Abstract
Infection and reinfection with multiple cytomegalovirus (CMV) strains have been shown to occur in immunocompromised individuals, sexually transmitted disease clinic attendees, and children attending day care centers. To characterize the CMV diversity in healthy seropositive individuals, 16 CMV PCR-positive specimens from 113 seropositive women were analyzed for glycoprotein gN and gB genotypes by cloning, followed by nucleotide sequencing of the plasmid DNA and/or restriction fragment length polymorphism (RFLP). The results showed that most (93.7%) of the PCR-positive specimens contained multiple gN and/or gB genomic variants, suggesting that the majority of women were infected with more than one virus strain. The results also showed that the RFLP technique might not be sufficiently sensitive to detect all of the genomic variants present in a sample.
Cytomegalovirus (CMV) species are important opportunistic agents in infection of immunocompromised individuals and a frequent cause of congenital infection. Infection with multiple strains of CMV has been shown to occur frequently in immunocompromised individuals and sexually transmitted disease (STD) clinic attendees (8, 10). In addition, reinfection with different CMV strains was documented to occur in children attending day care centers (2). More recently, CMV reinfections were demonstrated in seropositive women, and such reinfections can result in intrauterine transmission and damaging fetal infection (5).
Extensive genetic polymorphisms in envelope glycoproteins of CMV, including glycoprotein B (gpUL55), glycoprotein O (gpUL74), and glycoprotein N (gpUL73), have been demonstrated among clinical CMV isolates. Major envelope glycoprotein B (gB) of CMV has been demonstrated to elicit a strong neutralizing antibody response (6), and on the basis of restriction fragment length polymorphism (RFLP) analysis of clinical samples, four unique genomic variants, gB types 1 to 4, have been identified (9). Glycoprotein N has been shown to be highly polymorphic at the amino-terminal region, and most clinical CMV isolates have been shown to cluster into four distinct genomic variants, gN-1, gN-2, gN-3a, gN-3b, gN-4a, gN-4b, and gN-4c (11). Recent studies have shown that a significant proportion of the virus-neutralizing response was also directed against the gM/gN complex (17). No linkage between gN genotypes and gB genotypes has been observed (11).
Published studies using RFLP analyses to determine the gN genotypes have identified only a single gN type in a given sample (11, 13). Studies of glycoprotein B based on RFLP analyses showed the presence of a single genotype or a limited number of samples containing mixtures of two gB genotypes (3, 7). However, a recent study using hybridization with type-specific probes (10) showed mixtures of all genotypes. To determine the CMV strain diversity in healthy seropositive women, the presence of multiple gN and gB genomic variants in urine or peripheral blood was examined by two different methods, RFLP and cloning followed by nucleotide sequence analysis of the plasmid DNA.
(This research was presented in part at the 43rd Annual Meeting of the Infectious Diseases Society of America, San Francisco, CA, 7 October 2005, abstract 924.)
MATERIALS AND METHODS
Specimens and subjects.
The specimens studied consisted of 306 urine and 248 peripheral blood samples from 113 healthy, CMV-seropositive women who were tested for the presence of CMV immunoglobulin G antibodies in the postpartum period between February 2000 and June 2004. The women in the study were derived from a predominantly urban, low-income, African American population. Informed consent was obtained from all study participants, and the study was conducted in accordance with the guidelines of the Institutional Review Board for Human Use of the University of Alabama at Birmingham.
Detection of CMV DNA.
DNA was extracted from urine and peripheral blood specimens with a commercial spin column kit (Qiagen Inc., Chatsworth, CA). The samples were initially tested for the presence of CMV DNA by PCR with primers from the antigen domain 1 region of the gene encoding glycoprotein B as described previously (4). The antigen domain 1 region of gB has been shown to be highly conserved among clinic isolates of CMV (9). The PCR-positive samples were further analyzed to determine gN and gB genotypes.
Characterization of gN genomic variants. (i) Nucleotide sequence analysis following cloning of the PCR-amplified gN products.
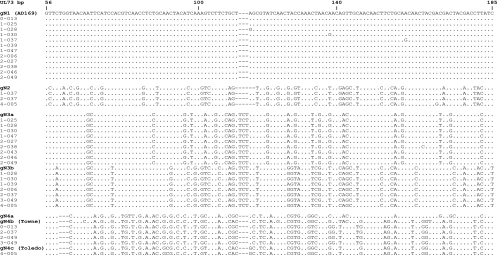
The samples that were CMV PCR positive were further subjected to PCR to amplify the gN region with primers gN-Fw (5′ GGC GGT GGT GTG ATG GAG TG) and gN-Rev (5′ AAT AGC CTT TGG TGG TGG TTG C). After an initial 2-min denaturation at 96°C, the samples underwent seven cycles of denaturation at 96°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 40 s and the annealing temperature was decreased by 1°C each cycle. The samples were further subjected to 28 cycles with an annealing temperature of 58°C and a final extension step at 72°C for 7 min. The PCR products were directly cloned into TOPO TA cloning vector pCR 2.1 (Invitrogen Inc., Carlsbad, CA). The colonies were screened for the presence of the gN gene insert by direct PCR amplification and then grown to an appropriate volume of culture medium. Initially, the plasmid DNA from five individual colonies was sequenced with the gN-Fw primer at the University of Alabama at Birmingham sequencing core facility. If two or more variants were found, no more colonies were screened. However, in the event that the first five colonies contained a single genotype, five additional colonies were sequenced. The nucleotide sequences were compared to the published gN subtype sequences of the four major gN genotypes (GenBank accession numbers AF309971, AF309976, AF309980, AF390773, AF309987, AF309997, and AF310004). A limited number of colonies from each recombinant were sequenced in both directions to confirm the sequence diversity and the genotype assignment (Fig. 1).
FIG. 1.
Alignment of gN (UL73) nucleotide sequences with only part of the variable region shown. Dots indicate identity, and dashes indicate deletions. Strains are grouped by comparing the sequences of the recombinants with the prototypic gN genotypes previously described (11). The sequences are listed with the unique study subject identifiers. Prototypical laboratory-adapted strains are in parentheses. The nucleotide sequences were aligned with that of AD169 (gN1 prototype) with the Vector NTI Advance software package V.10 (Invitrogen, Carlsbad, CA) and displayed as a printable output by the BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html).
(ii) RFLP analysis.
The PCR products amplified with the gN-Fw and gN-Rev primers described above were digested in four separate reactions with the restriction endonucleases SalI, SacI, BsaXI, and MmeI. The resulting restriction fragments were resolved by agarose gel electrophoresis, and the band patterns were analyzed in accordance with the restriction sites contained in different gN subtypes (12).
Characterization of gB genotypes by nested PCR followed by cloning.
The DNA samples were initially subjected to PCR to amplify the target gB region located in the variable region between bp 1138 and 1638 with gB primers gB1138 (5′ CAA GAR GTG AAC ATG TCC GA) and gB1638 (5′ GTC ACG CAG CTG GCC AG). The PCR products were diluted 1:10 and subjected to nested PCR with primers gB1319 (5′ TGG AAC TGG AAC GTT TGG C) and gB1604 (5′ GAA ACG CGC GGC AAT CGG), yielding a 285-bp product (3). The PCR products were gel purified and cloned into the pCR 2.1 TOPO TA cloning vector (Invitrogen Inc., Carlsbad, CA). The colonies were screened by direct PCR, and those containing the insert were grown in culture medium. Plasmid DNA from five individual colonies from each recombinant was sequenced with the M13 forward primer at the University of Alabama at Birmingham sequencing core facility, and if two or more variants were found, no more colonies were screened. However, in the event that the first five colonies contained a single genotype, five additional colonies were sequenced. The nucleotide sequences were compared to the published gB genotype sequences (GenBank accession numbers M60928, M60930, M609931, and M609933). As described above for gN genotype characterization, a limited number of colonies were sequenced in both directions to confirm the genotype assignment (Fig. 2).
FIG. 2.
Alignment of representative gB (UL55) sequences with the prototypes (only part of the variable region of gB is shown). Dots indicate identity, and dashes indicate deletions. Strains are grouped by comparing the sequences of the recombinants with the prototypic gB genotypes previously described (9). The sequences are listed with the unique study subject identifiers. The nucleotide sequences were aligned with the gB1 prototype with the Vector NTI Advance software package V.10 (Invitrogen, Carlsbad, CA) and displayed as a printable output by the BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html).
RESULTS
Of the 554 samples examined for the presence of CMV DNA, 16 (2.9%) were CMV PCR positive (9/306 urine samples and 7/248 blood samples). The positive samples were collected from 16 different study subjects. To determine the sensitivity of the PCR assay, the positive samples were subjected to a real-time PCR assay to estimate the amount of CMV DNA (4). This analysis showed that the sensitivity of the PCR assay was 300 copies/ml (data not shown).
The gpUL73 (gN) and gpUL55 (gB) diversity was examined in all 16 samples by RFLP and cloning of the gN gene and by cloning of the gB gene. Of the 16 samples, 15 (93%) were found to have more than one gN genomic variant (Table 1). The only sample that was found to contain a single gN genotype on cloning and nucleotide sequence analysis was also found to have a single gN genomic variant on RFLP examination (Table 2). Although the RFLP examination revealed that four additional samples contained a single gN genotype, nucleotide sequence analysis documented the presence of multiple gN genomic variants in the samples (Table 1).
TABLE 1.
Comparison of gN genotyping results by RFLP and the cloning techniques for each of the 16 PCR-positive samples
| Sample | Observed genotype(s)
|
|
|---|---|---|
| RFLP | Cloning | |
| 0-013 | 4 | 1, 4 |
| 1-025 | 1, 3 | 1, 3 |
| 1-028 | 1, 3 | 1, 3 |
| 1-030 | 1, 3 | 1, 3 |
| 1-037 | 1, 2 | 1, 2 |
| 1-039 | 1, 3 | 1, 3 |
| 1-047 | 1 | 1, 3 |
| 2-006 | 1, 3 | 1, 3 |
| 2-027 | 1, 3 | 1, 3 |
| 2-037 | 2, 3, 4 | 2, 3, 4 |
| 2-038 | 1, 3 | 1, 3 |
| 2-043 | 3 | 3 |
| 2-046 | 1 | 1, 3 |
| 2-049 | 1, 3 | 1, 3, 4 |
| 3-049 | 1, 3, 4 | 3, 4 |
| 4-005 | 4 | 2, 3, 4 |
TABLE 2.
Results of gN and gB genotyping of CMV in the urine and blood of CMV-seropositive women
| Glycoprotein | Technique | % of samples with indicated no. of genotypes (no.a of samples):
|
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| gN | RFLP | 31.2 (5) | 56.3 (9) | 12.5 (2) |
| gN | Cloning | 6.3 (1) | 75 (12) | 18.7 (3) |
| gB | Cloning | 31.3 (5) | 50 (8) | 18.7 (3) |
The total number of samples tested was 16.
Of the 16 samples analyzed for gB genotypes, 69% (11/16) were found to contain more than one gB genotype; eight samples had two genotypes, and three samples had three genotypes (Table 2). Among the 16 samples that underwent gN and gB genotype analysis by cloning, 15 (93.7%) samples contained multiple genotypes and only one sample had a single gN and gB genotype. The sample with only one gN genotype was from a blood specimen. Of the samples with a single gB genotype, two (40%) were from blood and three (60%) were from urine specimens.
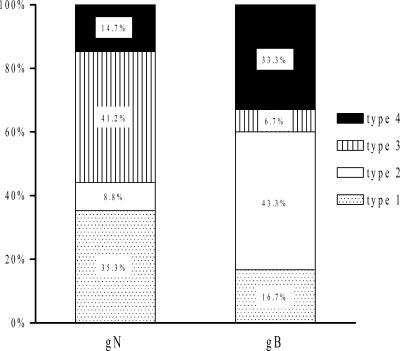
The most common glycoprotein N genotype in our study group was type 3 (41%), followed by type 1 (35%). Glycoprotein B genotype 2 (43%) was the most frequent, followed by type 4 (33%) (Fig. 3).
FIG. 3.
Relative frequencies of CMV gN and gB genotypes in urine and blood samples from CMV-seropositive women.
DISCUSSION
The findings of the present study clearly document the presence of multiple CMV strains in the majority of the CMV PCR-positive urine and peripheral blood specimens from the healthy seropositive women studied. The existence of multiple virus strains in the specimens was demonstrated by the presence of different and multiple gN and/or gB genomic variants. Similar findings were reported in a recent study in our laboratory in which multiple gN genotypes were detected in genital tract specimens from women who attended an STD clinic (15). In an earlier report, we documented the occurrence of CMV reinfection between pregnancies in seropositive women, and such reinfection could lead to intrauterine transmission and symptomatic congenital CMV infection (5). Several other studies have also documented CMV infection with multiple virus strains in a variety of population groups, including children attending day care centers, human immunodeficiency virus-infected individuals, allograft recipients, and infants with congenital CMV infection (1, 2, 10, 14). Together, the findings of these studies of different population groups suggest that infections with multiple CMV strains occur frequently.
It is possible that the observed strain diversity in our study population of healthy, seropositive women is due to an increased exposure to CMV, resulting in CMV reinfections. However, we could not determine the timing of reinfection(s) in our study. Previous studies have reported reinfection with multiple strains of CMV in immunocompetent hosts. Chandler et al. reported that four of eight women attending an STD clinic were found to be reinfected with a new strain of CMV (8). Bale et al. examined serial samples from 37 children attending day care centers and found that 19% had evidence of infection with more than one CMV strain (2). Although exposure to CMV through sexual activity or contact with young children could explain the strain diversity observed in our study, the association between increased CMV exposure and CMV strain diversity has not been documented.
Several previous studies that examined the genetic diversity of CMV glycoprotein gN only reported the presence of a single gN genotype in the specimens tested. Pignatelli et al. examined 223 viral isolates from a variety of patient populations, including allograft recipients, AIDS patients, congenitally infected children, and children with postnatally acquired CMV infection from four different geographic regions. The virus isolates examined in that study were obtained from urine, saliva, blood, amniotic fluid, and biopsy specimens (12). Although all four gN genotypes were present among the specimens tested, only one gN type was detected in a given specimen. In contrast, we could document the presence of multiple gN genomic variants in the majority of CMV PCR-positive specimens in our study. In the present study, we used two different techniques, RFLP and cloning of the amplified gN product, followed by nucleotide sequence analysis, to determine the gN genotypes, whereas the studies by Pignatelli et al. and others only used RFLP (16). It is likely that the addition of the cloning method may have improved our ability to distinguish different genomic variants present in a specimen. Additional factors, including the populations studied and the geographic origin of the specimens, may account for the differences between the present study and previously published reports. Furthermore, Pignatelli et al. studied virus isolates that had been propagated in cell culture rather than DNA extracted from the original specimen. The propagation of virus isolates in cell culture could have selected for a single virus strain.
Identification of gN and gB genotypes by RFLP is based on the ability to clearly visualize the restriction fragment band pattern, and therefore the amount of DNA in the samples tested is critical. Multiple gN genomic variants present in a sample may not be easily distinguished because the intensity of the fragments on gel electrophoresis depends on the amount and ratio of the type-specific DNA. Furthermore, because of overlapping and sometimes minute differences in band size, it may be difficult to properly identify all of the bands necessary for precise genotype identification, even on high-concentration agarose gel electrophoresis. The restriction sites could be affected by a single nucleotide change, which in turn makes it difficult to recognize correct genotypes. However, such a change in the restriction sites is unlikely to affect the genotype assignment because the grouping of genotypes is based on multiple nucleotide changes. The cloning and sequence analysis method is more accurate in determining the number of gN or gB genotypes because it is not limited by the relative amount of type-specific DNA contained in the sample. However, there are limitations to the use of cloning and sequence analysis in the detection of multiple strains in a sample. These include the sensitivity and specificity of the PCR assay used to amplify CMV DNA in the samples and the number of bacterial colonies that can be screened.
Like glycoprotein N, genotyping of glycoprotein B by RFLP has only shown one or two types present in a clinical sample. In a study based on samples from liver transplant patients, 87% (53/61) of the samples contained only one gB genotype (3). The remaining 13% of the samples contained two genotypes. This could be due to the fact that the use of the enzymes HinfI and RsaI produced indistinguishable restriction patterns in certain combinations of three and four different gB genotypes. Additionally, the above-described limitations of the ability of RFLP to detect multiple gN genotypes are also relevant to the detection of gB genotypes. However, mixtures of three or four different gB genotypes per sample were detected in a study using non-RFLP-based techniques. A recent study by Coaquette et al. used PCR amplification followed by hybridization with a single-stranded DNA probe specific for each CMV gB genotype detected by DNA enzyme immunoassay. In that study, of the 97 CMV isolates collected from 92 immunocompromised patients (transplant recipients and lymphoma and leukemia patients), 74% (72/97) contained a single gB genotype, 22% (21/97) contained two genotypes, 3% (3/97) contained three genotypes, and 1% (1/97) contained all four genotypes (10).
Similar to the results of previous studies, we did not observe a linkage between the gN and gB genotypes (11). However, the number of samples examined was small and we found only one sample that contained a single gN genotype and five samples containing a single variant of gB. Only one study sample contained a single genotype of both the gN and gB glycoproteins. Therefore, our study clearly did not have a sufficient sample size to examine the possible linkage between the gN and gB genotypes. In addition, our study was based on a predominantly African American, low-income population and therefore the findings of our study may not be representative of the general population.
Using RFLP and cloning followed by nucleotide sequence analysis, we could demonstrate infection with multiple CMV strains in most healthy seropositive women who had CMV DNA in their urine and/or peripheral blood. These findings need to be confirmed in a study with a larger number of samples and more diverse population groups.
Acknowledgments
Financial support for this study came from the National Institute on Deafness and Other Communication Disorders (R01 DC04163) and the General Clinical Research Center (M01 R00032).
We have no commercial conflicts of interest.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Arav-Boger, R., R. E. Willoughby, R. F. Pass, J. C. Zong, W. J. Jang, D. Alcendor, and G. S. Hayward. 2002. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J. Infect. Dis. 1861057-1064. [DOI] [PubMed] [Google Scholar]
- 2.Bale, J. F., Jr., S. J. Petheram, I. E. Souza, and J. R. Murph. 1996. Cytomegalovirus reinfection in young children. J. Pediatr. 128347-352. [DOI] [PubMed] [Google Scholar]
- 3.Binder, T., W. Siegert, A. Kruse, H. Oettle, F. Wilborn, R. Peng, H. Timm, P. Neuhaus, and C. A. Schmidt. 1999. Identification of human cytomegalovirus variants by analysis of single strand conformation polymorphism and DNA sequencing of the envelope glycoprotein B gene region-distribution frequency in liver transplant recipients. J. Virol. Methods 78153-162. [DOI] [PubMed] [Google Scholar]
- 4.Boppana, S. B., K. B. Fowler, R. F. Pass, L. B. Rivera, R. D. Bradford, F. D. Lakeman, and W. J. Britt. 2005. Congenital cytomegalovirus infection: association between virus burden in infancy and hearing loss. J. Pediatr. 146817-823. [DOI] [PubMed] [Google Scholar]
- 5.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 3441366-1371. [DOI] [PubMed] [Google Scholar]
- 6.Britt, W. J., L. Vugler, and E. B. Stephens. 1988. Induction of complement-dependent and -independent neutralizing antibodies by recombinant-derived human cytomegalovirus gp55-116 (gB). J. Virol. 623309-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraro, E., and C. F. Granato. 2003. Single human cytomegalovirus gB genotype shed in multiple sites at the time of diagnosis in renal transplant recipients. J. Med. Virol. 70240-243. [DOI] [PubMed] [Google Scholar]
- 8.Chandler, S. H., H. H. Handsfield, and J. K. McDougall. 1987. Isolation of multiple strains of cytomegalovirus from women attending a clinic for sexually transmitted diseases. J. Infect. Dis. 155655-660. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188388-390. [DOI] [PubMed] [Google Scholar]
- 10.Coaquette, A., A. Bourgeois, C. Dirand, A. Varin, W. Chen, and G. Herbein. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 39155-161. [DOI] [PubMed] [Google Scholar]
- 11.Pignatelli, S., P. Dal Monte, and M. P. Landini. 2001. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J. Gen. Virol. 822777-2784. [DOI] [PubMed] [Google Scholar]
- 12.Pignatelli, S., P. Dal Monte, G. Rossini, S. Chou, T. Gojobori, K. Hanada, J. J. Guo, W. Rawlinson, W. Britt, M. Mach, and M. P. Landini. 2003. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 84647-655. [DOI] [PubMed] [Google Scholar]
- 13.Pignatelli, S., P. Dal Monte, G. Rossini, T. Lazzarotto, M. R. Gatto, and M. P. Landini. 2003. Intrauterine cytomegalovirus infection and glycoprotein N (gN) genotypes. J. Clin. Virol. 2838-43. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen, L., C. Hong, D. Zipeto, S. Morris, D. Sherman, S. Chou, R. Miner, W. L. Drew, R. Wolitz, A. Dowling, A. Warford, and T. C. Merigan. 1997. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J. Infect. Dis. 175179-184. [DOI] [PubMed] [Google Scholar]
- 15.Ross, S. A., Z. Novak, G. Ashrith, L. B. Rivera, W. J. Britt, S. Hedges, J. R. Schwebke, and A. S. Boppana. 2005. Association between genital tract cytomegalovirus infection and bacterial vaginosis. J. Infect. Dis. 1921727-1730. [DOI] [PubMed] [Google Scholar]
- 16.Rossini, G., S. Pignatelli, P. Dal Monte, D. Camozzi, T. Lazzarotto, L. Gabrielli, M. R. Gatto, and M. P. Landini. 2005. Monitoring for human cytomegalovirus infection in solid organ transplant recipients through antigenemia and glycoprotein N (gN) variants: evidence of correlation and potential prognostic value of gN genotypes. Microbes Infect. 7890-896. [DOI] [PubMed] [Google Scholar]
- 17.Shimamura, M., M. Mach, and W. J. Britt. 2006. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J. Virol. 804591-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]