modulated by intramolecular disulfide bond formation (7) and Yap4 is modulated by protein phosphorylation (8). A variety of other mechanisms for achieving specificity are also likely, although they have not yet been systematically demonstrated for the Yap proteins. Such possibilities include cooperative binding with other transcriptional cofactors (9), TF homo- or heterodimerization (10) and differences in the kinetics of protein-DNA binding (11).
Functionally, the Yap family is involved in a variety of stress-related programs, including the response to DNA damage and oxidative, osmotic, and toxic metal stresses. As many as five Yaps (1, 2, 4, 5, and 6) have been implicated in the cellular response to methylmethanesulfonate (MMS), a DNA alkylating agent (12–16), and cis-diamminedichloroplatinum (CDDP), a DNA cross-linking agent (17, 18). With regard to other stresses, Yap TFs carry out overlapping but distinct biological functions with Yap1 being the major player in oxidative stress, Yap2 in cadmium stress, Yap4 and Yap6 in osmotic stress, and Yap8 in arsenic stress (5, 19). There is also evidence of cross-talk between Yap members. For instance, the yap1yap2 double mutant is more sensitive to oxidative stress than either single mutant alone, as is the yap1yap8 double mutant to arsenic stress (19).
Although these studies have established the strong role of the Yap family in the stress responses, a systematic examination of the different family members with regard to their specificities of transcriptional regulation has not yet been conducted. Toward this goal, we performed a series of systematic measurements to characterize the Yap transcriptional network in response to DNA damage, including genome-wide promoter binding profiles and mRNA expression patterns. Integration of these data yielded a genome-wide map of transcriptional regulation revealing both the cooperativity and specificity among different Yap transcription factors.
Results
Selection of Yaps Involved in Two Different DNA Damage Responses.
We first performed a genome-wide screen to identify Yap family members involved in the cellular response to MMS or CDDP. Yap TFs were selected for further study if they satisfied any one of four experimental criteria: (i) differential expression of the TF after exposure to MMS or CDDP; (ii) differential expression of genes whose promoters are bound by the TF after exposure to DNA damaging agents; (iii) growth sensitivity of the TF deletion strain under DNA damaging conditions; or (iv) Association of the TF with the DNA damage response in prior literature. Five family members (Yap1, Yap2, Yap4, Yap5, and Yap6) met one or more of these criteria [supporting information (SI) Fig. 6]. The agents MMS and CDDP were chosen in particular because they are widely used prototypical agents that induce distinct DNA damages. MMS is an alkylating agent that is widely used in basic research on DNA repair and stress response pathways (20), whereas CDDP is a chemotherapeutic used to treat various types of cancers (17).
Genome-Wide Promoter Binding Profiles of Yaps.
We used the technique of chromatin immunoprecipitation coupled with microarray chip (ChIP–chip) to profile the genome-wide DNA binding patterns of each of the five selected Yaps in both nominal and DNA-damaging conditions. Exponentially growing yeast cultures were exposed for 1 h to low or high concentrations of MMS (4.7 or 7.1 mM) or CDDP (0.4 or 1.4 mM), calibrated to induce 20% versus 50% cell killing, respectively (SI Fig. 7). Yap-associated chromatin was immunoprecipitated by using an anti-TAP antibody and analyzed with an Agilent yeast genome tiling array (Materials and Methods). The reproducibility of these data were ≈50% (average overlap of the sets of bound promoters identified by replicate experiments), which is comparable with other large-scale chIP and expression datasets (16, 21, 22) (SI Fig. 8).
At a significance threshold of P < 0.001, the five Yaps together were found to bind ≈400 gene promoters (430 in MMS, 381 in CDDP). Yap4 was found to bind the greatest number (268) of genes across the three growth conditions (Fig. 1). The average target gene overlap (intersection/union) among the five Yaps was 11.9%, 8.1%, and 8.3% under MMS, CDDP, and nominal conditions, respectively. (As a baseline comparison, the average overlap between the Yaps and an unrelated TF, Pdr1, was 2.9% under nominal conditions.) This result confirmed and expanded previous observations that Yaps have partially overlapping but nonredundant functions (5, 6, 19).
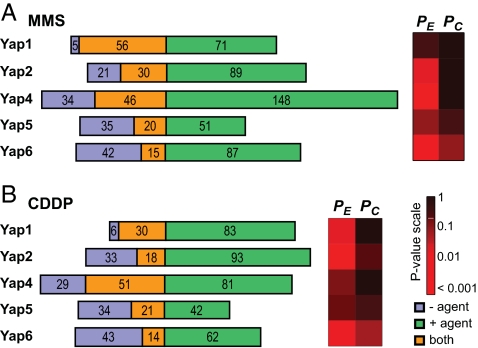
Fig. 1.
Summary of gene promoters bound by Yap TFs. (A) MMS response. (B) CDDP response. (Left) Number of promoters by each Yap. The three regions represent promoters bound exclusively in the absence of DNA damaging agent (blue), presence of DNA damaging agent (green), or in both conditions (orange). (Right) Changes in Yap promoter binding behaviors. PE, significance of expanded target set compared with negative control; PC, significance of contracted target set.
We also observed changes in Yap binding behavior after exposure to MMS or CDDP. After exposure to MMS, Yaps 1, 2, and 4 bound more genes (expansion, Fig. 1A; PE < 0.05, Fisher's exact test), whereas Yaps 5 and 6 bound different sets of genes compared with nominal conditions (shifted; PC < 0.05, PE < 0.05). After exposure to CDDP, Yaps 1 and 4 bound more genes, whereas Yaps 2, 5, and 6 bound different sets of genes compared with nominal conditions (Fig. 1B). Thus, the observed changes in global Yap binding patterns showed rough similarity across the two agents (i.e., compare Fig. 1 A with B).
Systematic Functional Validation of Yap Targets.
To validate the functional consequences of Yap binding at the level of gene expression, we next monitored MMS or CDDP induced expression changes in wild-type yeast and across gene knockout strains for each of the five Yap factors in our study. Normalized expression data were analyzed to identify regulatory relationships between Yaps and target genes. In this context, when a gene that is differentially expressed under MMS or CDDP becomes unresponsive in a specific Yap TF knockout background, the TF is said to be “regulatory epistatic” to the gene. For each possible TF/gene pair, we scored the significance of this effect, using a Bayesian scoring function (16). At P < 0.005, the five Yaps were epistatic to a total of 170 genes under MMS, corresponding to 49 genes on average and a range of 26 genes for YAP6 to 128 genes for YAP1 (Fig. 2A). Under CDDP, the five Yaps were epistatic to 85 genes, corresponding to 48 genes on average and a range of 37 genes for YAP1 to 60 genes for YAP5. The lists of genes identified for each Yap are provided in SI Table 2 (MMS) and SI Table 3 (CDDP).
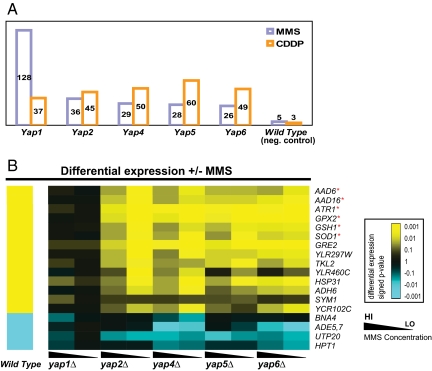
Fig. 2.
Regulatory epistasis. (A) Total number of genes epistatic to Yaps. (B) Representative epistatic genes involving YAP1 in MMS response. Expression changes are colored yellow for up-regulation or blue for down-regulation. *, genes previously known to function in the DNA damage response.
Fig. 2B shows examples of regulatory epistasis for YAP1 under MMS. As a positive control, these examples include well known stress response genes (GPX2, GSH1, SOD1, AAD6, AAD16, and ATR1); all of these genes were strong MMS-responders that became nonresponsive specifically in the yap1Δ strain. Our analysis also revealed many unknown epistatic relationships involving YAP1, especially the group of genes involved in purine biosynthesis (ADE1, ADE5/7, and ADE17) and nicotinic acid biosynthesis (BNA2 and BNA4) (SI Table 3). These functions are required by the cell to cope with DNA damaging agents. Both MMS and CDDP preferentially damage purines causing an imbalance of the cellular purine pool that needs to be replenished (23). Nicotinic acid is the substrate of poly(ADP-ribose) polymerase involved in repair of damaged DNA (24).
Divergence of DNA-Binding Sequences into Two Subfamilies.
Different binding specificities of homologous TFs can lead to marked differences in their regulatory functions. To chart the binding specificities for the Yap family, we searched for enriched DNA motifs within the promoters bound by each Yap factor in the ChIP–chip data. For Yap1, Yap2, Yap4, and Yap6, the motifs discovered were consistent with motifs reported in ref. 25 (Fig. 3). For Yap5, we identified a palindromic motif similar to that of Yap7 but different from the Yap5 motif reported by MacIsaac et al. (25), which is not palindromic and bears no similarity to the Yap family consensus [TTAn(x)TAA]. As supporting evidence for the newly discovered motif, the Yap5 DNA binding domain also shares higher sequence similarity with its counterpart in Yap7 than with other Yap factors (Fig. 3).
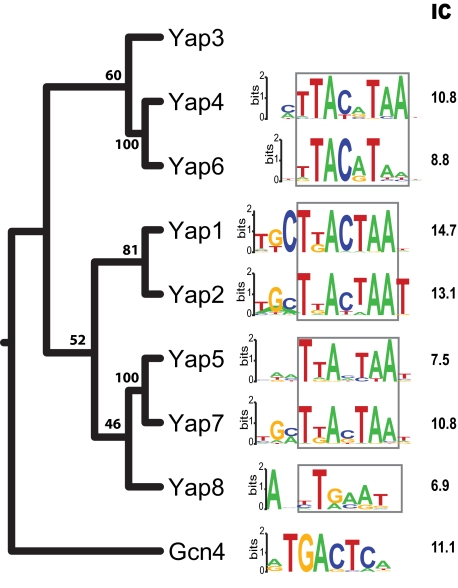
Fig. 3.
DNA binding motifs of five Yaps and their phylogeny. Core regions of the motifs are highlighted by a box. IC, information content. Numbers at each branch point are percentage bootstrap values. Motif resources: Yap1/2/4/5/6, this study; Yap8 and Gcn4, ref. 25; Yap7, ref. 51. Yap3 has no reported motif.
The binding specificity of a TF to its DNA sites can be measured by the information content (IC) of its DNA motif (26). Of the five Yaps in this study, Yap1 had the highest binding specificity whereas Yap5 had the lowest (Fig. 3), perhaps explaining why the Yap5 motif had been difficult to identify. Even for closely related paralogs such as Yap1 and Yap2, differences are apparent in their binding specificities. Although these TFs have very similar core motifs, Yap1 strongly prefers a C at position 3 (P = 9.0 × 10−5), whereas Yap2 strongly prefers a T at position 11 (P = 6.1 × 10−7) (SI Table 4 and SI Materials and Methods). More dramatic differences emerge as family members diverge further. Yap4 and Yap6 are closely related to each other but phylogenetically more distant from Yap1 and Yap2 (Fig. 3). Although all Yap DNA binding motifs share the same core half site (TTA), we found that Yap4 and Yap6 motifs had two base pairs in the spacer region compared with just one base pair for Yap1, Yap2, and Yap5 motifs. This difference in spacer length could be due to different dimerization properties of the leucine zipper domains.
Condition-Specific Combinatorial Regulation by Yaps.
Combinatorial regulation provides another mechanism to achieve target specificity. To better understand how Yap factors cooperate in response to different stress conditions, we computed the overlap in target genes for each pair of Yap TFs. As shown in Fig. 4A, Yap1 and Yap2 (21%, P = 2.3 × 10−38) and Yap2 and Yap6 (25%, P = 1.9 × 10−48) overlap very strongly in MMS, with lower but significant overlap observed between Yap1 and Yap6 (15%, P = 1.3 × 10−22) and several other TF pairs (SI Table 5). In CDDP, Yap1 and Yap2 (12%, P = 8.5 × 10−18) and Yap1 and Yap6 (11%, P = 6.8 × 10−14) also overlap significantly, but the fraction of coregulated genes between Yap2 and Yap6 is strikingly reduced (8%; Fig. 4B). In nominal conditions, we observed strong overlap between the targets of Yap4 and Yap 5 (15%, P = 1.4 × 10−19), but not Yap1 and Yap2 (9%; Fig. 4C). Thus, the pair Yap1/Yap2 appears to coregulate in response to both types of damage, the pair Yap2/Yap6 coregulates predominantly in MMS damage, and the pair Yap4/Yap5 coregulates only in nominal conditions. In addition, the overall number of overlapping TF pairs was lower in nominal than damaging conditions (6 nominal, 10 MMS, 8 CDDP). This increased target overlap among Yap factors could be due to the increased numbers of targets under DNA damage conditions. Alternatively, it could be due to increased cross-talk among Yap factors in response to stress.
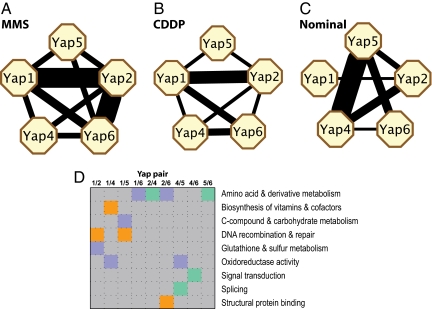
Fig. 4.
Significant overlaps between Yap target gene sets (P < 0.001). Line width is proportional to −log P. (A) MMS treatment. (B) CDDP treatment. (C) Nominal growth conditions. (D) Enriched functional categories of genes coregulated by pairs of Yaps. Blue, MMS response; orange, CDDP response; turquoise, nominal condition.
The gene functions targeted by each Yap pair also differed across the different agent exposures (Fig. 4D). For instance, genes targeted by both Yap1 and Yap2 under MMS were enriched for the functions of glutathione and sulfur metabolism (SAH1, GSH1, GTT2, P = 4.5 × 10−4), whereas genes targeted by the same TF pair under CDDP were enriched for functions in DNA recombination and repair (double strand break formation, REC114 and RAD50; P = 2.7 × 10−4). Studies suggest that MMS reduces the cellular glutathione pool (14, 27), which is important for sulfur metabolism. CDDP causes DNA interstrand cross-links, which requires double-strand break formation to repair this type of DNA damage (28).
Yaps 2, 4, 5, and 6 but Not Yap1 Can Function as Both Activators and Repressors.
Studies suggest that Yaps 2 and 6 can function either as activators or repressors, depending on the regulated genes (6, 29, 30). To further explore this phenomenon and its underlying mechanisms, we applied network component analysis (NCA) (31) to infer the regulatory directions (activating, repressing) of each Yap on its target genes. NCA infers the regulatory directions of TFs, using a linear model based on Hill's equation, which has been used to describe the relationship between promoter activity and transcription factor activities (32). The input to NCA consisted of expression profiles of genes for which a Yap TF showed regulatory epistasis (170 genes in MMS, 85 genes in CDDP) and TF-promoter binding data (817, 495, and 636 interactions in MMS, CDDP, and nominal conditions, respectively) from this study and from two large-scale studies (16, 21) (see SI Materials and Methods).
We inferred regulatory directions of the five Yaps on 64 genes (SI Fig. 9). Thirty-three of the 64 genes have been implicated in DNA damage response from previous microarray studies (14, 16, 23, 33). Of these 33 genes, all but one had inferred regulatory directions that were consistent with current knowledge about the DNA damage response. For instance, all genes involved in ribosomal biogenesis (RPF1, UTP20, UTP9, RPC82, and RPS4A) were repressed, and most genes involved in general stress response (SI Fig. 9) were activated, consistent with the hypothesis that protein synthesis is repressed while detoxification processes are activated after DNA damage (14, 23). Two other examples are CLB1 and CTS1, both of which were repressed in agreement with the assumption that cell cycle progression is arrested or slowed during damage repair (16, 34, 35).
Yap1 appears to function exclusively as an activator (P = 1.4 × 10−4, Fisher's exact test), whereas Yaps 2, 4, 5, 6 appear to function as either activators or repressors (P > 0.05), depending on the gene (SI Fig. 9). Thus, our results extend previous studies and suggest that all Yap TFs except for Yap1 can operate bidirectionally.
Yaps 1, 4, and 6 Interact with Chromatin Modification Factors.
Approximately 20% of yeast genes contain a TATA-box in their promoters (36). In contrast to genes without TATA elements, they are characterized as being stress-induced, expressed at extremely high or low levels, and under strong evolutionary selective pressure (36, 37). They are also tightly regulated by nucleosomes and chromatin remodeling factors (36). Of the five Yaps in our study, we found that the targets of Yaps 1, 4, and 6 were enriched for TATA-bearing genes (Table 1). Interestingly, these three were also the only Yaps (of eight) to have interactions with chromatin remodeling factors, as reported in a recent large-scale survey (38). This correlation suggests that Yaps 1, 4, and 6 indeed regulate expression of their target genes through interactions with chromatin remodeling factors and the TATA box.
Table 1.
Chromatin remodeling factors influence Yaps and TATA box binding
| Yap | Chromatin regulator partners | PHDAC | Fraction TATA genes | PTATA | PTBP |
|---|---|---|---|---|---|
| 4 | Hda1, Isw2, Set1, Sin3, Ssn6, Tup1 | 1.8 × 10−9/1.9 × 10−3 | 0.34 | 3.6 × 10−9 | 0.03 |
| 1 | Ino80 | 0.98/0.82 | 0.33 | 2.8 × 10−6 | 0.04 |
| 6 | Hda1, Isw2, Rpd3, Sin3, Sir2, Spt3, Ssn6, Tup1, Ubp8 | 0.56/2.7 × 10−3 | 0.28 | 1.4 × 10−3 | 0.04 |
| 5 | — | 0.31/0.11 | 0.24 | 0.07 | 0.36 |
| 2 | — | 0.79/0.10 | 0.21 | 0.27 | 0.23 |
Chromatin regulator interaction partners are from ref. 39. PHDAC, P values for overlaps between Yap targets and targets of either Hda1 (first number) or Rpd3 (second number); PTATA, P values for enrichment of TATA-bearing genes; PTBP, P values for increased TBP recruitment to promoters of Yap targets after MMS treatment.
To further test this hypothesis, we computed the target overlaps between Yaps and two histone deacetylases (HDACs), Hda1 and Rpd3, that had been predicted to interact with Yaps (Table 1). We found significant target overlaps between these two HDACs and Yaps 4 and 6 (Table 1), providing further evidence to support the predicted interactions. Yap1 targets did not overlap significantly with those of Hda1 or Rpd3, suggesting it interact with other chromatin remodeling factor(s) such as Ino80 (Table 1). To test the involvement of the TATA box, we examined the changes in TATA-box occupancy by TATA box binding protein (TBP) in promoters of Yap target genes after MMS treatment, as measured (39). We found that recruitment of TBP to Yaps 1, 4, and 6 target promoters significantly increased after MMS treatment, but not promoters bound by Yaps 2 and 5 (Table 1). Because both Hda1 and Rpd3 function as repressors, hypoacetylation, due to de-repression by Hda1 and Rpd3, could make the TATA box more accessible to TBP, resulting in its increased recruitment.
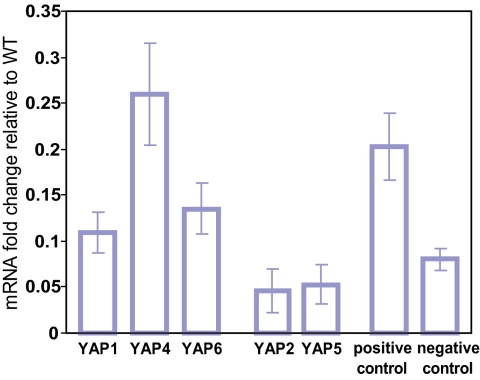
Finally, to still further corroborate the predicted interactions between Hda1 and Yaps 4 and 6, we measured the mRNA levels of Yap targets in hda1Δ cells, using genome-wide expression microarrays. Expression levels of the targets of Yaps 1, 4, and 6 were increased in the hda1Δ strain, whereas expression levels of Yap2 and Yap5 targets were unaffected (Fig. 5). Combined with the evidence of significant target overlap among Yaps 4 and 6 and Hda1 and Rpd3 (Table 1), these results establish that interacting with chromatin remodeling factors provides another means to confer target specificity to Yaps 1, 4, and 6.
Fig. 5.
Deletion of HDA1 increases Yap1, 4, and 6 target expression. Shown is the difference in fold change (hda1Δ+MMS/hda1Δ−MMS − WT+MMS/WT−MMS) averaged across all TF targets. Positive control, targets of two TFs (Sko1 and Sut1) known to interact with Hda1. Negative control, targets of three TFs (Dig1, Ecm22, and Rtg3) not predicted to interact with any chromatin remodeling factors in ref. 38.
Discussion
The Yap family of S. cerevisiae is likely to have been formed by six duplication events since the divergence of their common ancestor from that of Gcn4, including a whole-genome duplication and several segmental duplications (40) (SI Fig. 10). They can be divided into four related subfamilies based on their sequence similarities: Yap1 and Yap2, Yap4 and Yap6, Yap5 and Yap7, and Yap3 and Yap8. Based on their dN/dS ratios (the normalized ratio of amino acid-altering substitutions to silent substitutions, a traditional measure for evolutionary rate), Yaps 1, 4, 5, and 6 have evolved at a rate close to the average for the genome (0.11 ± 0.02), whereas Yaps 2, 3, 7, and 8 show accelerated evolution (0.21–0.54; SI Fig. 10) consistent with the hypothesis that they are under positive selection to fulfill a wider genetic program required to deal with new environmental stresses. In contrast to the DNA damage response, which is essential to maintain genome integrity in any organism and is thus a very ancient function, the heavy metal stress response mediated by Yap2 (cadmium) and Yap8 (arsenic) probably evolved later.
Four lines of evidence suggest a central regulatory role for Yap1 and Yap4 in the Yap-mediated network: (i) Phenotypically, the yap1Δ strain is the most sensitive among the Yap TFs to MMS treatment. Additionally, only Yap 4 and 6, when over-expressed, confer CDDP resistance (18). (ii) Yap4 targets the largest number of genes and Yap1 is epistatic to the largest number of genes in both DNA damaging conditions combined. (iii) Of the five Yaps in this study, Yap1 and Yap4 are the most conserved across six Sacchromyces genomes (41, 42). Their orthologs are detectable in five and six of these genomes, respectively, compared with two, four, or five orthologs for Yap2, Yap5, or Yap6, respectively. (iv) Our ChIP–chip data suggest that Yap6 is targeted by Yap1p under nominal conditions and by Yap1p and Yap4p under DNA damage conditions. Based on these observations, we propose a hierarchical model in which Yap1 and Yap4 together function as the major regulators in the Yap-mediated network, and other Yaps function as secondary regulators that expand the network's regulatory capacity.
Our results indicate functional linkages among three Yaps (Yap1/4/6), chromatin remodeling factors, and the TATA box in gene regulation after DNA damage. Recently, a chromatin-mediated mechanism for specification of conditional transcription factor targets has been proposed (43). In this model, a subset of a TF's binding sites could be made accessible by modifying nucleosome occupancy in the promoter region through the activities of chromatin remodeling factors. Based on analysis of existing data and new expression profiling of the hda1Δ strain in our study, we propose that such a mechanism is used by Yap1/4/6 to specifically regulate a subset of genes in response to DNA damage. According to this model, DNA damage results in the recruitment of chromatin remodeling factors to the vicinity of Yap binding sites by additional TFs. The nucleosomes around Yap1/4/6 binding sites and the TATA box are then rearranged or cleared by chromatin remodeling factors, which allow Yap TFs to bind and facilitate the assembly of RNA polymerase at the basal promoter. Furthermore, Yaps might interact with different remodeling factors. For instance, Yaps 4 and 6 work with HDACs, whereas Yap1 probably works with Ino80. It is also possible that Yap1/4/6 can operate independently of chromatin remodeling factors, depending on the function of the target genes.
In summary, we have described an integrated approach to delineate the complex regulatory network mediated by a large family of TFs. The strength of the approach lies in the integration of multiple types of measurement of the network with both environmental and genetic perturbations. In the future, network states measured under a diverse set of perturbations (both environmental and genetic) will enable further discrimination of the subtle functional differences among family members. The bZIP TF families are also pervasive in higher eukaryotes and are involved in a variety of processes that are critical to the function of the organism, such as embryogenesis, metabolism, and learning and memory (44–46). An integrative approach offers a powerful means to delineate the even more complicated regulatory networks in these organisms.
Materials and Methods
Strains and Media.
Strains used in ChIP–chip and expression were derived from haploid BY4741 (MATa his3D1 leu2D0 met15D0 ura3D0). Tandem affinity purification (TAP)-tagged strains used in ChIP–chip experiments were obtained from Open Biosystems. Yap deletion strains constructed by the Saccharomyces Gene Deletion Project were obtained from Research Genetics. Cells were cultured in standard yeast synthetic complete media (SC) at 30°C except when noted.
Agents.
CDDP and MMS were purchased from Sigma. CDDP and MMS stock solutions were freshly prepared in SC for each experiment. Treatment concentrations of each agent were determined by using colony survival curves. See SI Materials and Methods for details.
ChIP–chip Analysis.
Genome-wide TF binding locations were assayed as described in ref. 47. Briefly, samples were processed in parallel, using two distinct biological replicates from each TAP-tagged strain and grown to saturation in SC overnight at 30°C. Overnight cultures were then diluted in fresh SC and grown to an OD600 of 0.8–1.0. CDDP/MMS was added to the desired final concentration or left out for nominal conditions, and the cultures were grown for an additional hour. Cells were fixed with 1% formaldehyde, lysed, and sonicated to shear DNA. DNA fragments bound by the tagged TF were enriched by immunoprecipitation with an anti-TAP antibody (Open Biosystems). After reversal of cross-linking, the enriched DNA was amplified and Cy5-labeled by ligation-mediated PCR (LM-PCR). A non-antibody enriched DNA sample was amplified by LM-PCR, using a Cy3 label. IP-enriched and unenriched samples were cohybridized to a single yeast whole genome tiling array containing 44,290 60-mer oligonucleotides spaced ≈266 bp across 85% of the nonrepetitive part of the whole yeast genome (≈12 Mb).
Whole-Genome Expression Analysis.
Gene expression experiments were processed in parallel with at least two distinct biological samples from each strain grown to saturation in SC overnight at 30°C. Cell cultures were then treated in the same way as in the ChIP–chip experiments. Cells were harvested and immediately snap-frozen in liquid nitrogen to suspend gene expression and stored at −20°C before RNA extraction. Total RNA from each sample was isolated by hot acid phenol extraction and mRNA-purified via Poly(A)Pure kits (Ambion). Labeling of cDNA was performed in a dye-reversal scheme by direct incorporation, using a CyScribe First-Strand cDNA labeling kit (Amersham Biosciences). Corresponding Cy-3 and Cy-5 labeled samples were cohybridized to the yeast oligo microarray (G4140B; Agilent Technologies).
Data Postprocessing and Significance Assessment.
Scanned images were processed by using GenePix Pro 6.0 software (Axon Instruments) to obtain raw Cy-3 and Cy-5 foreground and background intensity measurements for each spot on the array. Raw spot intensities were normalized by using an in-house normalization pipeline (see SI Materials and Methods for details). Replicate arrays for each gene expression experiment were processed by using the VERA package (48) to estimate multiplicative and additive errors and to associate a P value of differential expression with each gene. Multiplicative and additive errors of replicate ChIP–chip experiments were estimated by using the Rosetta error model (49) and a P value of enrichment of binding was associated with each probe. A TF binding call at each probe is made by combining raw P values of adjacent probes.
Genome-Wide Epistasis Analysis.
Given a set of expression profiles, a TF is said to be epistatic to a gene if that gene is differentially expressed in the vast majority of profiles but does not change its expression in the knockout background of the TF in question. We used a Bayesian scoring scheme to score the significance of the regulatory epistasis as described in ref. 16. P values of epistasis are calculated by comparing the epistasis scores of a deletion strain to that of a null distribution derived from an additional independent wild-type experiment not used in the calculation of the epistasis scores of the deletion strain. See SI Materials and Methods for details.
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of the T.G.I. laboratory for helpful discussion. This work was funded by a postdoctoral fellowship from IBM Corporation (to K.T.) through its Institutes of Innovation program, National Institutes of Environmental Health Sciences Grant ES14811 and the Packard Foundation (T.G.I.), and National Institute of Mental Health Grant 2P30MH062261-07 (to T.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.A. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no.GSE10146).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708670105/DC1.
References
- 1.Ohno S. Evolution by Gene Duplication. London: Allen & Unwin; 1970. [Google Scholar]
- 2.Li WH, Yang J, Gu X. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Tupler R, Perini G, Green MR. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 4.Maconochie M, Nonchev S, Morrison A, Krumlauf R. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes L, Rodrigues-Pousada C, Struhl K. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen BA, Pilpel Y, Mitra RD, Church GM. Mol Biol Cell. 2002;13:1608–1614. doi: 10.1091/mbc.01-10-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuge S, Arita M, Murayama A, Maeta K, Izawa S, Inoue Y, Nomoto A. Mol Cell Biol. 2001;21:6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nevitt T, Pereira J, Rodrigues-Pousada C. Yeast. 2004;21:1365–1374. doi: 10.1002/yea.1188. [DOI] [PubMed] [Google Scholar]
- 9.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 10.Deppmann CD, Alvania RS, Taparowsky EJ. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- 11.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. J Biol Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 12.Begley TJ, Samson LD. DNA Repair (Amsterdam) 2004;3:1123–1132. doi: 10.1016/j.dnarep.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Mol Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelinsky SA, Estep P, Church GM, Samson LD. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM. Cancer Res. 2004;64:3940–3948. doi: 10.1158/0008-5472.CAN-03-3113. [DOI] [PubMed] [Google Scholar]
- 18.Furuchi T, Ishikawa H, Miura N, Ishizuka M, Kajiya K, Kuge S, Naganuma A. Mol Pharmacol. 2001;59:470–474. doi: 10.1124/mol.59.3.470. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigues-Pousada C, Nevitt T, Menezes R. Febs J. 2005;272:2639–2647. doi: 10.1111/j.1742-4658.2005.04695.x. [DOI] [PubMed] [Google Scholar]
- 20.Fry RC, Begley TJ, Samson LD. Annu Rev Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 21.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley R, Feizi H, Ideker T. Bioinformatics. 2007;24:71–77. doi: 10.1093/bioinformatics/btm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelinsky SA, Samson LD. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vispe S, Yung TM, Ritchot J, Serizawa H, Satoh MS. Proc Natl Acad Sci USA. 2000;97:9886–9891. doi: 10.1073/pnas.170280397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. BMC Bioinformat. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stormo GD, Fields DS. Trends Biochem Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 27.Wilhelm D, Bender K, Knebel A, Angel P. Mol Cell Biol. 1997;17:4792–4800. doi: 10.1128/mcb.17.8.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehoczky P, McHugh PJ, Chovanec M. FEMS Microbiol Rev. 2007;31:109–133. doi: 10.1111/j.1574-6976.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- 29.Dumond H, Danielou N, Pinto M, Bolotin-Fukuhara M. Mol Microbiol. 2000;36:830–845. doi: 10.1046/j.1365-2958.2000.01845.x. [DOI] [PubMed] [Google Scholar]
- 30.Yeang CH, Mak HC, McCuine S, Workman C, Jaakkola T, Ideker T. Genome Biol. 2005;6:R62. doi: 10.1186/gb-2005-6-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Proc Natl Acad Sci USA. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronen M, Rosenberg R, Shraiman BI, Alon U. Proc Natl Acad Sci USA. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caba E, Dickinson DA, Warnes GR, Aubrecht J. Mutat Res. 2005;575:34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Stuart D, Wittenberg C. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Racki WJ, Becam AM, Nasr F, Herbert CJ. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basehoar AD, Zanton SJ, Pugh BF. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 37.Tirosh I, Weinberger A, Carmi M, Barkai N. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 38.Steinfeld I, Shamir R, Kupiec M. Nat Genet. 2007;39:303–309. doi: 10.1038/ng1965. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Iyer VR. Mol Cell Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kellis M, Birren BW, Lander ES. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 41.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 42.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 43.Buck MJ, Lieb JD. Nat Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U, Wagner EF. Nature. 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- 45.Darlington GJ, Ross SE, MacDougald OA. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 46.Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- 47.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 48.Ideker T, Thorsson V, Siegel A, Hood L. J Comput Biol. 2000;7:805–817. doi: 10.1089/10665270050514945. [DOI] [PubMed] [Google Scholar]
- 49.Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 50.Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 51.Pachkov M, Erb I, Molina N, van Nimwegen E. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.