Abstract
Operons are widespread in prokaryotes, but are uncommon in eukaryotes, except nematode worms, where ∼15% of genes reside in over 1100 operons in the model organism Caenorhabditis elegans. It is unclear how operons have become abundant in nematode genomes. The “one-way street” hypothesis asserts that once formed by chance, operons are very difficult to break, because the breakage would leave downstream genes in an operon without a promoter, and hence, unexpressed. To test this hypothesis, we analyzed the presence and absence of C. elegans operons in Caenorhabditis briggsae, Caenorhabditis remanei, and Caenorhabditis brenneri, using Pristionchus pacificus and Brugia malayi as outgroups, and identified numerous operon gains and losses. Coupled with experimental examination of trans-splicing patterns, our comparative genomic analysis revealed diverse molecular mechanisms of operon losses, including inversion, insertion, and relocation, but the presence of internal promoters was not found to facilitate operon losses. In several cases, the data allowed inference of mechanisms by which downstream genes are expressed after operon breakage. We found that the rate of operon gain is ∼3.3 times that of operon loss. Thus, the evolutionary dynamics of nematode operons is better described as “easy come, slow go,” rather than a “one-way street.” Based on a mathematic model of operon gains and losses and additional assumptions, we projected that the number of operons in C. elegans will continue to rise by 6%–18% in future evolution before reaching equilibrium between operon gains and losses.
An operon is a cluster of linked genes that are under the control of a single promoter and are transcribed into one polycistronic mRNA (Jacob et al. 1960). Operons are prevalent in prokaryotic genomes (Salgado et al. 2000; Ermolaeva et al. 2001), but are uncommon in eukaryotes, with the exception of the phylum Nematoda (Spieth et al. 1993; Zorio et al. 1994; Evans et al. 1997; Blumenthal et al. 2002; Lee and Sommer 2003; Guiliano and Blaxter 2006). For example, about 15% of genes in Caenorhabditis elegans, a model organism belonging to Nematoda, reside in operons (Blumenthal et al. 2002; Blumenthal and Gleason 2003). For two reasons, nematode and prokaryotic operons are believed to have separate origins. First, gene compositions of nematode and prokaryotic operons are unrelated (Huynen et al. 2001). Second, in nematodes, the polycistronic pre-mRNA is processed by spliced-leader (SL) trans-splicing to generate monocistronic mRNAs (Spieth et al. 1993; Zorio et al. 1994; Blumenthal et al. 2002), which are then translated individually. In SL trans-splicing, SL RNA donates a short (∼15–50 nucleotides) leader sequence to pre-mRNA splice-acceptor sites and becomes the 5′ end of the mature mRNA (Krause and Hirsh 1987; Hastings 2005). In fact, 70% of C. elegans genes, including many genes outside operons, are trans-spliced (Blumenthal 2005). Within an operon, the most upstream gene is either not trans-spliced or trans-spliced by SL1 RNA, whereas the downstream genes are trans-spliced primarily by SL2 RNA, but occasionally by SL1 RNA (Spieth et al. 1993; Blumenthal and Steward 1997; Blumenthal et al. 2002). In contrast, in prokaryotes, polycistronic mRNAs are translated without being first cut into separate mRNAs.
The evolutionary dynamics of operons in prokaryotes has been well studied (Itoh et al. 1999; Lawrence 2002; Price et al. 2006), and several models have been proposed to explain the prevalence of operons in prokaryotes (Lawrence and Roth 1996; Lawrence 1999). However, these models are not applicable to nematode operons. For example, the natal model claims that genes found in operons are in situ duplicates (Lawrence 1999); but in C. elegans, most genes in the same operons are not paralogous to one another (Lercher et al. 2003), and most new duplicates are tandem inverted copies (Katju and Lynch 2003) that cannot form operons (Cavalcanti et al. 2006). The coregulation model postulates that coregulation of functionally related genes by a single promoter (as in operons) is beneficial. Indeed, prokaryotic operons often consist of genes involved in the same metabolic pathways that need to be coregulated (Lawrence 2002). But in C. elegans, constituent genes of an operon are rarely related in function and ubiquitously expressed housekeeping genes are over-represented in operons (Blumenthal 1998; Blumenthal and Gleason 2003), inconsistent with the coregulation model. The selfish operon model asserts that the organization of genes into operons is beneficial to constituent genes because proximity allows horizontal cotransfer of all genes required for a selectable phenotype (Lawrence and Roth 1996). Because nematodes experience much fewer, if any, horizontal gene transfers, the selfish operon model is apparently inapplicable to nematodes.
Then, why are there so many operons in nematodes? Lawrence proposed that the abundance of operons in nematodes is due to the parasitization of the trans-splicing machinery (Lawrence 1999). After the trans-splicing machinery invades a species and a few important genes adopt trans-splicing, the machinery can be stably retained. This is because in a gene that adopts trans-splicing, the DNA sequence between the promoter and the trans-splicing site becomes unconstrained, allowing out-of-frame ATGs to accumulate, which would prevent the correct reading frame from being translated if trans-splicing were lost (Blumenthal 2004). Because it is difficult for a gene to abandon trans-splicing once adopted by chance, trans-splicing gradually accumulates in the genome. Trans-splicing makes the origin of operons possible because it allows pre-mRNAs to escape exonucleolytic degradation (Blumenthal 2004). Subsequently, adjacent genes, regardless of their functional relationship, will form an operon by chance as long as the organism tolerates the coexpression of the constituent genes. Once formed, an operon is very difficult to break, because the downstream genes in the operon would have no promoters, and hence, become unexpressed upon breakage (Blumenthal and Gleason 2003). In other words, operons are protected from breakage by natural selection. If operons can only be gained but not lost, their abundance in the genome only requires sufficient evolutionary time. Thus, in theory, this “one-way street” hypothesis (Nimmo and Woollard 2002; Blumenthal 2004) can explain the prevalence and preservation of operons in nematodes. Although previous case studies identified several evolutionarily conserved (Evans et al. 1997) and nonconserved (Lee and Sommer 2003) operons, the general pattern of operon evolution is unclear. A recent genomic comparison showed that 96% of C. elegans operons are preserved in C. briggsae, much higher than the random expectation (Stein et al. 2003). However, it is unknown whether the 4% of the C. elegans operons that are not preserved in C. briggsae represent newly formed operons in C. elegans or newly broken operons in C. briggsae. In this study, we critically test the one-way street hypothesis by examining the evolutionary dynamics of nematode operons using the available genome sequences of C. elegans and five related nematodes (C. briggsae, C. remanei, C. brenneri, Pristionchus pacificus, and Brugia malayi) and uncover molecular mechanisms of operon gains and losses.
Results
Identification of operon losses
The phylogenetic relationships among C. elegans, C. briggsae, C. brenneri, C. remanei, P. pacificus, and B. malayi, the six nematode species with draft genome sequences, have been relatively well established (Blaxter et al. 1998; Cho et al. 2004; Kiontke et al. 2004) (Fig. 1). Furthermore, almost all operons of C. elegans have been identified, based on experimental examination of trans-splicing (Blumenthal et al. 2002). At the time of our study, 1133 C. elegans operons were annotated, making it possible to study the gains and losses of C. elegans operons using a comparative genomic approach. We first studied evolutionary losses of C. elegans operons in other nematodes. Because the DNA sequence divergences of P. pacificus and B. malayi from C. elegans are very large (Holterman et al. 2006), we limited the analysis of operon gains and losses primarily to the four Caenorhabditis species to ensure correct identification of orthologous genes and operons, but used P. pacificus, and B. malayi as outgroups when necessary.
Figure 1.
Gains and losses of operons in Caenorhabditis nematodes. The numbers of C. elegans operons showing various presence/absence phylogenetic distributions are given at the top of the figure, with the inferred numbers of operon gains (circled) and losses presented on tree branches. There are no C. elegans operons that are absent in all of the other three Caenorhabditis species, but present in one or two of the outgroups (P. pacificus and B. malayi). A pair of linked black circles shows the presence of an operon, while a pair of unlinked white circles shows the absence of the operon. Dashes show undetermined status.
If a C. elegans operon is also found in C. remanei and C. brenneri, but is broken in C. briggsae, we can infer, based on the parsimony principle, that the operon was lost in the C. briggsae lineage since its separation from the C. remanei lineage (Fig. 1). Because the probability that the same set of genes form an operon more than once during the evolution of Caenorhabditis is exceedingly small, use of the parsimony principle is justifiable here. Similarly, we can infer losses of C. elegans operons in the C. remanei lineage and the C. brenneri lineage, respectively (Fig. 1). Here, the presence and absence of a C. elegans operon in another species is based on a series of genomic sequence analyses detailed in the Methods section. Based on this analysis, we identified 13 operons that were lost in the C. briggsae lineage, nine operons lost in the C. remanei lineage, and nine operons lost in the C. brenneri lineage (Fig. 1).
If a C. elegans operon is present in C. brenneri, but is broken in C. briggsae and C. remanei, the most parsimonious scenario is that the operon was lost in the common ancestor of C. briggsae and C. remanei, and three such cases were identified (Fig. 1). We also observed four cases where a C. elegans operon is present in C. remanei, but is broken in C. brenneri and C. briggsae. To explain this pattern, we have to invoke two evolutionary events: two losses, two gains, or one loss and one gain. Because genes in the same operons are usually unrelated in function and because there are more than 19,000 genes in C. elegans, the probability is extremely low for the same two genes that are initially unlinked to form operons more than once. In fact, in none of the above four operons are the constituent genes linked in C. brenneri or C. briggsae. Therefore, the most probable explanation of such a pattern is independent losses of the operons in C. brenneri and C. briggsae (Fig. 1) (also see Discussion). Similarly, we identified three cases where a C. elegans operon is present in C. briggsae, but is broken in C. brenneri and C. remanei (Fig. 1). If a C. elegans operon is broken in C. briggsae, C. remanei, and C. brenneri, but is present in at least one of P. pacificus and B. malayi, we can infer that the operon was lost in the common ancestor of C. briggsae, C. remanei, and C. brenneri. But no such operons were identified (Fig. 1).
In the above analysis, an operon breakage in a species was inferred only when all of its constituent genes were still present in the genome, because we are interested in the separation of constituent genes of an operon, rather than gene loss. However, there is a small probability that although all of the constituent genes are present, some of them have become pseudogenes. For example, after the operon breakage, the downstream gene in a two-gene operon may be pseudogenized, equivalent to gene loss. Using GeneWise (Birney et al. 2004), we examined whether the open reading frame (ORF) was still intact for each downstream gene in the broken operons identified above, and found only two cases where the ORFs were disrupted. They are F46B6.6 in C. remanei and F37C12.2 in C. briggsae. Our resequencing of these genes showed that in both cases the disruptions were due to sequencing errors in the draft genome sequences. Thus, all operon losses we identified represented genuine separations of constituent genes. The list of all operon losses identified here is given in Supplemental Table S1.
Identification of operon gains
If a C. elegans operon is not found in any of C. briggsae, C. remanei, C. brenneri, P. pacificus, and B. malayi, it is most likely that it originated in the C. elegans lineage, since its separation from the common ancestor of C. briggsae, C. remanei, and C. brenneri. In total, 56 such newly formed C. elegans operons were identified (Fig. 1). Note that 15% of the constituent genes of these 56 C. elegans operons could not be identified in P. pacificus and B. malayi, due to the high divergences of these two species to C. elegans, but were assumed to be present in these outgroups, which may slightly inflate the estimated number of operon gains. We limited our analysis to C. elegans operons that were formed in the exterior C. elegans branch of the nematode phylogeny (Fig. 1), because the operon losses we identified above also took place within Caenorhabditis, and thus a comparison between the rates of operon gains and losses is possible. The list of all operon gains identified here is given in Supplemental Table S1.
Rates of operon gains and losses
Based on the presence and absence of each C. elegans operon in the other five nematode species, we mapped the operon gain and loss events onto the nematode phylogeny (Fig. 1). From the common ancestor of the four Caenorhabditis species to present, 56 operons were formed in the C. elegans lineage. During the same period of time, on average, {[(17 + 12)/2 + 3]+16}/2 = 16.75 operons were broken per lineage in the other three Caenorhabditis species (Fig. 1). If this breakage rate is similar to that in C. elegans, one can infer that the ratio between the numbers of operon gains and losses is 56/16.75 = 3.34, suggesting that the number of operons will continue to rise in Caenorhabditis evolution.
It has been estimated that the separation between C. elegans and C. briggsae occurred T = 80 to 110 million years ago (Mya) (Coghlan 2003; Stein et al. 2003; Hillier et al. 2007). Thus, the rate of operon formation is Rgain = 56/T = 0.51–0.70 operons per million years (Myr). Of the 1133 C. elegans operons annotated, 12 did not have protein sequences in at least one of the constituent genes, and therefore, were not used in our analysis. For an additional 98 operons, their gains and losses could not be unambiguously determined for various reasons (see Methods). Therefore, the actual number of operons examined for evolutionary dynamics was 1133 − 12 − 98 = 1023. Among them, 1023 − 56 = 967 were present in the most recent common ancestor of C. elegans and C. briggsae. Thus, the rate of operon loss is Rloss = (16.75/967)/T = 0.0173/T = 1.57–2.17 × 10−4 per operon per Myr.
It is conceivable that not all genes may be included in operons. If we assume that only a fraction (f) of all genes in the genome can potentially be included in operons, and that these genes form K operons, the rate of operon gain when there are already N operons in the genome may be modeled by the following function
where r is the intrinsic rate of operon gain. Assuming the constancy of r and Rloss, we have the differential equation
where Nt is the number of operons in the genome at time t. It can be shown that an equilibrium (i.e., dNt/dt = 0) will be reached when the number of operons in the genome becomes r/(Rloss+r/K) = 1/(0.00031 + 0.65/K). At that time, the numbers of operon gains and losses per Myr will be equal. Integrating Equation 2 gives
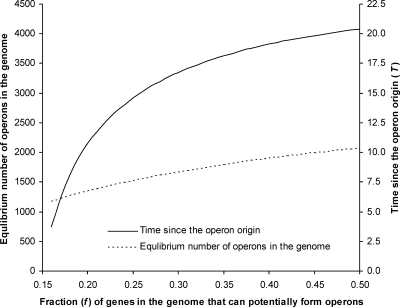
where N1 and N2 are the numbers of operons at time t1 and t2, respectively. Equation 3 can be used to estimate the time of operon origination (t1) when we plug in t2 = 0 (present), N1 = 0, N2 = 1133, and a K value. If we assume that the average size of 2.6 genes per operon (Blumenthal et al. 2002) in C. elegans remains unchanged, the proportion of genes that can potentially form operons is f = 2.6K/19,427, where 19,427 is the total number of genes in the C. elegans genome. Figure 2 shows the relationship between f and the equilibrium number of operons in the genome, and that between f and the estimated time of operon origin, respectively. For example, if f is 20%, the equilibrium number of operons is estimated to be 1344, ∼19% greater than the number of operons in present-day C. elegans, and operons are estimated to have originated 10.7T = 856–1177 Myr ago. The actual f value, however, is unknown (see Discussion).
Figure 2.
Relationship between the proportion (f) of genes that can potentially be included in operons and the projected number of operons in the genome at equilibrium (left Y-axis) and that between f and the predicted evolutionary time since the origin of operons (right Y-axis), in unit of T, the divergence time between C. elegans and C. briggsae.
Molecular evolutionary mechanisms of operon breakage
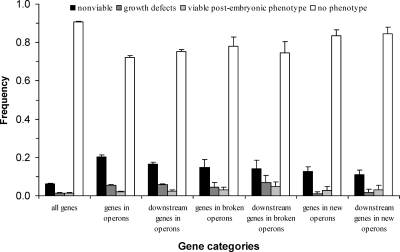
Although operon losses are indeed infrequent, a total of 48 losses (of 41 operons) have been observed in our analysis. It is interesting to ask whether the C. elegans operons that were lost in one or more other species are a special group of operons whose constituent genes are less important than those of other operons. To address this question, we used gene knockdown phenotypes from RNA interference (RNAi) experiments to measure gene importance, and then compared gene importance for the broken operons and all other operons of C. elegans. The knockdown phenotypes of 16,564 C. elegans genes (86% of the 19,427 predicted genes in the genome) have been examined in a systematic RNAi experiment (Kamath et al. 2003). Among them, 1813 genes belong to operons. We found that the frequencies of genes with the nonviable phenotype, growth defects phenotype, and viable post-embryonic phenotype are all higher for genes inside operons than outside of operons (Fig. 3). Thus, as shown in an earlier study (Blumenthal and Gleason 2003), genes within operons are more important than average genes in the genome. However, no significant difference in gene importance was observed between genes in broken operons and those in other operons (P = 0.10, χ2 test; Fig. 3). Because the breakage of an operon affects the downstream genes in the operon more than the upstream gene, we compared the downstream genes between broken operons and other operons, but again found no significant difference (P = 0.92; Fig. 3). Hence, constituent genes of broken operons are as important as those of other operons. In other words, the broken operons also contain many important genes, including essential genes. Because breakage of an operon would leave the downstream genes in the operon without promoters and unexpressed, the breakage should be deleterious and prohibited from fixation by purifying selection. It is thus of significant interest to identify the molecular mechanisms responsible for the successful operon breakages that occurred in evolution. Although many chromosomal rearrangements have happened in the evolution of Caenorhabditis and the flanking genes of an operon may have changed during evolution, it is still possible to infer the mechanisms of operon breakage accurately in a number of cases. Below we describe three mechanisms inferred from the observations.
Figure 3.
Proportions of genes showing different RNAi phenotypes in different gene categories. The phenotypic data are from Kamath et al. (2003). Error bars show one standard error.
Inversion
The C. elegans operon CEOP2520 is broken in C. briggsae (Fig. 4A). The breakage appears to be due to a chromosomal inversion that includes two genes, one of which (lsm-1) is the first gene in CEOP2520, while the other (rps-9) is outside of the operon. The inversion made the transcriptional direction of lsm-1 opposite that of F40F8.1 and F40F8.3, the downstream genes in the operon. Because lsm-1 is the first gene in CEOP2520 and has a promoter, the inversion should not affect its expression. But how can F40F8.1 and F40F8.3 still be expressed in C. briggsae? We hypothesized that after the inversion, rps-9 may become the first gene in the operon, so that its promoter is used to transcribe itself as well as F40F8.1 and F40F8.3. Consistent with this hypothesis, we were able to amplify C. briggsae F40F8.1 and F40F8.3 by RT–PCR using gene-specific primers and a SL2 primer (Fig. 5). Although we were also able to amplify F40F8.1 using a gene-specific primer and a SL1 primer, the amplification was much weaker (Fig. 5). These results indicate that C. briggsae F40F8.1 primarily uses SL2 spicing and F40F8.3 uses exclusively SL2 splicing. Although downstream genes in operons can use SL1 or SL2 splicing, SL2 is used solely in downstream genes. Thus, our data indicate that both C. briggsae F40F8.1 and F40F8.3 are downstream genes in an operon. The intergenic distances between constituent genes within an operon (i.e., from the stop codon of a gene to the translational start codon of the next gene) tend to be shorter than 2 kb (see Discussion and Supplemental Fig. S1). In the present case, the intergenic distance between rps-9 and F40F8.1 is 1.3 kb in C. briggsae, consistent with the general pattern. The rps-9 gene encodes a small ribosomal subunit S9 protein and lsm-1 encodes a small nuclear ribonucleoprotein splicing factor; both appear to be housekeeping genes. This fact explains why switching the first gene in the operon from lsm-1 to rps-9 was acceptable in evolution, although neither lsm-1 nor rps-9 has an apparent functional relationship with F40F8.1 (uridylate/adenylate kinase) or F40F8.3 (unknown function). Interestingly, lsm-1 and rps-9 have significantly different expression responses to various environmental stimuli in C. elegans (Kim et al. 2001). Thus, the expression responses of F40F8.1 and F40F8.3 in C. briggsae may differ from those in C. elegans. But this presumable expression change was apparently not too deleterious to prevent its fixation in C. briggsae. It seems likely that the first gene in operon CEOP2520 can be switched back from rps-9 to lsm-1 if a reverse inversion occurs in C. briggsae. Examination of additional species of Caenorhabditis such as C. japonica, C. drosophilae, and C. plicata, can test whether the first gene in this operon indeed oscillates between lsm-1 and rps-9 during evolution.
Figure 4.
Mechanisms of operon losses. Black arrows stand for genes belonging to operons in C. elegans or their orthologs in other species, with the circled numbers above the arrows showing the order of the genes in the C. elegans operon. Gray arrows show other genes. In C. elegans, horizontal solid lines link genes belonging to the same operon (boxed) and horizontal dash lines link genes not belonging to the same operon. In other species, genes inferred to be in the same operons are linked by horizontal solid lines; otherwise, they are linked by horizontal dash lines. Dotted lines show orthologous relationship between genes. In C. elegans, Caenorhabditis Genetics Center (CGC) names or sequence names are shown above the gene model, while in other species, the names are shown if they exist in WormBase release WS182. The operon breakage mechanisms shown here include inversion (A), relocation (B), and insertion (C).
Figure 5.
RT–PCR results showing the trans-splicing forms of various genes of C. briggsae. DNA bands are shown in white, while the background is black. Primers used are listed in Supplemental Table S3.
Relocation
Operon CEOP3416 represents the second mechanism of operon breakage. From the flanking genes, we infer that the first three genes in the operon relocated to another chromosome in C. briggsae, whereas the last gene in the operon remains in the original chromosome (Fig. 4B). The loss of the last constituent gene should not affect the expression of the first three genes. We confirmed that, in C. briggsae, the first gene (rpl-36) uses SL1 splicing, whereas the second and third genes (F37C12.3 and F37C12.2) use both SL1 and SL2 splicing (Fig. 5). Interestingly, we found that despite having lost the three upstream genes in the operon, the fourth gene in CEOP3416 (CBG16611) is still expressed in C. briggsae and uses both SL1 and SL2 splicing (Fig. 5). Because CBG16611 presumably does not have its own promoter, it most likely has formed an operon with F37C12.7 (CBG25148), the new upstream gene after the relocation of the three upstream genes in CEOP3416 (Fig. 4B). The distance between CBG25148 and CBG16611 is 1.8 kb (Fig. 4B), consistent with that in a typical operon.
Another case of breakage of operon by relocation is presented in Supplemental Figure S2. In this case, the gene cir-1 jumped out of operon CEOP1276 to a new chromosomal location in C. brenneri. Because cir-1 is an essential gene (Kamath et al. 2003), it has to be expressed. But the upstream gene of cir-1 in C. brenneri is on the other DNA strand. How cir-1 acquired its promoter in C. brenneri is unclear.
Insertion
In C. remanei, gene M01G5.3 is inserted into the intergenic region between Y50D7A.1 and Y50D7A.10, the two constituents of operon CEOP3022 (Fig. 4C). The intergenic distance between M01G5.3 and Y50D7A.10 is ∼4.3 kb, and it is not clear whether Y50D7A.1, M01G5.3, and Y50D7A.10 form a new operon in C. remanei (Fig. 4C). We cannot tell whether Y50D7A.10 is still in the operon, because a downstream gene in an operon tends to be SL1-spliced rather than SL2-spliced when it is far from its upstream gene (Blumenthal and Steward 1997; Blumenthal et al. 2002). Based on the distribution of intergenic distance in C. elegans operons, the probability that Y50D7A1.10 is a downstream gene of an operon is <1% (Supplemental Fig. S1). How it acquired its promoter remains unknown.
Molecular evolutionary mechanisms of operon formation and expansion
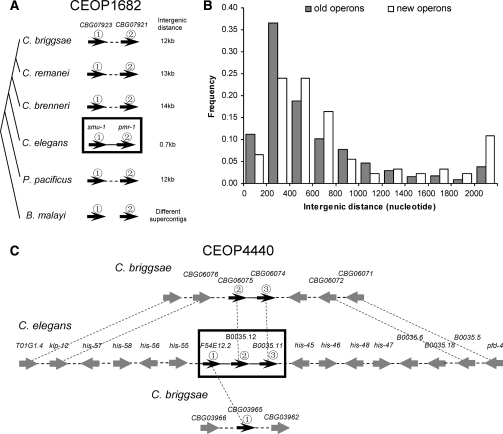
The prevailing view on the formation of new nematode operons is that it is a more or less neutral process, because constituent genes of most C. elegans operons have no functional relationship (Blumenthal et al. 2002; Blumenthal and Gleason 2003). Our examination of the 56 newly formed C. elegans operons confirmed this result (data not shown). The evolutionary process of operon gain is exemplified by C. elegans CEOP1682, which includes two genes, pmr-1 and smu-1 (Fig. 6A). The linkage patterns of the two genes in the six nematodes studied here allow us to infer that the two genes were not even linked in the common ancestor of the six nematodes, but became linked in the common ancestor of Caenorhabditis and P. pacificus. However, the intergenic distance between the two genes was probably too large for them to be in the same operon, as evident from the present-day intergenic distances in P. pacificus, C. briggsae, C. remanei, and C. brenneri. Only after C. elegans was separated from the other three Caenorhabditis species, the intergenic distance became reduced in the C. elegans lineage, allowing the formation of the operon following the switch from SL1-splicing to SL2-splcing in smu-1. The substantial reduction of the intergenic distance in C. elegans may be due to the acquisition of a new noncoding exon at the 5′ region of pmr-1.
Figure 6.
Mechanisms of operon gains. Black arrows stand for genes belonging to operons in C. elegans or their orthologs in other species, with the circled numbers above the arrows showing the order of the genes in the C. elegans operon. Gray arrows show other genes. In C. elegans, horizontal solid lines link genes belonging to the same operon (boxed) and horizontal dash lines link genes not belonging to the same operon. In other species, genes inferred to be in the same operons are linked by horizontal solid lines; otherwise, they are linked by horizontal dash lines. Dotted lines show orthologous relationship between genes. In C. elegans, Caenorhabditis Genetics Center (CGC), names or sequence names are shown above the gene model, while in other species, the names are shown if they exist in WormBase release WS182. (A) Formation of a new operon in C. elegans. (B) Comparison of intergenic distances between old operons and new operons. New operons are those formed after the divergence between C. elegans and C. briggsae, while old operons are those formed before that divergence. (C) Expansion of an existing operon by addition of F54E12.2 as the first gene in CEOP4440 in the C. elegans lineage.
If many operons are formed through reduction of intergenic distances of adjacent genes, then intergenic distances can further decrease but not increase after the establishment of the operons, because the increase would potentially break the operons and render the downstream genes in the operons unexpressed. Thus, we should expect that old operons have shorter intergenic distances than newly formed operons. Indeed, we found that the intergenic distances within C. elegans operons formed before the divergence of C. elegans, and the other three Caenorhabditis species are significantly shorter than those within operons formed after this divergence (P < 0.001, two-tail Mann-Whitney U-test; Fig. 6B).
We also observed several cases where an existing operon was expanded by inclusion of additional genes. A typical example is shown in Figure 6C. The C. elegans operon CEOP4440 consists of three genes. These three genes were not linked in P. pacificus and B. malayi. In all other Caenorhabditis species examined, the orthologs of the two downstream genes in CEOP4440 (B0035.12 and B0035.11) are linked, while the flanking genes are only partially conserved among the three species. We hypothesize that B0035.12 and B0035.11 formed an operon in the ancestor of C. briggsae and C. elegans; in C. elegans, F54E12.2 is added to the beginning of the operon, thus extending the operon from two to three genes. We found that CBG06074, the ortholog of B0035.11 in C. briggsae, uses both SL2 and SL1 splicing (Fig. 5), which is consistent with our hypothesis. The flanking genes of the operon suggest that the expansion of the operon was likely through the insertion of F54E12.2 (Fig. 6C). The functions of the three constituent genes in CEOP4440 appear to be related: F54E12.2 is an RNA polymerase II transcription termination factor, B0035.12 is an RNA-binding protein, and B0035.11 is an RNA polymerase II-associated protein.
Discussion
Operon gains and losses
Our comparative genomic analysis identified numerous cases of operon gains and losses in Caenorhabditis nematodes. We estimated that the rate of operon gain is ∼3.3 times that of operon loss in the past 80–110 Myr. In the very beginning of operon origin, the rate of operon loss is zero because there were no operons to lose. Hence, the rate ratio of operon gain to loss was infinity. A theoretical consideration shows that the rate ratio gradually declines over time, until reaching 1, when operon gains are offset by losses and the number of operons in the genome arrives at equilibrium (see Equations 2 and 3). Thus, strictly speaking, the one-way street hypothesis of operon evolution is incorrect, except at the time of operon origination. A more accurate description of the evolutionary dynamics of Caenorhabditis operons is “easy come, slow go,” at least for the past, present, and near future.
A recent phylogenetic survey of operons and trans-splicing among divergent lineages of nematodes suggested that nematode operons originated at least 500 Mya (Guiliano and Blaxter 2006). Because operons are uncommon outside of the phylum Nematoda (Nimmo and Woollard 2002; Blumenthal 2004) and because Nematoda originated ∼1000 Mya (Hedges 2002), it is likely that nematode operons originated between 500 and 1000 Mya, which is 5.3–10.5 times the midpoint divergence time between C. elegans and C. briggsae ([80 + 110]/2 = 95 Myr) (Coghlan 2003; Stein et al. 2003). Based on this information and Equations 2 and 3, we estimate that the equilibrium number of operons in the Caenorhabditis genome will be between 1200 and 1336 (Fig. 2), or 6%–18% greater than that in present-day C. elegans. It should be noted that the above estimate relies on a number of assumptions, including: (1) the date of operon origin relative to the date when C. elegans and C. briggsae diverged, (2) constancy of the intrinsic rate of operon gain per Myr (r) since the origin of nematode operons, (3) constancy of the rate of operon loss per operon per Myr (Rloss), and (4) the model describing the rate of operon gains (Equation 1). We think that the largest uncertainty is assumption 1, which was indirectly inferred based on several assumptions including the molecular clock. In the future, when the rate of operon gain (Rgain) is estimated from more than one point in the nematode phylogeny, we will be able to estimate K in Equation 1, which will allow us to estimate the equilibrium number of operons independent from assumption 1. Furthermore, we could date the origin of operons using K.
The most surprising finding of our study is numerous operon losses in evolution and a diversity of their underlying molecular evolutionary mechanisms. Three mechanisms, including inversion, insertion, and relocation, are observed. In a few cases, we can confidently infer how a downstream constituent gene of an operon acquired its promoter after the operon breakage. But, in at least 63% of the operon breakage cases, the downstream constituent genes of former operons are unlikely to have become the downstream genes in new operons, because they are on the opposite strand of DNA or have long intergenic distance from their upstream genes (>2 kb). How these genes acquired their promoters is difficult to infer without detailed experiments and analysis that would require additional work in the future.
Internal promoters in operons
Recently, Huang et al. (2007) reported that 27.7% of the 238 downstream genes in the C. elegans operons that they examined contain their own promoters. Thus, after the breakage of an operon, a downstream gene may still be expressed from its own promoter. If the presence of these internal promoters facilitates operon breakage, we should expect that (1) operons with internal promoters are more likely to be broken than those without internal promoters, and (2) the breakage point in an operon with an internal promoter is preferentially located immediately upstream of the gene with the internal promoter. Among the 65 C. elegans operons that Huang et al. reported to have internal promoters, 59 have all constituent genes in each of the other three Caenorhabditis species. Among these 59 operons, two were broken in C. briggsae, C. remanei, or C. brenneri, and two were recently formed in the C. elegans lineage. The breakage rate of these internal-promoter-containing operons (2/[59 − 2] = 3.51%) is even lower than that of other operons ([41 − 2]/{967 − [59 − 2]} = 4.29%), although the difference is not statistically significant (P > 0.5, two-tail Fisher’s exact test). Furthermore, between the two internal-promoter-containing operons that were broken in C. briggsae, C. remanei, or C. brenneri, one operon had a break point disagreeing with the internal promoter. Thus, our analysis suggests that internal promoters do not facilitate operon losses. This is probably because both operon promoters and internal promoters are needed for the expression of the downstream genes that have their own promoters. Interestingly, only 2/56 = 3.6% of operons that were newly formed in the C. elegans lineage were reported to contain internal promoters, compared with (59 − 2)/967 = 5.9% among old operons (P > 0.5, two-tail Fisher’s exact test). This suggests that internal promoters may be secondarily acquired after the formation of operons, rather than the original promoters of the downstream genes.
Independent operon losses versus ancestral polymorphisms
We identified four C. elegans operons that are present in C. remanei, but absent in C. briggsae and C. brenneri. The Caenorhabditis phylogeny suggests that each of these operons should have been lost once in C. briggsae and once in C. brenneri, given that independent origins of an operon comprising initially nonadjacent genes in C. elegans and C. remanei are improbable (Fig. 1). Because operon losses are also rare, the observation of multiple losses of an operon in the recent past seems odd. If operon losses are all random and independent, we expect to observe 17 × 16/967 = 0.28 operons that were lost twice, in C. briggsae and C. brenneri. The observed number is significantly greater than this expectation (P < 0.001, binomial test). Similarly, there are three C. elegans operons that have been lost twice, in C. remanei and C. brenneri, significantly more than the chance expectation (0.20) (P < 0.002). These observations suggest that these operon losses may not be random or independent. Two possibilities warrant discussion. First, operon losses may be beneficial if the constituent genes need to be differentially expressed, which is prohibited when the genes are in the same operon. Operon breakage makes differential expression possible. Second, it has been suggested that C. briggsae, C. remanei, and C. brenneri diverged in a relatively short time (Cho et al. 2004). Thus, it is possible that lineage sorting from ancestral polymorphisms in the common ancestor of the three species had not been completed in the most recent common ancestor of C. briggsae and C. remanei, causing the observed presence/absence pattern of an operon inconsistent with the species phylogeny. If the second possibility is correct, the breakage patterns of an operon inferred from flanking genes should be the same in the two species where the operon is broken. But in none of the seven cases were we able to find such evidence. As a control, we examined the three cases of operon losses that presumably occurred in the common ancestor of C. briggsae and C. remanei (Fig. 1). We found clear evidence that the breakage patterns are identical in the two species for one of the three cases (CEOP3780). Taken together, the analysis suggests that a small number of operon breakages might have been beneficial.
Potential caveats on the operational definition of operons
In this study, the presence and absence of C. elegans operons were examined in other nematodes. In C. elegans, these operons were identified and verified based on experimental evidence such as trans-splicing types and the presence of polycistronic pre-mRNA. But in other nematodes, we mainly relied on genomic DNA sequences to determine the presence and absence of C. elegans operons, although detection of trans-splicing by RT–PCR was also used in a few cases. Therefore, it is important to evaluate the criteria we set in the computational analysis of operon gains and losses.
The first criterion is the E-value cutoff in TBLASTN gene searches. We used 10−5 as the cutoff to search for C. elegans orthologs in C. briggsae, C. remanei, and C. brenneri genomes and identified 97 operons that were either gained or lost in Caenorhabditis (Fig. 1). This number, as well as the number of operons for each phylogenetic distribution shown in Figure 1, remained virtually unchanged when 10−10 or 10−15 were used as E-value cutoffs.
The second criterion is the upper limit of the intergenic distance between constituent genes that we used to define operons. If we set this limit too low, erroneous operon gains/losses may be inferred. But if the limit is set too high, many broken operons will be regarded as intact. In C. elegans, the largest observed distance is 8.189 kb (between kup-1 and pkc-1 in CEOP5312). Therefore, if the intergenic distance between two genes is >9 kb in a Caenorhabditis species, the two genes are not considered to be in the same operon in that species. In C. elegans, 96% of intergenic distances within operons are shorter than 2 kb. So, if two genes belonging to a C. elegans operon have an intergenic distance between 2 and 9 kb in another species, we treat the operon relationship between the two genes in that species to be ambiguous. The operon gains and losses we reported in this study do not involve ambiguous operons, with the exception of the operon gains where the ambiguous status in one Caenorhabditis species is allowed.
Conclusions
Using comparative genomics, we systematically examined operon gains and losses during the evolution of Caenorhabditis nematodes. Our results show that operons are “easy come, slow go” at this time, as the present rate of operon gain is ∼3.3 times that of operon loss. Our analysis projects that operons will continue to accumulate in the nematode genome in the future until equilibrium is reached. Contrary to the one-way street hypothesis, diverse molecular evolutionary mechanisms of operon breakage exist, and the expression mechanisms of downstream genes after operon breakage can be inferred in a few cases. However, our analysis is limited to gains and losses of C. elegans operons that occurred within Caenorhabditis, due to the lack of sufficient genomic sequence and trans-splicing data outside of Caenorhabditis. It will be of significant interest to examine whether the evolutionary dynamics of operons in other nematodes is similar to what have been uncovered here. Furthermore, the availability of additional Caenorhabditis genomes will aid in reconstruction of detailed processes of operon gains and losses, which will help understand both evolutionary forces and molecular mechanisms responsible for such events. As ∼10 additional nematode genomes are being sequenced (http://genomesonline.org/), we expect that a broader and more detailed picture of operon evolution will emerge.
Methods
Genomic data
We downloaded the C. elegans protein sequences from WormBase release WS182 (http://wormbase.org) and obtained the genome sequences of C. briggsae (Stein et al. 2003) (release Cb3), C. remanei (15.0.1), C. brenneri (4.0), and P. pacificus (5.0) from the Washington University School of Medicine Genome Sequencing Center (http://genome.wustl.edu/pub/organism/Invertebrates/; J. Spieth, pers. comm.). The genome sequence of B. malayi (Ghedin et al. 2004, 2007) was downloaded from TIGR ftp://ftp.tigr.org/pub/data/b_malayi/). The qualities of these genome sequences are excellent, as indicated by the high (>8.5) coverage (Supplemental Table S2). The operon annotations of C. elegans were obtained from WormBase WS182, with additional information from Dr. Tom Blumenthal that is projected to be published in WS185. In total, there were 1133 C. elegans operons, including 2817 constituent genes. Among these genes, 14 (in 12 operons) did not have protein sequences (either pseudogenes or microRNA genes). We thus analyzed the remaining 1121 operons, including 2777 genes. RNAi phenotypes of 16,563 genes in C. elegans (Kamath et al. 2003) were used in our analysis. Gene expression data were downloaded from Supplemental Table S1 of Kim et al. (2001) at http://www.sciencemag.org/feature/data/kim1061603/gl/gene_list.html. The list of downstream genes with internal promoters were downloaded from the Supplemental Table S1 of Huang et al. (2007) at http://www.genome.org/content/vol0/issue2007/images/data/gr.6824707/DC1/Supplementary_Table_1.doc.
Gene identification
We used protein sequences of constituent genes in the annotated operons of C. elegans as queries to search for homologous genes in the genome sequences of C. remanei, C. briggsae, and C. brenneri, respectively, by TBLASTN (E-value cutoff = 10−5). In total, 2685 operon genes of C. elegans had at least one hit in C. briggsae, 2703 had at least one hit in C. remanei, and 2699 had at least one hit in C. brenneri. The results were virtually identical when an E-value of 10−10 or 10−15 was used. Because P. pacificus and B. malayi are highly divergent from C. elegans, an E-value of 1 was used in TBLASTN searches of C. elegans homologs in these two species. Of the constituent genes of C. elegans operons, 2424 and 2299 genes had at least one hit in P. pacificus and B. malayi, respectively. If a TBLASTN hit covered <50% of the length of a protein, we lowered the neighborhood word threshold score from 12 to 6 (the -f option) and adjusted the E-value cutoff to 1 to confirm the existence of the gene. That is, the hit was still considered genuine if the match covered >50% under the new parameters.
Operon identification
If (1) all of the genes in a C. elegans operon have homologs in species X, (2) these homologous genes are on the same strand of the same chromosome (or supercontig) in X, and (3) each intergenic region among these genes is shorter than 2 kb in X, we consider the operon to be present in X. Otherwise, we consider the operon to be absent in X, except that when (1) and (2) hold and one or more intergenic distances are between 2 and 9 kb, the situation is considered to be ambiguous in X. Eighty-eight C. elegans operons each have at least one gene that cannot be identified in at least one of the other three Caenorhabditis species. For 10 additional C. elegans operons, their gains or losses could not be unambiguously determined, because each of them is broken into two pieces, located at the ends of two supercontigs in at least one of the other three Caenorhabditis species. After excluding these cases, 1121 − 88 − 10 = 1023 operons were analyzed in this study.
When a C. elegans operon is inserted with one or more genes in another species, it is possible that the inserted genes are not identified and the intergenic distance within the operon is overestimated. We examined all such potential operons that have constituent genes located on the same DNA strand and have intergenic distances between 9 and 20 kb, but found only one case where an inserted gene was missed. This case was subsequently recovered and presented in Figure 4C.
Confirmation of the integrity of downstream genes in broken operons
Using GeneWise (Birney et al. 2004), we examined whether a downstream gene in a operon still has an intact ORF in the species where the operon is broken. Among 61 genes that were tested, two lacked ORFs. To confirm this result, we extracted genomic DNA from strains AF16 of C. briggsae (Fodor et al. 1983) and EM464 of C. remanei (Baird et al. 1992) by TRIzol (Invitrogen), used PCR to amplify gene F37C12.2 in AF16 and gene F46B6.6 in EM464, and sequenced the PCR products by the dideoxy method at the University of Michigan DNA Sequencing Core. The strains used here were the same as used in genome sequencing.
Examination of trans-spicing forms
Total RNA was purified from adults of C. briggsae strain AF16 using TRIzol, reverse-transcribed using the RETROscript Kit (Ambion Inc.), and PCR-amplified using primers listed in Supplemental Table S3. For examining SL1 splicing, the 3′ primer used was a gene-specific primer and the 5′ primer used was a SL1 primer. For examining SL2 splicing, the 3′ primer used was a gene-specific primer and the 5′ primer used was a mixture of six SL2-like primers, because SL2 sequences are variable.
Acknowledgments
We thank Soochin Cho for technical assistance and consultation; Tom Blumenthal for sharing new C. elegans operon data before publication; Meg Bakewell, Tom Blumenthal, Wendy Grus, Ben-Yang Liao, Xionglei He, and two anonymous reviewers for valuable comments; the Caenorhabditis Genetics Center for supplying the AF16 and EM464 worm strains; WormBase for invaluable source of data; and the Washington University School of Medicine Genome Sequencing Center and Institute of Genomic Research for making the various nematode genome sequences available before publication. This work was supported by research grants from National Institutes of Health to J.Z.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7112608
References
- Baird S.E., Sutherlin M.E., Emmons S.W., Sutherlin M.E., Emmons S.W., Emmons S.W. Reproductive isolation in Rhabditidae (Nematoda: Secernentea): Mechanisms that isolate six species of three genera. Evolution Int. J. Org. Evolution. 1992;46:585–594. doi: 10.1111/j.1558-5646.1992.tb02067.x. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M.L., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Mackey L.Y., Dorris M., Frisse L.M., Dorris M., Frisse L.M., Frisse L.M., et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Operons in eukaryotes. Brief Funct. Genomic Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Trans-splicing and operons. In: The C. elegans Research Community, editor. Wormbook. 2005. http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Gleason K.S., Gleason K.S. Caenorhabditis elegans operons: Form and function. Nat. Rev. Genet. 2003;4:110–118. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Steward K., Steward K. RNA processing and gene structure. In: Riddle D.L., et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 117–145. [PubMed] [Google Scholar]
- Blumenthal T., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Chiu W.L., Duke K., Kiraly M., Duke K., Kiraly M., Kiraly M., et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Cavalcanti A., Stover N., Landweber L., Stover N., Landweber L., Landweber L. On the paucity of duplicated genes in Caenorhabditis elegans operons. J. Mol. Evol. 2006;62:765–771. doi: 10.1007/s00239-005-0203-3. [DOI] [PubMed] [Google Scholar]
- Cho S., Jin S.-W., Cohen A., Ellis R.E., Jin S.-W., Cohen A., Ellis R.E., Cohen A., Ellis R.E., Ellis R.E. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A. “Evolution of the genomes of two nematodes in the genus Caenorhabditis.”. Department of Genetics, University of Dublin; Ireland: 2003. Ph.D. thesis. [Google Scholar]
- Ermolaeva M.D., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D., Zorio D., MacMorris M., Winter C.E., Lea K., Blumenthal T., Zorio D., MacMorris M., Winter C.E., Lea K., Blumenthal T., MacMorris M., Winter C.E., Lea K., Blumenthal T., Winter C.E., Lea K., Blumenthal T., Lea K., Blumenthal T., Blumenthal T. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc. Natl. Acad. Sci. 1997;94:9751–9756. doi: 10.1073/pnas.94.18.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A., Riddle D.L., Nelson F.K., Golden J.W., Riddle D.L., Nelson F.K., Golden J.W., Nelson F.K., Golden J.W., Golden J.W. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica. 1983;29:203–217. [Google Scholar]
- Ghedin E., Wang S., Foster J.M., Slatko B.E., Wang S., Foster J.M., Slatko B.E., Foster J.M., Slatko B.E., Slatko B.E. First sequenced genome of a parasitic nematode. Trends Parasitol. 2004;20:151–153. doi: 10.1016/j.pt.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Guiliano D.B., Miranda-Saavedra D., Miranda-Saavedra D., et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiliano D.B., Blaxter M.L., Blaxter M.L. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genet. 2006;2:e198. doi: 10.1371/journal.pgen.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings K.E.M. SL trans-splicing: Easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hedges S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Fulton L.A., Koboldt D.C., Waterston R.H., Koboldt D.C., Waterston R.H., Waterston R.H. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman M., van der Wurff A., van den Elsen S., van Megen H., Bongers T., Holovachov O., Bakker J., Helder J., van der Wurff A., van den Elsen S., van Megen H., Bongers T., Holovachov O., Bakker J., Helder J., van den Elsen S., van Megen H., Bongers T., Holovachov O., Bakker J., Helder J., van Megen H., Bongers T., Holovachov O., Bakker J., Helder J., Bongers T., Holovachov O., Bakker J., Helder J., Holovachov O., Bakker J., Helder J., Bakker J., Helder J., Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Huang P., Pleasance E.D., Maydan J.S., Hunt-Newbury R., O'Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Pleasance E.D., Maydan J.S., Hunt-Newbury R., O'Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Maydan J.S., Hunt-Newbury R., O'Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Hunt-Newbury R., O'Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., O'Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J., Marra M.A., Moerman D.G., Jones S.J., Moerman D.G., Jones S.J., Jones S.J. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res. 2007;17:1478–1485. doi: 10.1101/gr.6824707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M.A., Snel B., Bork P., Snel B., Bork P., Bork P. Inversions and the dynamics of eukaryotic gene order. Trends Genet. 2001;17:304–306. doi: 10.1016/s0168-9525(01)02302-2. [DOI] [PubMed] [Google Scholar]
- Itoh T., Takemoto K., Mori H., Gojobori T., Takemoto K., Mori H., Gojobori T., Mori H., Gojobori T., Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- Jacob F., Perrin D., Sanchez C., Monod J., Perrin D., Sanchez C., Monod J., Sanchez C., Monod J., Monod J. Operon: A group of genes with the expression coordinated by an operator. C. R. Hebd. Seances Acad. Sci. 1960;250:1727–1729. [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Le Bot N., Moreno S., Sohrmann M., Moreno S., Sohrmann M., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Katju V., Lynch M., Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Lund J., Kiraly M., Duke K., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Lund J., Kiraly M., Duke K., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Kiraly M., Duke K., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Duke K., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S., Eizinger A., Wylie B.N., Davidson G.S., Wylie B.N., Davidson G.S., Davidson G.S. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Gavin N.P., Raynes Y., Roehrig C., Piano F., Fitch D.H.A., Gavin N.P., Raynes Y., Roehrig C., Piano F., Fitch D.H.A., Raynes Y., Roehrig C., Piano F., Fitch D.H.A., Roehrig C., Piano F., Fitch D.H.A., Piano F., Fitch D.H.A., Fitch D.H.A. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Hirsh D., Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Lawrence J.G. Shared strategies in gene organization among prokaryotes and eukaryotes. Cell. 2002;110:407–413. doi: 10.1016/s0092-8674(02)00900-5. [DOI] [PubMed] [Google Scholar]
- Lawrence J.G., Roth J.R., Roth J.R. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-Z., Sommer R.J., Sommer R.J. Operon structure and trans-splicing in the nematode Pristionchus pacificus. Mol. Biol. Evol. 2003;20:2097–2103. doi: 10.1093/molbev/msg225. [DOI] [PubMed] [Google Scholar]
- Lercher M.J., Blumenthal T., Hurst L.D., Blumenthal T., Hurst L.D., Hurst L.D. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R., Woollard A., Woollard A. Widespread organisation of C. elegans genes into operons: Fact or function? Bioessays. 2002;24:983–987. doi: 10.1002/bies.10181. [DOI] [PubMed] [Google Scholar]
- Price M.N., Arkin A.P., Alm E.J., Arkin A.P., Alm E.J., Alm E.J. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Smith T.F., Collado-Vides J., Collado-Vides J. Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J., Brooke G., Kuersten S., Lea K., Blumenthal T., Brooke G., Kuersten S., Lea K., Blumenthal T., Kuersten S., Lea K., Blumenthal T., Lea K., Blumenthal T., Blumenthal T. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chinwalla A., Clarke L., Clee C., Coghlan A., Clarke L., Clee C., Coghlan A., Clee C., Coghlan A., Coghlan A., et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A.R., Cheng N.N., Blumenthal T., Spieth J., Cheng N.N., Blumenthal T., Spieth J., Blumenthal T., Spieth J., Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]