Abstract
Primates have evolved a variety of restriction factors that prevent retroviral replication. One such factor, TRIM5α, mediates a postentry restriction in many Old World primates. Among New World primates, Aotus trivirgatus exerts a similar early restriction mediated by TRIMCyp, a TRIM5–cyclophilin A (CypA) chimera resulting from a CypA retrotransposition between exons 7 and 8 of the TRIM5 gene. Macaca nemestrina do not express TRIM5α; therefore, we asked whether these animals and related Old World primates express TRIMCyp. RT-PCR of total RNA from M. nemestrina and Macaca fascicularis yielded three TRIMCyp amplification products, one of which is predicted to encode a TRIMCyp chimera containing a full-length CypA. Unlike A. trivirgatus, genomic sequencing of M. nemestrina and M. fascicularis identifies a CypA retrotransposition in the 3′ untranslated region of the TRIM5 locus. There is ≈78% homology between the predicted protein sequences of Old World and New World primate TRIMCyp, with most of the differences found in the TRIM5-derived sequence. Notably, exon 7 is absent from both M. nemestrina and M. fascicularis TRIMCyp. Neither M. nemestrina nor M. fascicularis TRIMCyp could restrict HIV-1 or simian immunodeficiency virus SIVmac in an in vitro infectivity assay. The discovery of TRIMCyp in both M. nemestrina and M. fascicularis indicates that TRIMCyp expression may be more common among Old World primates than previously believed. Convergent evolution of TRIMCyp in both Old World and New World primates suggests that TRIMCyp may have provided evolutionary advantages.
Keywords: convergent evolution, cyclophilin A, HIV, retroviruses, TRIM5α
The current HIV pandemic resulting from cross-species transmissions of simian immunodeficiency viruses SIVcpz or SIVsmm to humans has been well documented (1). However, the mechanisms enabling such transmissions are not yet fully understood. Mammals have evolved several restriction factors capable of inhibiting the replication of certain retroviruses in a species-specific manner. One of the best described primate host-restriction factors is TRIM5α, which is expressed in most Old World primates.
Macaca mulatta TRIM5α exerts an early, postentry block to HIV-1 replication (2). TRIM5α is a member of the tripartite motif family of proteins, characterized by the ordered N-terminal to C-terminal expression of a RING domain, B-Box, and coiled coil, also known as an RBCC domain (3). The TRIM5α isoform also expresses a B30.2 domain at the C terminus, which is required for recognition of the incoming retroviral capsid (4–7). Changes to the B30.2 domain have been shown to dramatically affect the breadth and potency of TRIM5α-mediated anti-retroviral activity. For instance, the ability to restrict HIV-1 replication may be conferred to Homo sapiens TRIM5α by changing a single residue to the amino acid found in M. mulatta TRIM5α, R332P (8). Similarly, site-directed mutagenesis studies have demonstrated that mutations around the cyclophilin A (CypA) binding loop of HIV-1 capsid effect the potency of TRIM5α-mediated restriction, suggesting that the B30.2 domain interacts with or near the CypA binding loop (5, 9, 10).
New World primates Aotus trivirgatus exert a postentry restriction to HIV-1 mediated by a TRIM5–CypA chimera. Sequencing the A. trivirgatus TRIM5 gene identified a LINE-1-mediated retrotransposition of CypA into intron 7, resulting in the expression of a fusion protein called TRIMCyp, which is unique to the Aotus genus (11–13). TRIMCyp retains the N-terminal tripartite motif of all TRIM family members, but the B30.2 domain of TRIM5α is replaced by the CypA domain. Functionally, TRIM5α and TRIMCyp are similar, preventing reverse transcription of incoming viruses at an early postentry stage. However, TRIMCyp exerts a more potent restriction to incoming retroviruses than TRIM5α, and unlike TRIM5α, TRIMCyp-mediated restriction is sensitive to cyclosporin A. A. trivirgatus has been shown to express only TRIMCyp, not TRIM5α (11–12).
We recently demonstrated that the Old World primates Macaca nemestrina do not express TRIM5α. Instead, they transcribe novel isoforms TRIM5θ and TRIM5η (14). These isoforms likely arise because of a single-nucleotide polymorphism (SNP) at the intron 6 splice acceptor. TRIM5θ is truncated proximal to the B30.2 domain, and TRIM5η lacks 9 aa encoded by exon 7 located at the N terminus of the B30.2 domain. However, neither isoform restricts HIV-1 in vitro. We therefore asked whether M. nemestrina or related macaque species express TRIMCyp to compensate for the loss of TRIM5α-mediated retroviral restriction.
Results
Old World Primates M. nemestrina and Macaca fascicularis both Express TRIMCyp.
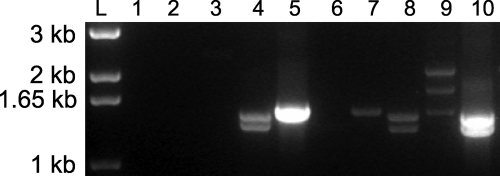
Total RNA was isolated from M. nemestrina, M. mulatta, and M. fascicularis. We performed RT-PCR by using a forward TRIM5-specific primer and a reverse CypA-specific primer. To facilitate later functional analyses, the forward primer was designed to replace the start codon with a hemagglutinin (HA) epitope tag. As expected, no amplification products were identified in M. mulatta total RNA. However, multiple amplification products were found in both M. nemestrina and M. fascicularis (Fig. 1).
Fig. 1.
RT-PCR amplification of TRIM5α and TRIMCyp from Old World primate total RNA. Total RNA was isolated from Owl monkey kidney (OMK) cells, M. mulatta PBMC, M. fascicularis PBMC, and M. nemestrina PBMC. Primers were designed to amplify either TRIM5α (odd lanes) or TRIMCyp (even lanes). Lane L, kb+ ladder; lane 1, dH2O TRIM5α; lane 2, dH2O TRIMCyp; lane 3, OMK TRIM5α; lane 4, OMK TRIMCyp; lane 5, M. mulatta TRIM5α; lane 6, M. mulatta TRIMCyp; lane 7, M. fascicularis TRIM5α; lane 8, M. fascicularis TRIMCyp; lane 9, M. nemestrina TRIM5; and lane 10, M. nemestrina TRIMCyp.
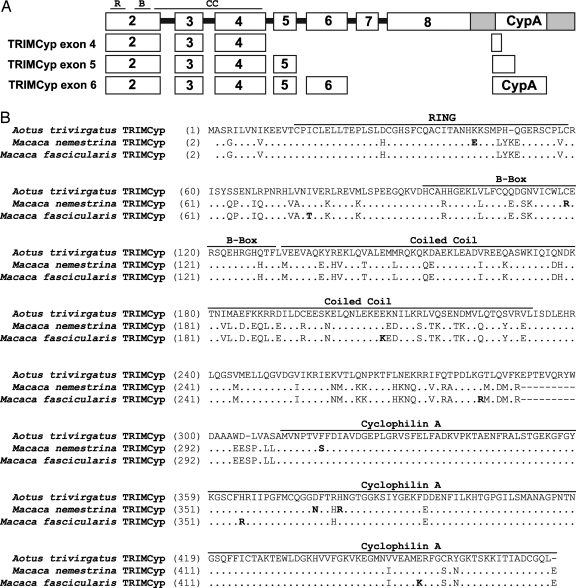
The amplification products from each species were gel-purified, cloned, and sequenced. Sequence analysis of each cDNA species confirmed that animals from both species expressed three TRIMCyp transcripts, with a CypA-coding region joined to exon 4, exon 5, or exon 6 of TRIM5. Because the 5′ portion of the CypA gene is 2 nt out of frame with its coding region, the TRIMCyp exon 4 and TRIMCyp exon 5 transcripts are predicted to encode fusion proteins with a truncated CypA domain (Fig. 2A). Only the TRIMCyp exon 6 transcript is predicted to express a TRIMCyp protein of 469 aa in M. nemestrina and M. fascicularis, comparable to the 475-aa TRIMCyp from A. trivirgatus (Fig. 2B). Therefore, although these observations are consistent with the description of multiple TRIMCyp isoforms in A. trivirgatus, the identities of the isoforms are different. The TRIMCyp isoform with the full-length CypA joined to exon 6 will be referred to as TRIMCyp.
Fig. 2.
Alignment of A. trivirgatus, M. nemestrina, and M. fascicularis TRIMCyp predicted amino acid sequences. (A) Schematic diagram (not to scale) of the genomic organization of the TRIM5 and CypA loci on chromosome 14 (top). The shaded region indicates the exon 8 3′ UTR. Exons encoded by each TRIMCyp splice variant are indicated below. R, RING; B, B-Box; CC, coiled coil. (B) Alignment of TRIMCyp exon 6 predicted amino acid sequences from M. nemestrina and M. fascicularis and the published A. trivirgatus TRIMCyp sequence. The N-terminal methionine is omitted in this alignment because the start codon was replaced with a 5′ HA epitope tag during cDNA amplification. The RING, B-Box, coiled coil, and CypA domains are indicated by labeled bars over the sequence. Amino acid differences between M. nemestrina and M. fascicularis are indicated by bold letters.
Old World Primate TRIMCyp Results from a Retrotransposition of CypA in the 3′ Untranslated Region (UTR) of the TRIM5 Gene.
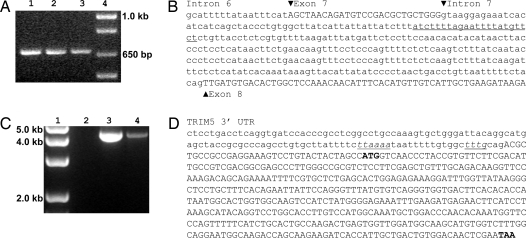
To determine the origin of the TRIMCyp transcripts observed in the Old World primates, we looked for a CypA retrotransposition in TRIM5. Genomic DNA was isolated from M. mulatta, M. nemestrina, and M. fascicularis peripheral blood monocytes (PBMC). We amplified a fragment from TRIM5 exon 6 to exon 8. The amplification product from all three species was identical in size (≈650 bp, Fig. 3A), indicating the absence of additional DNA sequences inserted in the M. fascicularis and M. mulatta TRIM5 intron 7. Sequence analysis of the TRIM5 locus shows that, unlike A. trivirgatus, there is no CypA retrotransposition in Old World primate TRIM5 intron 7 (Fig. 3B). We also identified a G-to-T SNP at the M. fascicularis TRIM5 intron 6 splice acceptor identical to a SNP that we previously found in M. nemestrina (14).
Fig. 3.
Genomic characterization of Old World primate TRIMCyp. (A) PCR amplification of the TRIM5 gene from M. mulatta, M. nemestrina, and M. fascicularis PBMC genomic DNA using a forward primer specific for exon 6 and a reverse primer specific for exon 8. Lane 1, M. mulatta; lane 2, M. fascicularis; lane 3, M. nemestrina; and lane 4, kb+ DNA ladder. (B) Sequence of the M. fascicularis TRIM5 gene from intron 6 to exon 8. The underlined sequence indicates the location corresponding to the site of CypA retrotransposition in A. trivirgatus. (C) PCR amplification of TRIM5 exon 8 and CypA. M. nemestrina and M. fascicularis PBMC genomic DNA was PCR-amplified with a forward primer specific for TRIM5 exon 5 and a reverse primer specific for CypA. Lane 1, kb+ DNA ladder; lane 2, dH2O; lane 3, M. fascicularis; and lane 4, M. nemestrina. (D) Sequence of the M. fascicularis TRIM5 3′ UTR and retrotransposed CypA. Lowercase letters indicate the TRIM5 3′ UTR. Italicized and underlined letters identify the conserved retrotransposition sequence. Capital letters identify the retrotransposed CypA sequence in TRIMCyp. Bold letters highlight the start and stop codons of the CypA coding sequence.
PCR amplification using a forward TRIM5 exon 5-specific primer and a reverse CypA-specific primer yielded an amplification product of ≈4.5 kb in M. nemestrina and M. fascicularis (Fig. 3C). Sequence analysis of this amplification product identifies a CypA gene in the 3′ UTR of the TRIM5 locus (Fig. 3D). Nucleotide sequences directly upstream of the CypA gene are consistent with consensus sequences used by LINE-1-mediated retrotransposition (15).
Alignment of Old World and New World TRIMCyp.
Alignment of the predicted amino acid sequences of A. trivirgatus, M. nemestrina, and M. fascicularis TRIMCyp identifies 97% homology between the two macaque species and 78–79% homology between the Old World and New World primates. The majority of the sequence diversity between A. trivirgatus and the macaque TRIMCyp is located in the RBCC region, with the coiled coil and the downstream linker 2 region accumulating most of the amino acid changes (Fig. 2B).
A comparison of Old World and New World primate TRIMCyp revealed two notable features between the linker 2 region and the CypA domain. One is the absence in Old World primate TRIMCyp of 9 aa (amino acids 231–240, EPTEVQRYW in A. trivirgatus) encoded by TRIM5 exon 7. This sequence is predicted to encode an α-helix, which is postulated to play an important role in maintaining the restriction function of TRIM5α (14, 16, 17). The other notable feature is the region immediately upstream of the CypA coding sequence. Alignment of the nucleotide sequences in this region with the CypA gene from H. sapiens (GenBank accession no. NM_021130) or from M. mulatta chromosome 14 (GenBank accession no. NW_001100384.1, nucleotides 666274–676274) (18, 19) indicates that this junction sequence is most likely derived from the sequences upstream of the CypA coding regions in their respective species, similar to what has been postulated for A. trivirgatus TRIMCyp (11, 12).
In contrast to the TRIM5-derived sequences, the CypA domain is well conserved between Old World and New World primate TRIMCyp. We compared these TRIMCyp CypA domains with the crystal structure of CypA complexed with 25-mer of HIV-1 Gag polyprotein, which includes the CypA binding loop (20). This crystal structure identifies two primary interacting regions on CypA equivalent to amino acids 357–375 and amino acids 404–428 in TRIMCyp. The C-terminal interacting region of the CypA domain is conserved among the three species. There are four amino acid differences in the N-terminal interacting region of the CypA domain. However, all of the residues that are postulated to interact directly with Gag (20, 21) are conserved in all three primate species (Fig. 2B). Comparison of CypA sequences in M. fascicularis TRIMCyp identifies differences at two amino acid residues, 356 (H/R) and 399 (I/T) (Figs. 2B and 3D), which most likely represents intraspecies polymorphism because these sequences were derived from different animals.
Neither M. nemestrina TRIMCyp nor M. fascicularis TRIMCyp Restrict HIV-1 in Vitro.
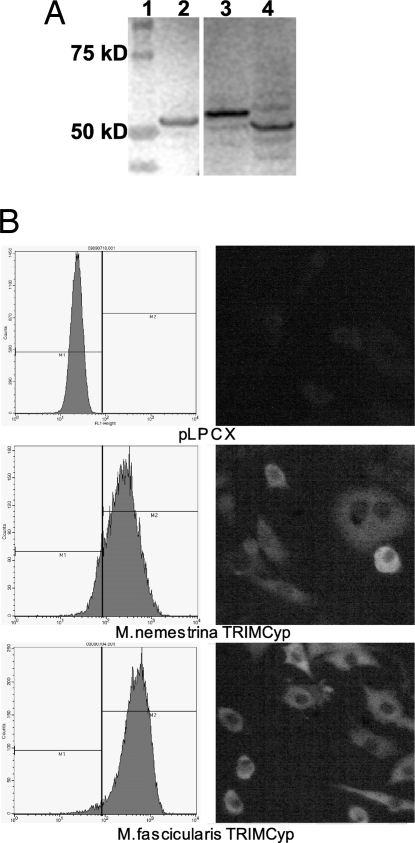
To evaluate the ability of these Old World primate TRIMCyp proteins to restrict HIV-1, TRIMCyp from M. nemestrina and M. fascicularis was N-terminally HA-tagged and stably transduced into CrFK cells. Expression and intracellular localization of each HA-tagged TRIMCyp isoform was examined by immunoblotting, flow cytometry, and immunofluorescence analyses. Immunoblotting analysis confirmed TRIMCyp expression at similar levels for each isoform tested (Fig. 4A). Flow cytometry confirmed that the majority of CrFK cells express the transduced gene, and immunofluorescence analysis established that each TRIMCyp isoform was expressed in the cytoplasm (Fig. 4B).
Fig. 4.
Expression of TRIMCyp by stably transformed CrFK cells. (A) Immunoblotting analysis of HA-tagged TRIMCyp proteins in CrFK whole-cell lysates. Lane 1, Protein Plus ladder; lane 2, M. nemestrina TRIMCyp; lane 3, M. mulatta TRIM5α; and lane 4, M. fascicularis TRIMCyp. (B) Analysis of stably transduced CrFK cells by flow cytometry (Left) and immunofluorescence assays (Right). The stably transduced TRIMCyp protein is indicated below each image. A monoclonal anti-HA Alexa Fluor conjugate 488 antibody was used in both assays to label the HA-tagged TRIMCyp in each cell line. The percentages of cells expressing M. nemestrina and M. fascicularis TRIMCyp are 85% and 94%, respectively.
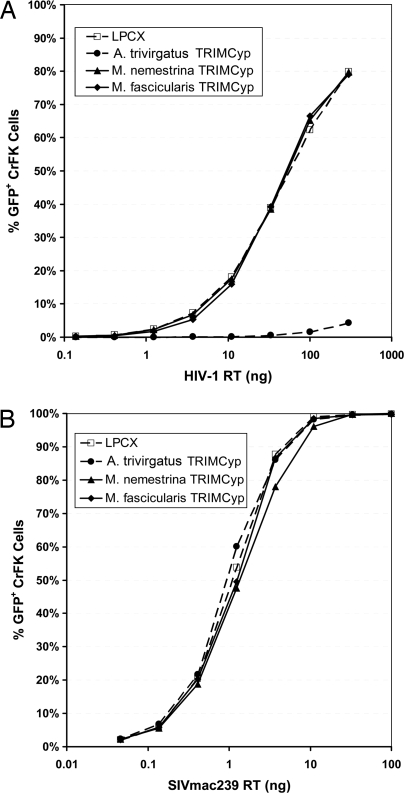
We infected these TRIMCyp-expressing CrFK cells with HIV-1 pNL4–3Δenv-eGFP or env-minus SIVmac239 enhanced GFP (eGFP) pseudotyped with vesicular stomatitis virus (VSV)-G. GFP-positive CrFK cells were quantified 48 h postinfection by flow cytometry analysis. As expected, A. trivirgatus TRIMCyp inhibited HIV-1 replication in vitro ≈100-fold. However, neither M. nemestrina TRIMCyp nor M. fascicularis TRIMCyp inhibited the replication of HIV-1 in these cell lines (Fig. 5A). As expected, none of the TRIMCyp isoforms tested restricted SIVmac239 in vitro (Fig. 5B).
Fig. 5.
Infectivity of HIV-1 and SIVmac in CrFK cells stably transduced with TRIMCyp. Serial dilutions of VSV-G pseudotyped HIV-1 pNL4–3-Δenv-eGFP (A) or env-minus SIVmac239-eGFP (B) were used to infect stably transduced CrFK cells. Forty-eight hours after infection, the percentage of GFP-positive cells was determined by flow cytometric analysis.
Discussion
We report the expression of TRIMCyp molecules in two Old World primates, M. nemestrina and M. fascicularis. Previously, the only published description of TRIMCyp expression was from the Aotus genus of New World primates. Subsequent to our initial finding, we became aware of M. nemestrina TRIMCyp cDNA sequences deposited in GenBank by Su and colleagues (GenBank accession nos. DQ308404–DQ308406). In this communication, we corroborate the expression of TRIMCyp not only in M. nemestrina but also in M. fascicularis. We also examine the genetic structure of the TRIMCyp transcript and the functionality of TRIMCyp proteins from both species. Unlike New World primates, in which TRIMCyp arose through CypA retrotransposition into intron 7 of the TRIM5 gene, the CypA domain in Old World primates resulted from retrotransposition into the 3′ UTR of the TRIM5 locus. The discovery of TRIMCyp in these animals raises the possibility that expression of these molecules may be more prevalent in primate species than originally thought.
Coexpression of TRIM5 and TRIMCyp transcripts in M. nemestrina and M. fascicularis most likely results from alternative splicing. We previously reported that the M. nemestrina TRIM5 gene contains a G-to-T SNP altering the canonical splice acceptor of intron 6. The use of alternative downstream splice acceptors results in two novel TRIM5 isoforms (14). In this article, we report that the same SNP exists in the intron 6 splice acceptor of M. fascicularis TRIM5. This polymorphism therefore appears to be more common among Old World monkeys than previously thought, because the same G-to-T SNP also has been observed in a minority of M. mulatta (W. Johnson, personal communication). Alternative splicing also may occur in animals that retain the canonical intron 6 splice acceptor. Because this acceptor sequence (AG) is followed by another AG dinucleotide in the exon 7 of most macaque TRIM5 genes examined, the tandem repeat of AG dinucleotides may cause ambiguity in splice acceptor site utilization, resulting in alternative splicing. In support of this notion, a 2-nt deletion has been identified in TRIM5α transcripts from M. mulatta that are homozygous in their AG intron 6 sequences (T. Kodama, personal communication).
In contrast to A. trivirgatus TRIMCyp, macaque counterparts identified in this study failed to restrict HIV-1. Alignment of the predicted protein sequences identifies significant sequence diversity between Old World and New World primate TRIMCyp, with most differences found in the TRIM5-derived sequence. It remains to be determined whether and to what extent sequence diversity in this region contributes to the difference in the ability of New World and Old World primate TRIMCyp to restrict HIV-1. However, TRIMCyp chimerae containing non-TRIM5 RBCC domains have been shown to restrict HIV-1 despite the low homology between the RBCC domains of the chimerae and the A. trivirgatus TRIMCyp (22), suggesting the structure of the RBCC domain may be more important than the primary sequence. Old World primates also differ from A. trivirgatus by the lack of exon 7 in their TRIMCyp sequence. This exon is predicted to encode an α-helix that may play an important role in the structure and function of TRIM5α (16, 17). M. nemestrina TRIM5η excludes exon 7 and is incapable of restricting HIV-1 in vitro; however, a chimera composed of H. sapiens exons 2–7 and M. nemestrina exon 8 acquires the ability to restrict HIV-1 (14, 23). Similarly, the linker 2 region, which includes exon 7, also has been shown to contribute to the ability of CypA constructs to restrict HIV-1 (24) Therefore, the role of exon 7 sequences in maintaining the functionality of TRIM5 and TRIMCyp proteins warrants further examination.
Although we have yet to confirm the in vivo expression of Old World primate TRIM5 and TRIMCyp at the protein level, the possibility of coexpression of these proteins in M. nemestrina and M. fascicularis may have interesting biological implications. A previous study has demonstrated that coexpression of New World primate TRIMCyp and human TRIM5α prevents TRIM5α-mediated restriction of “N-tropic” strains of the murine leukemia virus (N-MLV) (25). This inhibition requires interactions between the two proteins, suggesting heteromultimerization disrupts the structure necessary for recognition of HIV-1 capsid. For this reason, it will be of interest to determine whether both TRIM5α and TRIMCyp are indeed expressed in M. fascicularis and, if so, whether coexpression of these molecules may inhibit or modulate TRIM5α-mediated restriction.
The functional significance of TRIMCyp in Old World primates described here is unknown. Despite their failure to restrict HIV-1 or SIV replication, it remains to be seen whether macaque TRIMCyp will restrict other retroviruses. Alternatively, Old World primate TRIMCyp may represent an evolutionary vestige resulting from ancient retroviral challenges, similar to a function recently ascribed to H. sapiens TRIM5α (26). If so, the different strategies used by Old World and New World primates to express TRIMCyp molecules may represent an example of convergent evolution that resulted in competitive advantage for these very divergent primate species against retroviral challenges. Further studies will be needed to elucidate the evolutionary and functional role of these molecules.
Materials and Methods
Animals.
Three M. nemestrina, two M. fascicularis, and two M. mulatta housed at the Washington National Primate Research Center were used in this study. Whole blood was collected from these animals in heparinized Vacutainer tubes in accordance with an Institutional Animal Care and Use Committee approved protocol.
RNA/DNA Samples.
The TRIMCyp gene from M. nemestrina and M. fascicularis was cloned and sequenced at genomic and cDNA levels. PBMC were isolated from heparinized whole blood by Lymphoprep density centrifugation. Total RNA was isolated from 1 × 107 PBMC by using the RNeasy Mini Kit (Qiagen) following the manufacturer's suggested protocol. DNA was isolated from 5 × 106 PBMC with the Puregene DNA Purification Kit (Gentra Systems) following the manufacturer's suggested protocol.
PCR and Cloning: cDNA and Genomic DNA Amplification.
Amplification of TRIMCyp cDNA from total RNA was performed by using the SuperScript III One-Step RT-PCR with Platinum Taq kit (Invitrogen) using primers XhoIHA5TRIM (14) and CypA*RMCSNotI (5′-GTATATGCGGCCGCTTATTCGAGTTGTCCACAGTCAG-3′). Primers MneT5Aex51.23 (5′-GGTGTGGATGGCATCATTAAAAG-3′) and CypA*RMCSNotI were used to amplify a fragment of the TRIM5 gene from PBMC genomic DNA with Platinum PCR Super Mix High Fidelity (error rate: 1.8 ± 0.4 × 10−6; Invitrogen). Approximately 100 ng of total RNA or DNA from each macaque and 0.4 μM concentrations of each primer were used in each reaction. The RT-PCR assay was incubated at 55°C for 30 min, then 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 90 s, then a 68°C hold for 5 min and storage at 4°C. The PCR assay was incubated at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, and 68°C for 10 min, then a 68°C hold for 10 min and storage at 4°C. One microliter of undiluted PCR product from each amplification was cloned by using the Strataclone PCR cloning kit (Stratagene) following the manufacturer's suggested protocol.
Sequencing and Data Analysis: cDNA and Genomic Sequencing.
Nine cDNA clones from three M. nemestrina, three cDNA clones from one M. fascicularis, and one genomic clone from each species were sequenced by the DNA Sequencing and Gene Analysis Center (Department of Pharmaceutics, University of Washington) using the Big Dye V 3.1 reaction mix (Applied Biosystems) on an Applied Biosystems 3100 Genetic Analyzer. Primers M13F, T583.100, T5F356.375, MneT5ex4F, MneT5ex6F, T5R217.198, MneT5ex4R, MneT5ex6R (14), MneT5CyF1 (5′-GTTCTTCGACATTGCCGTCG-3′), MneT5CyR1 (5′-TTCTGTGAAAGCAGGAACCC-3′), and MneT5CyR2 (5′-CTCTCCTGAGCTACAGAAGG-3′) were used to sequence cDNA clones, and primers T5Aex51.23, MneT5Int5F2 (5′-CCTCTCTTGATATGTCTCAG-3′), MneT5ex6F, T5F1205.1224 (5′-GCTTCCAACCTGATGCAATG-3′), MneT5ex8F, MneT5CyF1, MneT5CyF2, CypA*RMCSNotI, MneT5CyR1, MneT5CyR2, MneT5ex8R1, and MneT5ex6R were used to sequence genomic clones (14).
Sequencing products were assembled with Sequencher 4.1 (Gene Codes). The resulting genetic and predicted amino acid sequences were analyzed by using Vector NTI Advance 10.1.1 (Invitrogen).
Stable Transduction of CrFK Cells with TRIMCyp Genes.
CrFK and 293T cells were cultured in DMEM/10% FBS. TRIMCyp cDNA with a 5′ HA tag was cloned into the pLPCX retroviral expression vector. 293T cells were used to produce retroviral vectors as described in ref. 27. These supernatants were used to infect CrFK cells, and 24 h after infection, 4 μg/ml puromycin was added to the medium to select for cells stably transduced with the gene of interest.
Immunoblotting.
HA-tagged TRIMCyp proteins expressed by stably transduced CrFK cells were detected by Western blotting of whole-cell lysates. Cells were lysed in Laemlli-SDS sample buffer (Bio-Rad). Samples were normalized by total protein (OD280) concentration. The SDS/PAGE was performed on a 4–12% NuPAGE denaturing gel (Invitrogen), and separated bands were transferred onto a nitrocellulose membrane (Bio-Rad). The primary antibody for detection of the HA tag was HA.11 (Covance). After the binding of primary antibody, the membrane was incubated with a goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Sigma–Aldrich).
Flow Cytometry and Immunofluorescence Assay.
Expression and localization of HA-tagged TRIMCyp proteins inside the expressing cells was analyzed by staining with anti-HA Alexa Fluor 488 conjugate (Invitrogen). Stably transduced CrFK cells were permeabilized by using the BD Cytofix/Cytoperm Plus intracellular staining kit (Becton Dickinson). An aliquot of 106 cells was analyzed by flow cytometry with a FACScan (Becton Dickinson) to enumerate cells expressing the HA-tagged TRIM5 protein. Another aliquot of cells was grown on glass slides, permeabilized, and stained as described above, then analyzed and photographed on an inverted microscope (Leica Microsystems) with a green fluorescence filter set.
Infectivity Assays.
CrFK cells stably transduced with TRIMCyp or empty vector were seeded in 6-well culture plates in DMEM, 10% FBS, and 4 μg/ml puromycin at a density of 2 × 105 cells per well. After 24 h, the cells were washed in PBS and infected with 3-fold dilutions of VSV-G pseudotyped HIV-1 pNL4–3 or SIVmac239, prepared as described in ref. 28, in 1 ml of DMEM, 10% FBS, and 5 μg/ml polybrene. Two hours postinfection, the virus-containing supernatant was replaced with 2 ml of DMEM and 10% FBS. At 48 h postinfection, infected cells expressing GFP were enumerated by flow cytometric analysis using a FACScalibur (Becton Dickinson) and analyzed by FlowJo software (Tree Star, Inc.).
ACKNOWLEDGMENTS.
We thank Michael Emerman (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing the A. trivirgatus TRIMCyp CrFK cells and plasmids mGP, pL-VSV-G, and pCMV tat; Welkin Johnson and Ruchi Newman for their help with the establishment of stably transduced CrFK cells; and David Evans (New England National Primate Research Center, Southborough, MA) for providing the env-minus SIVmac239 eGFP plasmid. This work was supported by National Institutes of Health Grants P51 RR00016 (to the Washington National Primate Research Center) and K08 AI061738 (to G.B.).
Footnotes
References
- 1.Sharp PM, et al. The origins of acquired immune deficiency syndrome viruses: Where and when? Philos Trans R Soc London B. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 3.Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens CM, et al. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owens CM, Yang PC, Gottlinger H, Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human TRIM5α leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Ylinen LM, Keckesova Z, Wilson SJ, Ranasinghe S, Towers GJ. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5α alleles. J Virol. 2005;79:11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, et al. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5α. J Virol. 2007;81:7280–7285. doi: 10.1128/JVI.00406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro IP, et al. Evolution of cyclophilin A, TRIMCyp retrotransposition in New World primates. J Virol. 2005;79:14998–15003. doi: 10.1128/JVI.79.23.14998-15003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan G, Kodama T, Kozyrev Y, Hu SL. Novel TRIM5 isoforms expressed by Macaca nemestrina. J Virol. 2007;81:12210–12217. doi: 10.1128/JVI.02499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo JS, et al. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24:967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. Cyclophilin A complexed with a fragment of HIV-1 gag protein: Insights into HIV-1 infectious activity. Structure (London) 1997;5:139–146. doi: 10.1016/s0969-2126(97)00172-x. [DOI] [PubMed] [Google Scholar]
- 21.Braaten D, Ansari H, Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap MW, Dodding MP, Stoye JP. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J Virol. 2006;80:4061–4067. doi: 10.1128/JVI.80.8.4061-4067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javanbakht H, et al. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. 2007;367:19–29. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berthoux L, Sebastian S, Sayah DM, Luban J. Disruption of human TRIM5α antiviral activity by nonhuman primate orthologues. J Virol. 2005;79:7883–7888. doi: 10.1128/JVI.79.12.7883-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5α antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 27.Bartz SR, Vodicka MA. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 28.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]