Abstract
The linear, double-stranded DNA genome of vaccinia virus contains covalently closed hairpin termini. These hairpin termini comprise a terminal loop and an A+T-rich duplex stem that has 12 extrahelical bases. DeMasi et al. have shown previously that proteins present in infected cells and in virions form distinct complexes with the telomeric hairpins and that these interactions require the extrahelical bases. The vaccinia virus I6 protein was identified as the protein showing the greatest specificity and affinity for interaction with the viral hairpins (J. DeMasi, S. Du, D. Lennon, and P. Traktman, J. Virol. 75:10090-10105, 2001). To gain insight into the role of I6 in vivo, we generated eight recombinant viruses bearing altered alleles of I6 in which clusters of charged amino acids were changed to alanine residues. One allele (temperature-sensitive I6-12 [tsI6-12]) conferred a tight ts phenotype and was used to examine the stage(s) of the viral life cycle that was affected at the nonpermissive temperature. Gene expression, DNA replication, and genome resolution proceeded normally in this mutant. However, proteolytic processing of structural proteins, which accompanies virus maturation, was incomplete. Electron microscopic studies confirmed a severe block in morphogenesis in which immature, but no mature, virions were observed. Instead, aberrant spherical virions and large crystalloids were seen. When purified, these aberrant virions were found to have normal protein content but to be devoid of viral DNA. We propose that the binding of I6 to viral telomeres directs genome encapsidation into the virus particle.
Vaccinia virus, the prototypic member of the Poxvirus family, contains a 192-kb double-stranded DNA genome encoding approximately 200 proteins involved in DNA replication, gene expression, morphogenesis, and virus-host interaction (reviewed in reference 27). The genome is a linear duplex with covalently closed hairpin termini. These termini exist in two 104-nucleotide (nt) isoforms (termed flip and flop) which are inverted complements of one another (2, 3). The hairpin comprises a 4-nt terminal loop and a 44-bp highly A+T-rich duplex stem with 12 extrahelical bases, 10 on one strand and 2 on the other. Extrahelical bases in the termini are maintained during the viral life cycle in all poxviruses, although the positions and numbers of extrahelical bases differ.
We have hypothesized that the viral telomeres play a role in DNA replication. The current replication model proposes that a nick proximal to the terminus is introduced, exposing a 3′ hydroxyl group that serves as the primer terminus for the initiation of DNA synthesis (reviewed in reference 35). Evidence supporting this model was obtained by labeling synchronized infections with [3H]thymidine; radiolabel was first incorporated within 200 bp of the telomere region (30, 31). Additional evidence was provided by studies in our laboratory that used a minichromosome replication assay (8). Minichromosomes contain a plasmid-derived stuffer sequence flanked by viral telomeric sequences of various lengths. When minichromosomes were introduced into infected cells by transfection, the minimal telomeric region necessary and sufficient for efficient replication of the minichromosome comprised 150 to 200 bp. This region includes the 104-nt hairpin and an additional 87 bp of a unique sequence, which encompasses but extends beyond the sequence elements required for resolution of concatemeric replication intermediates.
Given the importance of telomeres in viral replication, a considerable effort was made to identify proteins that might interact with the telomeres while promoting the initiation of DNA replication. Using electrophoretic mobility shift assays, we demonstrated that each isoform of the telomeric hairpin was able to form two distinct complexes with proteins present in virions and infected cells (6). These interactions required the extrahelical bases that are so characteristic of the viral hairpins. Using biotinylated hairpins immobilized on streptavidin-conjugated magnetic beads, our laboratory purified and identified the I1 and I6 proteins as those responsible for forming these complexes.
Fortuitously, our laboratory had previously studied the I1 protein, showing it to be expressed during late times of infection and encapsidated in the virion core (21). Purified recombinant I1 protein possesses promiscuous DNA-binding activity but prefers viral hairpins as a substrate. Repression of I1 (enabled by the construction of an IPTG [isopropyl-β-d-thiogalactopyranoside]-inducible recombinant virus) resulted in a specific and tight arrest in viral morphogenesis. Viral assembly appeared to progress normally through the formation of immature virions with nucleoids (IVN), but no further maturation into infectious, mature virions was seen.
In contrast to I1, no studies analyzing the structure or function of the I6 protein have been published to date. The molecular mass of I6 is predicted to be 43.5 kDa, and apart from counterparts in other poxviruses, no significant homology to any other proteins was found by computer analysis. Our laboratory confirmed that recombinant I6 protein specifically recognizes extrahelical bases and binds to the viral hairpins with great specificity and stability (6). The work described herein was designed to elucidate the role of I6 in vivo. We describe the generation of eight recombinant viruses bearing altered alleles of I6 and demonstrate that one of these alleles (I6-12) confers a tight ts phenotype. Analysis of the conditionally lethal phenotype of this virus has uncovered a key role for the I6 telomere-binding protein, and by extrapolation, for the viral telomeres, in the poorly understood process of DNA encapsidation.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, Taq DNA polymerase, T4 DNA ligase, calf intestinal alkaline phosphatase, pancreatic RNase, Escherichia coli DNA polymerase I, DNase I, and DNA molecular weight standards were purchased from either Roche Molecular Biochemicals (Indianapolis, Ind.) or New England Biolabs (Beverly, Mass.). [35S]Met and 32P-labeled nucleoside triphosphates were purchased from PerkinElmer Life Sciences (Boston, Mass.). Lipofectamine Plus, geneticin (G418 sulfate), and 14C-labeled protein molecular weight markers were purchased from Invitrogen (Carlsbad, Calif.). Rifampin (RIF) and 1-β-d-arabinofuranosylcytosine (araC) were obtained from Sigma (St. Louis, Mo.). Isatin-β-thiosemicarbazone (IBT) was obtained from Pfaltz and Bauer (Waterbury, Conn.). Oligonucleotides were purchased from IDT (Coralville, Iowa).
Cells and viruses.
BSC40 African green monkey kidney cells were maintained as monolayer cultures in Dulbecco modified Eagle medium (DMEM) (Invitrogen) containing 5% fetal bovine serum (FBS). Stocks of wild-type (wt) vaccinia virus (WR strain) and the viruses described herein, which contained mutant alleles of the I6 gene, were prepared by ultracentrifugation of cytoplasmic extracts through a 36% sucrose cushion.
Site-directed mutagenesis and cloning of the I6 gene into pUC/Neo.
Eight regions of the I6 gene were chosen for clustered charge-to-alanine mutagenesis (Fig. 1) (7, 32). Mutations were generated by overlap PCR using the HindIII I fragment of the vaccinia virus genome as the template. In the first round of PCR, one reaction was performed using an upstream primer (U) and a downstream primer carrying the engineered nucleotide substitutions (#mut-3); a second reaction was performed using an upstream primer carrying the desired substitutions (#mut-5) and a downstream primer (D). The two PCR products from this first round of amplification overlapped by 17 bp. A mixture of the two products served as the template for a second round of PCR performed with primers U and D. The final PCR product contained the desired mutations flanked by 300 bp of upstream and downstream sequences. Primers U and D introduced BamHI recognition sites at the termini of the amplified fragments. The final PCR products were gel purified, digested with BamHI, and cloned into pUC/Neo DNA that had been previously digested with BamH1 and treated with calf intestinal alkaline phosphatase. This plasmid carries the neomycin resistance gene (Neo) under the control of the constitutive viral p7.5 promoter (21, 24). The constructs were subjected to automated DNA sequencing to ensure the presence of the desired nucleotide substitutions and the absence of any additional mutations. The primers used are listed in Table 1.
FIG. 1.
Clustered charge-to-alanine mutagenesis of the vaccinia virus I6L gene. The predicted amino acid sequence of the I6 protein of the WR strain is shown. A total of 13 clusters of charged residues were found (underlined); eight of these regions (numbered) were selected for mutagenesis. For each of the mutant alleles, the codons encoding the charged residues (shaded in gray) were mutated to encode alanine. Viruses in which the endogenous I6 gene was replaced with the individual mutant alleles were obtained by transient dominant selection.
TABLE 1.
Primers used in the studya
| Upstream primer
|
Downstream primer
|
||
|---|---|---|---|
| Primer nameb | Sequence | Primer nameb | Sequence |
| U (1, 2, 4) | 5′ CAGGATCCAGCTGTTGGAATCTGA | D (1, 2, 4) | 5′ CGGGATCCGGTAGTCTCTCTTTCG |
| U (5, 6) | 5′ CAGGATCCGATGGACTGTTTACTA | D (5, 6) | 5′ CGGGATCCTATAGATACCCTTCC AG |
| U (9, 12, 13) | 5′ CAGGATCCTTTCCCATTCGATACAG | D (9, 12, 13) | 5′ CGGGATCCATTTGACGTATGTAGCG |
| #1-5 | 5′ TAGCACCTACCGCAGCATTGTCTCCGTCAGATG | #1-3 | 5′ TGCTGCGGTAGGTGCTAGAGACTTTGAAGCTA |
| #2-5 | 5′ GGCGGCTGTTCTTGCATCACTCTTGATCATG | #2-3 | 5′ ATGCAAGAACAGCCGCCGGAGTATTAATGGGC |
| #4-5 | 5′ CGTCGCCGCTGCTCCTGAAACACACGTGG | #4-3 | 5′ CAGGAGCAGCGGCGACGCCCGTCAACCAA |
| #5-5 | 5′ CACAGCAGCAGCATTAAGTATTAATACGTT | #5-3 | 5′ TTAATGCTGCTGCTGTGGTAAGTACCGAT |
| #6-5 | 5′ GTCCGGCAGCGGCACTACCGTATATCATCG | #6-3 | 5′ GTAGTGCCGCTGCCGGACTTATATAGCTTA |
| #9-5 | 5′ ATAATGCGGCTCTAAACACAATCTTGG | #9-3 | 5′ GTTTAGAGCCGCATTATTAATATGAGTAA |
| #12-5 | 5′ CTAATGCGGCAGCAATATGTAAGA AACATTG | #12-3 | 5′ CATATTGCTGCCGCATTAGTAACTGACTTAA |
| #13-5 | 5′ TATGTGCGGCACATTGTTTCGTCAATTC | #13-3 | 5′ ACAATGTGCCGCACATATTTTTTTCTTAT |
BamHI sites are underlined; the mutated nucleotides are shown in boldface characters.
For the U and D primers, the numbers in parentheses designate the alleles for which each primer was used.
Transient dominant selection and isolation of I6 recombinant viruses.
Transient dominant selection was used to replace the endogenous I6 gene with the mutant allele (10, 21, 24, 32). Dishes (35-mm diameter) of BCS40 cells were infected with wt virus at a multiplicity of infection (MOI) of 0.03 PFU/cell at 37°C. At 3 h postinfection (hpi), 3.5 μg of the desired pUC/Neo-I6 plasmid was introduced into cells by transfection using Lipofectamine Plus reagent (Invitrogen) and cells were transferred to 32°C. 32°C was utilized for all subsequent steps in the generation of the recombinant viruses. At 16 hpi, geneticin (G418 sulfate) was added to a final concentration of 3 mg per ml to select for the spread and replication of virus that had incorporated the plasmid into the genome. Cells were harvested 2 days after infection, and virus was released by three cycles of freezing and thawing and two 15-s bursts of sonication. After two rounds of plaque purification in the presence of geneticin, incorporation of the plasmid into the drug-resistant viral isolates was confirmed by PCR using primers specific for the Neo gene. These “parental” viruses have the entire plasmid incorporated into the I6 locus and therefore have tandem repeats of a portion of the I5, I6, and I7 regions flanking the Neor gene. Drug selection was then removed, and several rounds of plaque purification were performed to enable recombination to occur within these repeats. Such recombination leads to the loss of the Neor gene and the retention of either the wt or mutant allele of I6. The loss of the Neor gene was monitored by PCR. The I6 allele was amplified by PCR from those viruses shown to have undergone recombinational resolution, and automated DNA sequence analysis was used to distinguish viruses carrying the desired mutant I6 alleles from wt isolates. The appropriate plaques were expanded, and purified viral stocks (designated vI6-1, vI6-2, vI6-4, vI6-5, vI6-6, vI6-9, vI6-12, and vI6-13) were prepared by ultracentrifugation of cytoplasmic extracts through a 36% sucrose cushion.
Determination of 24-h viral yields.
Dishes (35-mm-diameter) of confluent BSC40 cells were infected at an MOI of 2 with wt or vI6 viruses and incubated at either 32 or 40°C. At 24 hpi, cells were harvested by scraping, collected by centrifugation, resuspended in phosphate-buffered saline (PBS) (140 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4 [pH 7.4]), and disrupted by three cycles of freezing and thawing and two 15-s bursts of sonication. Viral yields were determined by plaque assays performed at 32°C on BSC40 monolayers.
Marker rescue.
Dishes (35-mm-diameter) of BSC40 cells were infected with tsI6-12 at an MOI of 0.006 and incubated at 32°C. At 3 hpi, 5 μg of linearized plasmid DNA was introduced into cells with Lipofectamine Plus reagent. Plasmids contained no insert, the wt I6 open reading frame (ORF), the genomic HindIII I fragment, or the genomic HindIII G fragment (negative control). At 6 hpi, dishes were fed with fresh DMEM-5%FBS and transferred to 40°C. At 2 dpi, cells were harvested and the yield of temperature-insensitive virus was determined by titration at 40°C.
Analysis of viral protein synthesis.
Confluent BSC40 cells were infected with wt virus or tsI6-12 at an MOI of 10 and incubated at 32 or 40°C. At the appropriate times, cells were rinsed with prewarmed methionine-free DMEM, incubated for 45 min in the same medium supplemented with 100 μCi of [35S]Met/ml, and then harvested. Cells were harvested at 2, 3.5, 5, 6.5, and 8 hpi; the proteins within whole-cell lysates were resolved by electrophoresis on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and then visualized by autoradiography.
Analysis of the proteolytic processing of viral structural proteins.
Cells were infected with wt virus or tsI6-12 at an MOI of 5 in the presence or absence of RIF (100 μg/ml) and incubated at 32 or 40°C. At 8 hpi, cells were labeled as described above for 45 min; samples were either harvested immediately (pulse) or rinsed and fed with prewarmed DMEM-5%FBS and harvested at 24 hpi (chase).
Analysis of viral DNA synthesis by Southern dot blot hybridization.
Dishes (35-mm-diameter) of confluent BSC40 cells were infected with wt virus or tsI6-12 at an MOI of 5 and incubated at 32 or 40°C. At 3, 6, 9, 12, and 24 hpi, cultures were harvested and analyzed in duplicate by dot blot DNA hybridization as previously described (32). Viral DNA was detected by hybridization with a radiolabeled probe representing the HindIII E and HindIII G fragments of the vaccinia virus genome. Data were quantitated by phosphorimager analysis and plotted using Sigma Plot software.
Genome resolution assay.
Cells were infected with wt virus or tsI6-12 at an MOI of 2 and incubated at 32 or 40°C. As controls, cells were also infected with wt virus (MOI of 2) in the presence of 60 μM IBT or with vROG8 virus (a recombinant in which expression of the G8 gene is dependent upon inclusion of IPTG in the culture medium) (41) at an MOI of 15. At 18 hpi, cells were harvested and cytoplasmic DNA was prepared, digested with BstEII, and resolved on a Tris-acetate-EDTA-agarose gel. The DNA was transferred to a ZetaProbe membrane, and Southern blot hybridization was performed using a radiolabeled probe representing a 1.2-kb PvuII-EcoRI fragment released from the pSV9 plasmid (25, 26). This probe hybridizes to the genomic telomeres and adjacent sequences. The blot was then visualized by autoradiography.
Electron microscopy.
Dishes (60-mm-diameter) of BSC40 cells were infected with tsI6-12 virus at an MOI of 2 and incubated at 32 or 40°C. At 17 hpi, cells were rinsed with PBS and fixed in situ with 1% glutaraldehyde-0.1 M PBS (pH 7.4) on ice for 30 min. Cells were scraped gently, collected by centrifugation (800 × g for 10 min), and then processed for conventional electron microscopy as described previously (7, 36). Purified viral particles were concentrated by sedimentation (13,000 × g for 5 min) and then processed as described above. Samples were examined on a Hitachi H-600 transmission electron microscope.
Purification of virus particles.
Four 15-cm-diameter dishes of BSC40 cells were infected with either wt virus or tsI6-12 at an MOI of 2 and kept at 32 or 40°C. At 24 hpi, cells were harvested and disrupted by Dounce homogenization. Virus particles were purified from cytoplasmic extracts by ultracentrifugation through a 36% sucrose cushion (43,000 × g for 80 min) followed by banding on a 25 to 40% sucrose gradient (6,000 × g for 45 min). After the gradients were photographed, 20 fractions (250 μl each) were collected beginning at the bottom of the tube. Aliquots from each fraction were solubilized with 0.25% NP-40-0.25% deoxycholate-250 mM NaCl-0.5 mM dithiothreitol, and the protein content was quantitated by a Bradford assay (Bio-Rad, Hercules, Calif.). Fractions containing detectable amounts of protein which corresponded to those containing a light-scattering band were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and silver staining.
Immunoblot analysis.
Protein samples were resolved by SDS-PAGE and electrophoretically transferred to nitrocellulose membranes in CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] buffer (10 mM CAPS [pH 11.3], 10% methanol). Membranes were incubated with sera generated in our laboratory which react with the L4, F18, and I1 proteins (21, 24) or with sera directed against the I7 and L1 proteins, which were generous gifts from S. Shuman (Sloan-Kettering Institute, New York, N.Y.) and D. Hruby (Oregon State University, Corvallis, Oreg.), respectively. Horseradish-peroxidase-conjugated goat anti-rabbit secondary antibody was used, and immunoreactive proteins were visualized after chemiluminescent development.
Southern dot blot analysis of purified viral particles.
Fractions 7 to 18 (5 μl each) from the 25 to 40% sucrose gradient (see above) were diluted in 50 μl of water, boiled for 5 min, and applied (using a Bio-Dot microfiltration apparatus [Bio-Rad]) to a hydrated ZetaProbe membrane (Bio-Rad). Samples were denatured and renatured in situ, and the blot was then subjected to Southern dot blot hybridization using 106 cpm of radiolabeled probe/ml representing the vaccinia virus HindIII E and HindIII F genomic fragments. Data were analyzed on a phosphorimager.
Computer analysis.
Autoradiographic films and electron micrographic negatives were scanned using a SAPHIR scanner (Linotype-Hell Co., Hauppauge, N.Y.). Plaque assays were photographed using an AlphaImager (Alpha Innotech, San Leandro, Calif.) documentation system. Data for the Southern dot blot analyses were acquired using a Storm PhosphorImager (Molecular Dynamics, Sunnydale, Calif.) and quantitated using ImageQuant software (Molecular Dynamics). Images were adjusted with Adobe Photoshop 5.0 software (Adobe Systems Inc., San Jose, Calif.), data were plotted using SigmaPlot 7.1 software (SSPS, Chicago, Ill.), and final figures were assembled and labeled with Canvas 8.0 software (Deneba Systems, Miami, Fla.).
RESULTS
We implemented a genetic analysis of the telomere-binding I6 protein to study its function in vivo. Clustered charge-to-alanine mutagenesis was used to generate several altered alleles of I6 that might engender a ts phenotype within the context of viral infection. This technique has been successfully used in our laboratory and others to generate viruses with conditionally lethal mutations in such viral genes as H5, A20, and D1 (7, 15, 16, 18, 32). Clusters of charged amino acids are thought to have a high likelihood of lying on the surface of a protein and are therefore likely to serve as interaction sites with other molecules. When these residues are changed to alanine, these interactions may weaken, with the decrement of function being temperature dependent (38). If the stability and/or activity of the protein were essential for virus viability, such a mutation would thus cause a conditional lethality that would manifest itself at 40 but not at 32°C, the temperatures chosen for experimental use.
Replacement of the endogenous I6 allele with alleles carrying clustered charge-to-alanine substitutions.
A total of 13 clusters of charged residues were identified in the predicted amino acid sequence of the I6 protein (Fig. 1). Of these, eight clusters spread throughout the I6 ORF were chosen for alteration. Overlap PCR was used to change the codons for these residues (shaded) in regions 1, 2, 4, 5, 6, 9, 12, and 13 so that they would instead encode alanine. Mutant alleles were then inserted into the pUC/Neo plasmid, which contains the neomycin resistance gene (Neor) under the control of a constitutive vaccinia virus promoter (24), to facilitate their placement within the endogenous I6 locus of the viral genome.
Allelic replacement of the endogenous (wt) I6 gene was accomplished with the use of transient dominant selection (10, 21, 24). pUC/Neo plasmids containing the mutant I6 alleles were introduced into cells infected with wt virus, and following infection-transfection, the cells were maintained at 32°C in the presence of G418 for 2 days. Viruses that had incorporated the plasmid into their genomes via homologous recombination were selected by two rounds of plaque purification performed in the presence of G418. Insertion of the plasmid was confirmed by screening viral isolates for the presence of the Neor gene by PCR. These G418r viruses contain an unstable, tandem duplication of the I6 region flanking the Neor gene. When selective pressure is removed by performing additional rounds of plaque purification in the absence of G418, the duplicated sequences readily undergo homologous recombination, resulting in the loss of the Neor gene. This resolution reaction was monitored by PCR; viruses in which the resolved genome retained the mutant, rather than the wt, I6 allele were identified by DNA sequence analysis. All eight recombinant viruses, vI6-1, vI6-2, vI6-4, vI6-5, vI6-6, vI6-9, vI6-12, and vI6-13, were successfully isolated at 32°C. Thus, each of the alleles generated could support virus viability at this permissive temperature.
Phenotypic analysis: several I6 alleles affect plaque size and/or viral yield.
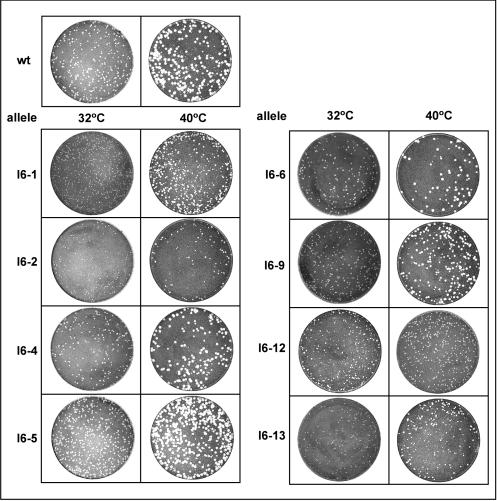
The eight viruses generated were tested for their ability to form plaques at 32 and 40°C (Fig. 2). All mutant viruses were able to form plaques at 32°C, although the plaques observed for vI6-1, vI6-2, and vI6-13 were somewhat smaller than those formed by wt virus. The viruses showed distinct differences at 40°C. vI6-4, vI6-5, vI6-6, and vI6-9 formed large plaques comparable in size to those formed by wt virus, whereas vI6-1, vI6-2, vI6-12, and vI6-13 formed smaller plaques. This temperature-dependent decrease in plaque size might reflect a reduction in infectious viral yield during the first and subsequent rounds of infection or might instead reflect a decrease in the ability of the mutant virus to spread to adjacent cells after the first round of infection.
FIG. 2.
Comparison of the plaque-forming abilities of wt virus and the vI6 viruses at 32 and 40°C. Confluent monolayers of BSC40 cells were infected with 150 PFU of virus for 48 h at the indicated temperature and then fixed and stained with crystal violet. All the mutant viruses formed plaques at both 32 and 40°C. At 32°C, viruses carrying alleles 1, 2, and 13 formed somewhat smaller plaques than did wt virus. At 40°C, viruses carrying alleles 1, 2, 12, and 13 formed significantly smaller plaques than did wt virus.
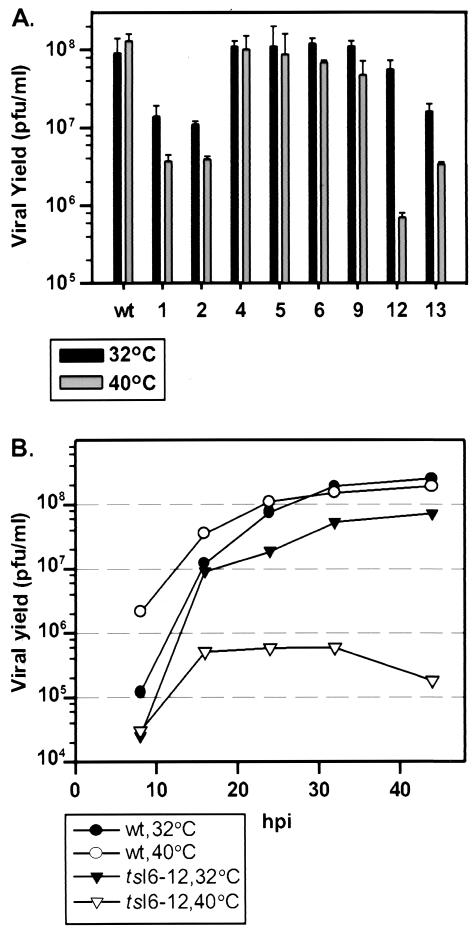
To quantitate the total yield of infectious virus from a single round of infection, BSC40 cells were infected with wt or mutant viruses at an MOI of 2 and maintained at 32 or 40°C for 24 h. As shown in Fig. 3A, wt infections produced similar amounts of infectious virus at both temperatures; similar results were obtained for vI6-4, vI6-5, vI6-6, and vI6-9. vI6-1, vI6-2, vI6-12, and vI6-13 were ts. For vI6-1, vI6-2, and vI6-13, the viral yield at 32°C was 6- to 8-fold lower than that obtained from wt infections performed in parallel and 30- to 40-fold lower than that obtained from wt infections at 40°C. vI6-12 exhibited the tightest ts phenotype; the yield at 32°C was comparable to that obtained with wt virus, whereas a 100-fold reduction in viral yield was seen at 40°C, the nonpermissive temperature. This virus, denoted as tsI6-12, was chosen for further characterization, as described below.
FIG. 3.
Comparison of virus production levels during synchronous infections with wt virus and the vI6 mutants at 32 and 40°C. (A) Quantitation of 24-h infectious viral yield: alleles 1, 2, 12, and 13 affected virus production, with allele 12 causing a tight ts phenotype. Confluent monolayers of BSC40 cells were infected with wt virus or vI6 viruses (MOI of 2) and incubated at 32 or 40°C. At 24 hpi, cells were harvested and disrupted and the yield of cell-associated, infectious virus was measured by plaque assay at 32°C. (B) Time course of virus production in tsI6-12-infected cultures: virus production was diminished, not delayed. Confluent monolayers of BCS40 cells were infected with wt virus or tsI6-12 (MOI of 2), incubated at 32 or 40°C, and harvested at 8, 16, 24, 32, or 44 hpi. Cells were disrupted, and the viral yield was measured by plaque assay at 32°C.
Surprisingly, tsI6-12, which shows a 2-log reduction in 24-h viral yield at 40°C, was still able to form small plaques at this temperature. We therefore monitored the time course of virus production in cells infected with tsI6-12 to more clearly distinguish between reduced and delayed virus production (Fig. 3B). In wt-infected cells, virus production was more rapid at 40 than at 32°C but reached an equivalent plateau at 24 hpi. A similar growth curve was observed for the tsI6-12 infection performed at 32°C, although the viral yield was slightly lower than that obtained for wt virus at 32°C. In contrast, cells infected nonpermissively with tsI6-12 showed an increase in viral yield from 8 to 16 hpi, after which no further virus production was seen. This result demonstrates that the production of infectious virus during a nonpermissive tsI6-12 infection is blocked rather than delayed.
Marker rescue was performed to confirm that the ts phenotype of tsI6-12 was indeed caused by the engineered mutation and was not due to an unintended, nonfortuitous change in the genome of this virus. Cells were infected at 40°C with tsI6-12 and transfected with linearized plasmid DNA with or without a wt I6 ORF insert; at 48 hpi, the yield of temperature-insensitive virus was titrated at 40°C. Plasmid DNA containing the wt I6 gene, but not empty plasmid, resulted in a significantly increased yield of virus able to form large plaques at 40°C (data not shown), verifying that the ts phenotype was caused solely by the amino acid substitutions within the I6 gene.
Neither viral DNA synthesis nor concatemer resolution is defective in nonpermissive tsI6-12 infections.
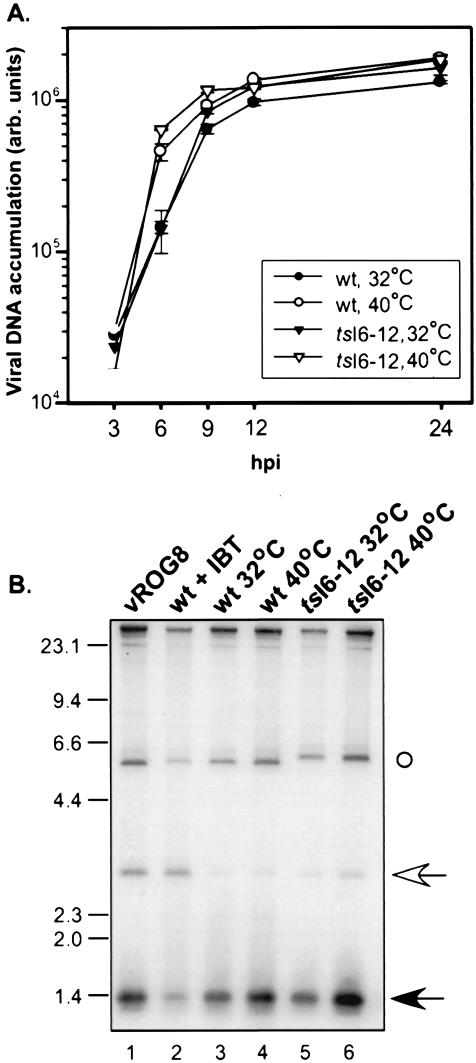
Given the affinity of the I6 protein for the viral telomeric hairpins and the importance of these structures for minichromosome replication, we hypothesized that I6 might play a role in DNA replication. To examine this possibility, we quantitated the accumulation of viral DNA in cells infected with wt virus or tsI6-12 at 32 or 40°C. Cell lysates were prepared at 3, 6, 9, 12, and 24 hpi and analyzed by Southern dot blot analysis. As shown in Fig. 4A, the profiles of DNA accumulation were comparable in all of the samples, indicating that the tsI6-12 allele does not cause a ts defect in viral DNA synthesis. We also examined the temporal profile of viral protein synthesis in cells infected with wt virus or tsI6-12 at both 32 and 40°C. Lysates were prepared from cultures metabolically labeled with 35S-Met at various times postinfection, resolved by SDS-PAGE, and visualized by autoradiography; the patterns of protein expression directed by the two viruses were indistinguishable (data not shown). Because DNA synthesis is a prerequisite for the switch to intermediate and late transcription, these data confirm that the tsI6-12 allele does not affect the progression of DNA replication and also indicate that it has no impact on viral gene expression.
FIG. 4.
DNA replication and concatemer resolution are not impaired in nonpermissive tsI6-12 infections. (A) Cells were infected with wt virus or tsI6-12 (MOI of 5) and incubated at 32 or 40°C. Cells were harvested at 3, 6, 9, 12, or 24 hpi, and the levels of intracellular, viral DNA (expressed in arbitrary [arb.] units) were determined by Southern dot blot hybridization (in duplicate) and quantitated by phosphorimager analysis. (B) Cells were infected with wt virus (lanes 3 and 4) or tsI6-12 (lanes 5 and 6) (MOI of 2) and incubated at 32°C (lanes 3 and 5) or 40°C (lanes 4 and 6). As controls, cells were infected with vROG8 (MOI of 15) in the absence of IPTG (lane 1) or with wt virus (MOI of 2) in the presence of 60 μM IBT (lane 2) (conditions which lead to the accumulation of unresolved concatemers). At 18 hpi, cells were harvested and cytoplasmic DNA was extracted and digested with BstEII. DNA was resolved by electrophoresis, transferred to a ZetaProbe membrane, and hybridized with a radiolabeled probe that annealsto the termini of mature genomes (1.3-kb predicted fragment [filled arrow]) and the junction fragment of unresolved concatemeric intermediates (2.6-kb fragment [open arrow]). (The probe also hybridizes to the BstEII fragment proximal to the telomeric regions ○.) The electrophoretic migration of DNA standards is shown at the left, with sizes indicated in kilobases.
Next, we asked whether viral replication intermediates (concatemers) were efficiently resolved into monomeric genomes. Viral DNA was extracted from cells infected with wt or tsI6-12 virus at 32 or 40°C and analyzed by Southern blot hybridization using a probe that differentiates between concatemeric and monomeric genomes. As controls, cells were infected under conditions that compromised late gene expression and hence led to diminished concatemer resolution. These conditions are engendered by infection with wt virus in the presence of the drug IBT or with vROG, a recombinant virus in which the late-transcription factor G8 is repressed when IPTG is omitted from the culture medium (29, 41). As shown in Fig. 4B, the samples prepared from cells infected with vROG (lane 1) or with wt virus in the presence of IBT (lane 2) both accumulated unresolved concatemers (2.6-kb BstII band) whereas monomers (1.3-kb predicted band) predominated in the samples prepared from wt and tsI6-12 infections performed at either 32 or 40°C (lanes 3 to 6). We conclude that genome resolution proceeds normally during nonpermissive infections performed with tsI6-12.
Proteolytic processing of late viral core proteins is incomplete in tsI6-12.
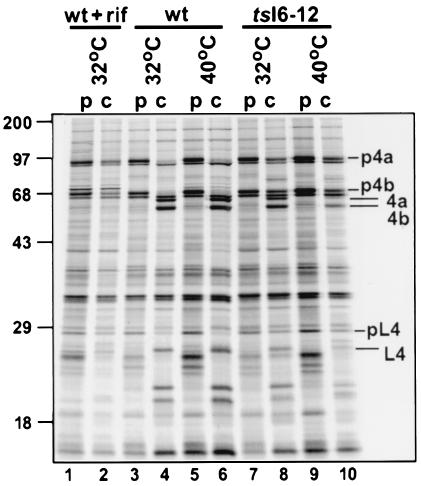
The data described above indicate that the replication cycle of tsI6-12 proceeds normally through the biochemical processes of viral gene expression, DNA replication, and genome resolution. Therefore, the defect must lie in a subsequent stage of the viral life cycle—affecting either virion morphogenesis or virion infectivity. As an initial test of whether morphogenesis might be affected, we examined the proteolytic processing of the major core proteins. Three abundant viral structural proteins (p4a, p4b, and preL4) are proteolytically cleaved in a manner that is tightly linked to intracellular mature virus (IMV) formation (37). Cells were infected with wt virus or tsI6-12 at 32 or 40°C; as a control, cells were also infected with wt virus in the presence of RIF, a drug that inhibits viral morphogenesis (14, 28). At 8 hpi, cells were pulsed with [35S]-Met for 45 min; half of the cultures were harvested immediately (Fig. 5, lanes p) and the other half were rinsed and fed with medium containing unlabeled methionine and incubated for an additional 16 h (lanes c). During wt infection, cleavage of the three viral precursors into the products 4a, 4b, and L4 was nearly complete at both temperatures (compare lanes 3 and 4 to lanes 5 and 6). In the presence of RIF, however, only minimal cleavage is observed (compare lanes 1 and 2). Processing is somewhat diminished in the tsI6mut-12 infection performed at 32°C, as evidenced by the persistence of detectable levels of uncleaved precursors in the sample harvested after the 16-h chase (compare lanes 7 and 8 to lanes 3 and 4). This diminution of processing is more pronounced at 40°C, at which temperature only ∼30% of the precursor is processed (compare lanes 9 and 10 to lanes 5 and 6). This observation implies that viral morphogenesis is compromised during nonpermissive tsI6-12 infections.
FIG. 5.
Proteolytic processing of viral structural proteins is impaired during nonpermissive tsI6-12 infections. Cells were infected with wt virus (lanes 3 to 6) or tsI6-12 (lanes 7 to 10) (MOI of 10) and incubated at 32°C (lanes 3, 4, 7, and 8) or 40°C (lanes 5, 6, 9, and 10). As a control, cells were infected with wt virus in the presence of RIF (100 μg/ml) (lanes 1 and 2), which causes morphogenesis to arrest at an early stage prior to the proteolytic processing of core proteins. At 8 hpi, cells were metabolically labeled with [35S]-Met for 45 min; cells were then either harvested immediately (pulse [p] lanes [lanes 1, 3, 5, 7, and 9]) or refed with DMEM-5% FBS, returned to 32 or 40°C, and harvested at 24 hpi (chase [c] lanes [lanes 2, 4, 6, 8, and 10]). Samples were resolved by SDS-PAGE and visualized by autoradiography. Short dashes indicate the precursor form of the virion core proteins (p4a, p4b, and preL4); longer lines indicate the proteolytically processed forms (4a, 4b, and L4). Protein standards are shown at the left; their molecular masses are shown in kilodaltons.
Morphogenesis is blocked at the IV-to-IMV transition during nonpermissive tsI6-12 infections.
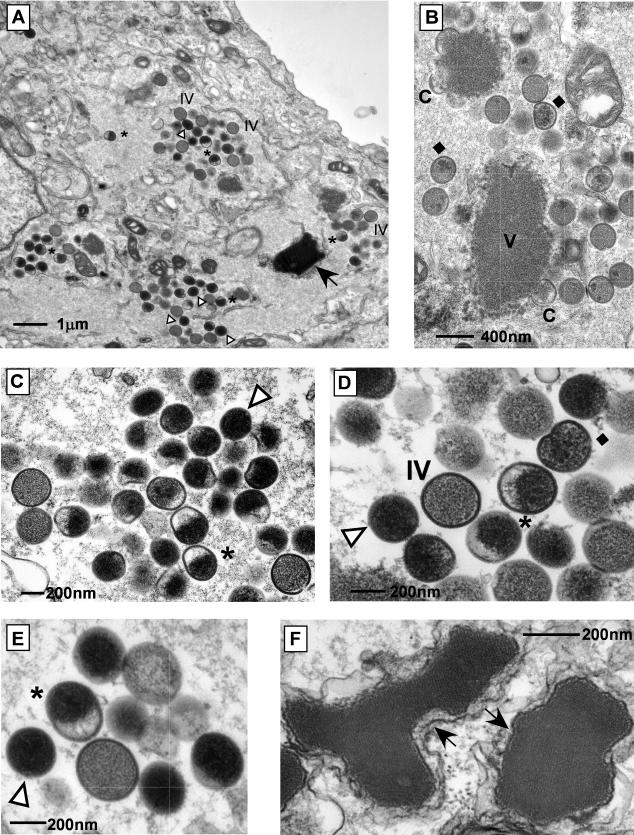
Virus assembly proceeds through several visually distinct stages: the appearance of electron-dense regions (termed virosomes) in the cytoplasm, the formation of crescents at the virosomal periphery, the maturation of these crescents into viroplasm-containing immature virions (IV), the development of nucleoid-containing IVN, and transformation into the infectious form known as IMV (reviewed in reference 27). Transmission electron microscopy was used to investigate the progression of virion morphogenesis in cells infected with tsI6-12. In cultures infected at the permissive temperature, all of the typical hallmarks of morphogenesis were observed (data not shown). We also noted the presence of some additional, unusual structures (see below), which were more dramatic and predominant in cells infected at the nonpermissive temperature. The images shown in Fig. 6 illustrate the range of structures seen in cells infected with tsI6-12 at 40°C. Cells contained normal virosomes, crescents, and IV. However, there was a virtual absence of IVN and IMV. A few IV contained several dark internal foci but nothing comparable to a nucleoid. In other IV, the internal electron-dense material was distributed asymmetrically to one hemisphere. The most abundant and distinct structures observed were spherical, electron-dense particles. Another unusual feature was the presence of large crystalloids, often surrounded by membrane. Brick-shaped crystalloids (thought to represent viral DNA) have been documented in infected cells treated with RIF, a drug that arrests morphogenesis prior to IV formation (14). The crystalloids seen here were several times larger than those observed in the presence of RIF and also lacked a precise rectangular shape; however, the internal structure of parallel filaments was retained.
FIG. 6.
During nonpermissive infections with tsI6-12, virion morphogenesis proceeds through IV formation but arrests prior to IMV formation. BSC40 cells were infected with tsI6-12 (MOI of 2) and maintained at 40°C for 17 h. Cells were fixed in situ and processed for transmission electron microscopy. Structures are indicated as follows: V, virosome; C, crescent; ♦, IV with electron-dense foci; *, half-filled particle; Δ, electron-dense spherical particle; →, crystalloid. Note the absence of IVN and IMV. Final magnifications: (A) ×9,000, (B) ×25,000, (C) ×30,000, (D) ×45,000, (E) ×48,000, (F) ×75,000.
The execution point of the I6-12 allele occurs at late times of infection.
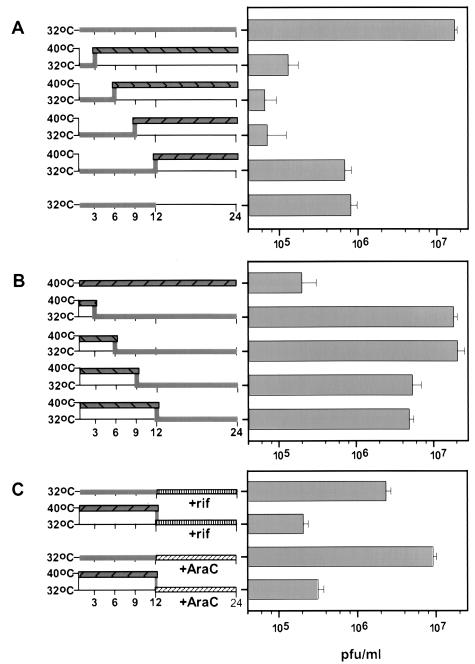
Temperature shift experiments represent classical genetic tools for the determination of the execution point of conditionally lethal mutants. To determine when the tsI6-12 allele exerts its effect during the infectious cycle in a manner that causes an arrest in virus maturation, we performed a series of shift-up and shift-down experiments. When infections were initiated at 32°C and cultures were then shifted up to 40°C at 3, 6, or 9 hpi, the 24-h yield was approximately 2 logs lower than that obtained from cells maintained at the permissive temperature for the full 24 h (Fig. 7A). When cultures were shifted to 40°C at 12 hpi, the 24-h yield was no higher than that obtained from the 32°C cultures harvested at 12 hpi. Thus, even when infections were maintained at the permissive temperature for the first 12 h of infection, the shift to 40°C blocked further production of virus. The execution point was therefore very late in the infectious cycle, during the crucial stages of virus assembly.
FIG. 7.
The execution point of the tsI6-12 allele occurs late during infection (>12 hpi). Confluent monolayers of BSC40 cells were infected with tsI6-12 (MOI of 2) and harvested at 24 hpi unless otherwise indicated. Viral yield was measured by a plaque assay at 32°C. In each case, the experimental strategy is depicted at the left, with the periods at 32°C shown with grey bars and periods at 40°C shown with hatched grey bars. The data are shown at the right; all experiments were performed in triplicate and the average titers are shown (with error bars). (A) Shift-up experiments. All infections were initiated at 32°C. Two cultures were maintained at the permissive temperature and harvested at 12 or 24 hpi. The remaining cultures were shifted to the nonpermissive temperature at 3, 6, 9, or 12 hpi and then harvested at 24 hpi. (B) Shift-down experiments. All infections were initiated at 40°C. One culture was maintained at 40°C for 24 h. The remaining cultures were shifted to the permissive temperature at 3, 6, 9, or 12 hpi and then harvested at 24 hpi. (C) Reversal of the ts arrest by a shift to 32°C at 12 hpi requires de novo DNA synthesis and virion morphogenesis. Confluent monolayers of BSC40 cells were infected at 32 or 40°C. At 12 hpi, araC (20 μM) (diagonally hatched bars without shading) or RIF (100 μg/ml) (vertically hatched bars without shading) was added to the medium and cultures were shifted to or maintained at 32°C. Data shown in panels A and B (e.g., the results for the last experiment shown at the bottom of panel B [shift-down at 12 hpi]) were obtained in parallel and served as the controls for these experiments.
The converse experiment was also performed: infections were initiated at 40°C and cultures were shifted down to 32°C at various times postinfection (Fig. 7B). When infected cultures were shifted down at 3 or 6 hpi, the viral yield was equivalent to that obtained from fully permissive infections. Even when cultures were shifted down at 9 or 12 hpi, only a threefold decrease in viral yield was seen. These shift-down experiments confirm that the function of the I6 protein is most crucial at late times of infection.
We next asked whether the recovery of virus production seen in cultures shifted down to 32°C in the midst of infection depended on de novo morphogenesis and/or DNA synthesis. When RIF was added to the cultures as they were shifted down to the permissive temperature at 12 hpi, the viral yield was reduced to the levels obtained from cultures maintained at 40°C (Fig. 7C). Since RIF blocks morphogenesis at crescent formation prior to the block seen in tsI6-12 infections, we conclude that the viral structures that accumulate during nonpermissive tsI6-12 infections are dead-end products and that de novo morphogenesis is required to reverse the ts arrest. An analogous experiment was performed using araC, an inhibitor of DNA synthesis. Addition of araC to permissive infections at 12 hpi did not reduce the 24-h viral yield, indicating that the pool of DNA synthesized prior to 12 h was sufficient to support normal levels of virus production. However, when araC was added to the 40°C cultures as they were shifted to the permissive temperature at 12 hpi, the viral yield was reduced to the levels obtained from cultures maintained at 40°C. These data show that reversibility of the ts arrest is also dependent on de novo DNA synthesis and suggest that the DNA crystalloids that accumulate at 40°C cannot provide viral genomes for encapsidation into fresh virions.
Morphology, protein composition, and specific infectivity of viral particles purified from nonpermissive tsI6-12 infections.
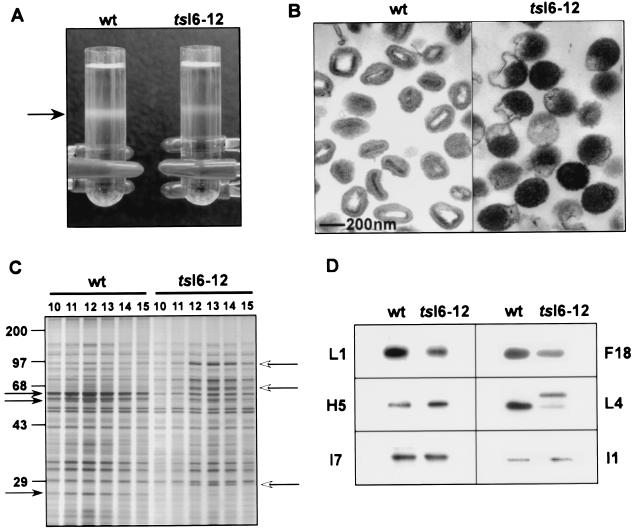
IMVs produced during wt infections can be purified by ultracentrifugation through a 36% sucrose cushion followed by banding on a 25 to 40% sucrose gradient. We attempted to apply the same protocol to the purification of viral particles from cells infected with tsI6-12 at the nonpermissive temperature. In the tsI6-12 sample, a light-scattering band was observed at a position slightly above that seen with the wt sample (Fig. 8A). Fractions were collected from each gradient, and those containing the highest optical density were assayed for protein content and composition, infectivity (viral titer), and ultrastructural morphology. Protein concentrations were determined on fractions solubilized with mild detergent to ensure complete dissociation of viral particles; the peak tsI6-12 fractions contained one-third to one-half of the total protein obtained from the peak wt fractions. To quantitate the infectivity of the samples, monolayers were infected at 32°C with an equivalent number of particles (normalized by protein content) from the ts or wt peak fractions and plaques were counted after 48 h. The particle/PFU ratio of the tsI6-12 preparations was 80-fold higher than that of the wt preparations. The determination that the specific infectivity of tsI6-12 particles was 1.25% of that of wt particles corresponded well to our initial demonstration that the viral yield from tsI6-12-infected cultures was 2 logs lower than that obtained from wt-infected cultures (Fig. 2). When we examined pooled purified preparations by conventional transmission electron microscopy, the wt sample was shown to consist mainly of IMV whereas the tsI6-12 sample contained deformed particles corresponding to the half-filled and dense spherical particles observed in the infected cells (Fig. 8B; compare with Fig. 6).
FIG. 8.
Purification and characterization of wt and tsI6-12 particles produced during 40°C infections. (A) Sucrose gradient purification. Cells were infected with wt virus or tsI6-12 (MOI of 2) for 24 h at 40°C. Virus particles were purified from cytoplasmic extracts by sedimentation through a 36% sucrose cushion followed by banding on a 25 to 40% sucrose gradient. The arrow points to light-scattering bands containing viral particles. (B) Transmission electron microscopy of wt and tsI6-12 particles. Peak fractions from the sucrose gradients were retrieved, concentrated, and subjected to analysis by conventional transmission electron microscopy. (C) Comparison of the overall protein profiles of the wt and tsI6-12 particles. Gradient fractions 10 to 15 corresponded to the light-scattering bands and contained the peak protein content. Equal volumes of the indicated fractions were resolved by SDS-PAGE and developed by silver staining. Closed arrows (left side of panel) indicate proteolytically processed viral core proteins, and open arrows (right side of panel) point to the uncleaved precursor form of these core proteins. Protein standards are shown at the left, with their molecular masses indicated in kilodaltons. (D) Detection of selected viral proteins in purified wt and tsI6-12 particles. Samples of purified wt and tsI6-12 particles containing equal amounts of protein, as measured by a Bradford assay, were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Filters were probed with antisera specific for the L1, H5, I7, F18, L4, and I1 proteins. Immunoreactive proteins were visualized by chemiluminescence after development with appropriate secondary antisera.
Aliquots from the gradients corresponding to the light-scattering band were also resolved by SDS-PAGE and visualized by silver staining (Fig. 8C). wt and tsI6-12 fractions were found to have similar protein profiles once the processing status of the structural proteins was taken into account. p4a, p4b, and pre-L4 predominated in the fractions from the tsI6-12 gradient, whereas the cleaved products predominated in the fractions obtained from the wt sample. To obtain a more refined view of the protein composition of the two particle preparations, equivalent amounts of protein were resolved electrophoretically and subjected to immunoblot analysis using antisera generated against multiple viral proteins (Fig. 8D). All the proteins surveyed (L1, H5, I7, I1, F18, and L4) were present at similar levels in the wt and tsI6-12 samples, except the L4 protein was present primarily as an uncleaved precursor in the ts-I6 sample.
tsI6-12 particles are devoid of viral DNA.
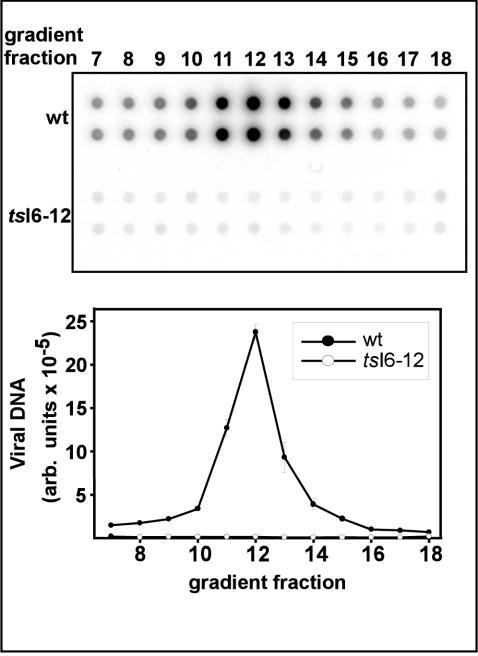
The gradient fractions described above were also subjected to Southern dot blot analysis to quantitate the amount of viral DNA encapsidated in the purified particles (Fig. 9). In the wt sample, a characteristic profile of DNA content was seen, with peak levels found in fraction 12. A dramatically different result was seen for the tsI6-12 sample: no peak of DNA was found. Background levels of hybridization were detected for the whole range of fractions tested. The mutant particles, therefore, appear to be devoid of viral DNA. We conclude that at nonpermissive temperatures, the tsI6-12 mutation leads to a failure in genome encapsidation.
FIG. 9.
Purified tsI6-12 particles are devoid of viral DNA. Equal volumes of fractions 7 to 18 of the sucrose gradients used to purify wt and tsI6-12 particles were spotted in duplicate onto ZetaProbe membranes. Southern dot blot analysis was utilized to quantitate the levels of viral DNA in each fraction. A digital image from a phosphorimager is shown in the top portion of the figure; hybridization signal levels (in arbitrary [arb.] units) were quantitated by phosphorimager analysis and a graphical representation of the data are shown in the bottom portion of the figure.
DISCUSSION
The vaccinia virus I6 protein first came to our attention because of its ability to bind to the telomeric hairpins of the viral genome with high specificity and stability; this interaction is dependent upon the extrahelical bases that distinguish these unusual telomeres (6). We hypothesized that this protein would be involved in an important facet of genome metabolism, either in the initiation of DNA synthesis or the encapsidation of viral DNA. We chose a genetic approach to the deciphering of the role of I6 in vivo, attempting to generate I6 alleles that would confer a conditionally lethal phenotype. Clusters of charged residues in eight regions of the I6 ORF were mutated to alanine, and recombinant viruses containing these mutations at the endogenous locus were successfully isolated and analyzed for temperature sensitivity. Our initial screening of comparative viral replication at 32 and 40°C involved two assays: (i) a plaque assay and (ii) the quantitation of viral yield from a single, synchronous infectious cycle. The phenotypes of vI6-4, vI6-5, vI6-6, and vI6-9 were indistinguishable from that of wt virus in both experiments. In contrast, vI6-1, vI6-2, vI6-12, and vI6-13 exhibited a small plaque phenotype at the nonpermissive temperature. A reduction in plaque size can reflect either a diminution in total virus production or a specific defect in viral spread to neighboring cells. To distinguish between the two possibilities, the amount of total cell-associated virus produced in a single infectious cycle was measured. vI6-1, vI6-2, and vI6-13 showed a decrease in viral yield of 6- to 8-fold relative to that of wt virus at 32°C and a 30- to 40-fold decrease compared to that of wt virus at 40°C. vI6-12 exhibited the tightest ts phenotype, producing wt levels of infectious virus at 32°C and 100-fold less virus at 40°C. This mutant (denoted tsI6-12) was chosen for further characterization.
We next examined several parameters of the viral life cycle to determine which processes were impaired during nonpermissive infections with tsI6-12. No defect was observed in viral gene expression, DNA replication, or concatemer resolution. These data indicated that the arrest occurs at a late stage of infection and involves either viral morphogenesis or virion infectivity. Proteolytic processing of several major core proteins known to undergo cleavage during virion maturation was diminished, suggesting that morphogenesis was likely to be affected by the tsI6-12 mutation. We therefore pursued a more thorough analysis of virion morphogenesis by electron microscopy. tsI6-12-infected cells harvested at either 12 or 17 hpi showed a clear and dramatic perturbation in virus assembly. The hallmarks of the earlier stages of viral morphogenesis, such as electron-dense virosomes, membrane crescents, and IV, were observed. However, the normal progression of morphogenesis was then blocked, since there were few, if any, IVN or IMV. Instead, we observed several types of aberrant particles: IV containing small electron-dense foci, half-filled particles with electron-dense material distributed asymmetrically at one pole, and spherical, electron-dense particles. In addition, we saw large membrane-enclosed crystalloids, which were comprised of an array of parallel filaments. Crystalloids (thought to comprise viral DNA) have been observed previously in cells infected in the presence of RIF (14), which blocks morphogenesis prior to IV formation, and in the absence of functional A32 or I7 proteins (4, 9). Whereas the typical crystalloids seen in the presence of RIF have a fairly uniform, oblong appearance, the crystalloids seen in tsI6-12-infected cells were much larger and more irregular, as if several crystalloids with different orientations had adhered together. As has been documented for wt infections performed in the presence of RIF (data not shown) and for nonpermissive ts16 infections (9), the crystalloids we observe during nonpermissive tsI6-12 infections appear to be surrounded by membranes. The presence of membranes surrounding the crystalloids is particularly interesting in the light of recent evidence that DNA synthesis occurs in membrane-delimited regions of the cytoplasm (34).
Consistent with the observed defect in virion morphogenesis, temperature-shift experiments demonstrated that the execution point of I6 during a tsI6-12 infection occurs during very late stages of the virus life cycle. These experiments also demonstrated that de novo morphogenesis is required to reverse the ts arrest, since no recovery of virus production was seen when cultures were shifted down to 32°C at 12 hpi in the presence of RIF. Apparently the aberrant particles that accumulate at 40°C are dead-end products rather than functional intermediates, since they are unable to mature into IMV upon a shift to the permissive temperature. A restoration of virus production was also not seen when cultures were shifted to the permissive temperature in the presence of araC, indicating that de novo DNA synthesis is also needed for reversal of the ts phenotype. The crystalloids that accumulated in tsI6-12-infected cells must have represented viral DNA that had assumed an aggregated, unusable conformation. In contrast, the crystalloids that accumulated in the presence of RIF appeared to be depots of functional DNA, since the RIF-induced arrest to virus production can be reversed at 12 hpi in the presence of araC (data not shown).
Our electron microscopic analysis indicated that in nonpermissive tsI6-12 infections, morphogenesis arrested at a point beyond the formation of IVs. We were interested in purifying the aberrant particles that seemed to accumulate to further analyze their structure and function. Cytoplasmic extracts from nonpermissively infected cultures were therefore sedimented through a sucrose cushion, and the pellet was applied to a sucrose gradient. A light-scattering band was observed at a position slightly above that seen with the wt sample. The contents of the band were retrieved and concentrated; electron microscopic analysis indicated that the preparation consisted almost exclusively of the half-filled and electron-dense spherical particles described above. The wt sample, as expected, contained almost exclusively IMV; the specific infectivity (in PFU per microgram) of the tsI6-12 particles was 1.25% of that of wt virus. The overall protein profile of the tsI6-12 particles appeared similar to that of the wt IMV, with the exception that precursor, rather than processed, forms of such core proteins as 4a, 4b and L4 were seen. More specifically, the I7, I1, H5, L1, L4, and F18 proteins were found at similar levels in the two preparations, although L4 was primarily unprocessed in the tsI6-12 particles.
The most dramatic characteristic of the tsI6-12 particles was the absence of any detectable viral DNA. We conclude that the primary defect in nonpermissive tsI6-12 infections is the inability of the developing virions to package viral DNA. This encapsidation defect is certainly consistent with the two-log reduction in infectious virus production that we observed. However, the fact that tsI6-12 formed small but detectable plaques at 40°C is puzzling. These plaques may have arisen due to limited spread of the residual number of infectious particles that were produced. Alternatively, the plaques may reflect the delivery of aberrant particles into neighboring cells, with concomitant induction of cytopathic effect due to viral entry and/or delivery of viral proteins. In support of the latter hypothesis, we have infected monolayers with the purified, aberrant particles produced at 40°C from tsI6-12-infected cells and shown that the encapsidated F18 protein is in fact delivered into these cells (data not shown) (24).
The phenotype of tsI6-12 at nonpermissive temperature is remarkably similar to that seen when expression of the A32 protein is repressed (vA32i-IPTG) (4). A32 is a putative ATPase with limited sequence homology to the products of gene I of filamentous, single-stranded DNA bacteriophages and to the IVa2 gene of adenovirus, both shown to be ATPases involved in DNA packaging (22). In cells infected nonpermissively with vA32i (-IPTG), morphogenesis arrests after IV formation; crystalloids and spherical electron-dense particles were observed, although the half-filled particles described here were not seen. The purified, electron-dense particles had a protein profile similar to that of wt IMV, but the viral DNA content was only 18% of that of wt IMV.
Thus, A32 and I6 are now the two vaccinia virus proteins implicated directly in the process of DNA encapsidation. Interestingly, the DNA-deficient particles obtained in both cases still contain wt levels of the F18 protein. F18 is an abundant, basic, 11-kDa phosphoprotein that has been described as part of the nucleoprotein complex and is the most abundant component of the virion core (19, 20). Repression of F18 leads to a block in morphogenesis at the stage of IVN formation (42). The observation that F18 enters viral particles at wt levels in the absence of DNA encapsidation certainly argues against a conventional role for F18 as an integral component of the nucleoprotein complex. It remains possible, however, that F18 might coat the DNA after entry during the process of core formation and virion maturation. Given the finding that the particles purified from tsI6-12-infected cells lack viral DNA, it was initially surprising that they formed a light-scattering band with a density that was only slightly different from that of wt particles. However, it was reported some years ago that the viral DNA accounts for only 3% of the mass of the virion (43).
The complex process of viral DNA packaging has been best studied in bacteriophage models such as φ29, λ, T4, and P22 (reviewed in references 5, 11, and 13). DNA is packaged into the procapsid head, a spherical intermediate that transforms into the mature capsid following DNA encapsidation. DNA entry occurs at a single opening termed the connector or portal. In the case of λ phage, the terminase binds to DNA cos sites found at the termini of concatemeric intermediates and then associates with docking proteins located on the procapsid connector. The ATPase activity of the terminase provides energy needed for DNA translocation through the channel. Once an internal cos site marking the boundary of the monomeric genome is reached, the terminase cleaves the concatemer so that only one copy of the genome is packaged.
DNA encapsidation in animal viruses is less well understood, although there are many parallels between herpes simplex virus type 1 (HSV-1) and adenovirus packaging and the bacteriophage pathway. Namely, key proteins binds to specific sequences in the DNA and then localize to a site in the procapsid as a consequence of DNA binding and prior to DNA entry. Seven proteins in HSV-1 have been implicated in DNA packaging: UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (reviewed in reference 17). UL28 recognizes and binds to terminal DNA sequences (pac sites) (1), which are important for packaging and cleavage of concatemeric intermediates into monomeric genomes. UL28 binds UL15, a putative ATPase, and the heterodimer associates with the procapsid possibly by direct interaction with the UL6 portal protein (23, 33, 39, 40). The current hypothesis proposes that the UL28/UL15 heterodimer acts as the terminase, with UL28 binding to specific sites on the viral DNA and UL15 providing the energy for genome resolution and translocation.
It is tempting to speculate whether a related packaging mechanism is utilized by vaccinia virus. Unlike that of HSV-1, resolution of the vaccinia virus concatemeric intermediates is uncoupled from DNA encapsidation, although it is a prerequisite step. In the absence of the Holliday junction resolvase A22, branched and concatemeric genomes accumulate and a morphogenesis defect similar to that observed with vA32-IPTG and tsI6-12 is seen (12). Apparently, only monomeric forms of the genome are packaged and concatemers are excluded; a defect in resolution indirectly results in a DNA packaging defect. However, concatemer resolution is not impaired in the absence of functional A32 or I6 (conditions under which genome encapsidation is abolished). We propose the following working model for vaccinia virus genome encapsidation. We hypothesize that I6 binds to the telomeres of monomeric genomes and then interacts with A32 found on the surface of IV. We propose that once binding by I6 occurs, the ATPase activity of A32 becomes activated and provides the energy needed for DNA translocation into the maturing virion. DNA translocation may take place through a pore or an unsealed gap in the immature virion that closes once the translocation process is complete. Alternatively, the entry of the genome may lead to invagination of the IV membrane such that an internal membrane surrounds the core of IMV. With the identification of the I6 protein as an essential component of the encapsidation machinery, testing and refinement of this working model of poxvirus DNA encapsidation should prove to be feasible and intriguing.
Acknowledgments
This work was supported by NIH R01 AI 21758 awarded to P.T.
We thank B. Moss, S. Shuman, and D. Hruby for providing viruses and antisera mentioned in the text. We also thank Joseph DeMasi for his input during the early stages of the work and Gang Ning for help with the electron microscopy.
REFERENCES
- 1.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baroudy, B. M., S. Venkatesan, and B. Moss. 1982. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 28:315-324. [DOI] [PubMed] [Google Scholar]
- 3.Baroudy, B. M., S. Venkatesan, and B. Moss. 1983. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47(Pt. 2):723-729. [DOI] [PubMed] [Google Scholar]
- 4.Cassetti, M. C., M. Merchlinsky, E. J. Wolffe, A. S. Weisberg, and B. Moss. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMasi, J., S. Du, D. Lennon, and P. Traktman. 2001. Vaccinia virus telomeres: interaction with the viral I1, I6, and K4 proteins. J. Virol. 75:10090-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMasi, J., and P. Traktman. 2000. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J. Virol. 74:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du, S., and P. Traktman. 1996. Vaccinia virus DNA replication: two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc. Natl. Acad. Sci. USA 93:9693-9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. K. Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkner, F. G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 64:3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, A. D., and B. Moss. 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 75:6460-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimes, S., P. J. Jardine, and D. Anderson. 2002. Bacteriophage phi 29 DNA packaging. Adv. Virus Res. 58:255-294. [DOI] [PubMed] [Google Scholar]
- 14.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett, D. E., and R. C. Condit. 1994. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc. Natl. Acad. Sci. USA 91:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassett, D. E., J. I. Lewis, X. Xing, L. DeLange, and R. C. Condit. 1997. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology 238:391-409. [DOI] [PubMed] [Google Scholar]
- 17.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 18.Ishii, K., and B. Moss. 2001. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J. Virol. 75:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, S. Y., and W. R. Bauer. 1987. Biosynthesis and phosphorylation of vaccinia virus structural protein VP11. Virology 159:399-407. [DOI] [PubMed] [Google Scholar]
- 20.Kao, S. Y., E. Ressner, J. Kates, and W. R. Bauer. 1981. Purification and characterization of a superhelix binding protein from vaccinia virus. Virology 111:500-508. [DOI] [PubMed] [Google Scholar]
- 21.Klemperer, N., J. Ward, E. Evans, and P. Traktman. 1997. The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol. 71:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin, E. V., T. G. Senkevich, and V. I. Chernos. 1993. Gene A32 product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes 7:89-94. [DOI] [PubMed] [Google Scholar]
- 23.Koslowski, K. M., P. R. Shaver, J. T. Casey, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, K., B. Lemon, and P. Traktman. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69:7823-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merchlinsky, M., C. F. Garon, and B. Moss. 1988. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J. Mol. Biol. 199:399-413. [DOI] [PubMed] [Google Scholar]
- 26.Merchlinsky, M., and B. Moss. 1986. Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell 45:879-884. [DOI] [PubMed] [Google Scholar]
- 27.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2884. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 28.Moss, B., E. N. Rosenblum, E. Katz, and P. M. Grimley. 1969. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature 224:1280-1284. [DOI] [PubMed] [Google Scholar]
- 29.Pacha, R. F., and R. C. Condit. 1985. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J. Virol. 56:395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogo, B. G., E. M. Berkowitz, and S. Dales. 1984. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology 132:436-444. [DOI] [PubMed] [Google Scholar]
- 31.Pogo, B. G., M. O'Shea, and P. Freimuth. 1981. Initiation and termination of vaccinia virus DNA replication. Virology 108:241-248. [DOI] [PubMed] [Google Scholar]
- 32.Punjabi, A., K. Boyle, J. DeMasi, O. Grubisha, B. Unger, M. Khanna, and P. Traktman. 2001. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J. Virol. 75:12308-12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tolonen, N., L. Doglio, S. Schleich, and J. Krijnse-Locker. 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12:2031-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traktman, P. 1996. Poxvirus DNA replication, p. 775-798. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 36.Traktman, P., A. Caligiuri, S. A. Jesty, K. Liu, and U. Sankar. 1995. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanSlyke, J. K., and D. E. Hruby. 1990. Posttranslational modification of vaccinia virus proteins. Curr. Top. Microbiol. Immunol. 163:185-206. [DOI] [PubMed] [Google Scholar]
- 38.Wertman, K. F., D. G. Drubin, and D. Botstein. 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., J. G. Keck, and B. Moss. 1992. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J. Virol. 66:6470-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y. F., and B. Moss. 1991. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J. Virol. 65:6101-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwartouw, H. T. 1964. The chemical composition of vaccinia virus. J. Gen. Microbiol. 34:115-123. [DOI] [PubMed] [Google Scholar]