Abstract
Recent studies have characterized significant differences in the cis-regulatory sequences of related organisms, but the impact of these differences on gene expression remains largely unexplored. Here, we show that most previously identified differences in transcription factor (TF)-binding sequences of yeasts and mammals have no detectable effect on gene expression, suggesting that compensatory mechanisms allow promoters to rapidly evolve while maintaining a stabilized expression pattern. To examine the impact of changes in cis-regulatory elements in a more controlled setting, we compared the genes induced during mating of three yeast species. This response is governed by a single TF (STE12), and variations in its predicted binding sites can indeed account for about half of the observed expression differences. The remaining unexplained differences are correlated with the increased divergence of the sequences that flank the binding sites and an apparent modulation of chromatin structure. Our analysis emphasizes the flexibility of promoter structure, and highlights the interplay between specific binding sites and general chromatin structure in the control of gene expression.
Keywords: chromatin structure, evolution, gene expression
Introduction
The unique phenotype of each organism is defined by a combination of its gene content and the regulation of these genes. Evolution of protein sequence and its contribution to phenotypic adaptation has been studied extensively (Pal et al, 2006), while most of what we know about regulatory evolution comes from the study of individual genes (Wray, 2007). Regulatory evolution reflects changes in genomic sequences that influence (either directly or indirectly) gene expression. Among the multiple mechanisms that control gene expression, the binding of transcription factors (TFs) to sequence-specific binding sites within the upstream promoters is arguably the best-characterized regulatory scheme. The short lengths of TF-binding sites, and their sensitivity to even a small number of mutations, make them ideal candidates for driving gene expression divergence (ED) in cis.
Recent studies have begun to compare the promoters of related organisms in search for such regulatory differences. These analyses are hindered by the difficulty to distinguish differences in functional (e.g. TF-binding sites) from non-functional promoter elements. Nonetheless, leveraging on prior knowledge of TF-binding motifs, several studies predicted the gain and loss of thousands of TF-binding sites both in yeast and mammalian species (Donaldson and Gottgens, 2006; Doniger and Fay, 2007). Furthermore, two very recent studies have used chromatin immunoprecipitation to directly identify differences in TF binding among related species (Borneman et al, 2007; Odom et al, 2007).
While these studies rely on the premise that observed differences in TF-binding sequences represent gene ED and ultimately phenotypic evolution, this was not directly examined. In fact, various evidences have indicated that extensive promoter divergence, including differences in TF-binding sequences, may evolve neutrally, with no influence on gene expression. First, several studies found that promoters from different species which have extensively diverged still drive the same reporter expression patterns (Ludwig et al, 1998; Takahashi et al, 1999; Romano and Wray, 2003; Ruvinsky and Ruvkun, 2003; Oda-Ishii et al, 2005; Fisher et al, 2006; Wang et al, 2007). Second, it was shown that changes in TF-binding sequences are poorly correlated with divergence of gene expression among yeast paralogs (Zhang et al, 2004). Third, Doniger and Fay (2007) introduced mutations in binding sequences that were found in Saccharomyces paradoxus and S. mikatae to the orthologous promoters in S. cerevisiae and examined their effect on gene expression using reporter assays. The expected effects on gene expression were found only in 3 out of the 11 cases that were examined. Finally, a comprehensive analysis of ∼1% of the human genome (The ENCODE Project Consortium, 2007) has shown that a large percentage of the functional regulatory elements are not conserved among mammals, suggesting the neutral evolution of these sequences. Thus, it appears that promoters are highly flexible, and are capable of maintaining stabilized gene expression pattern through many realizations of sequences, even when binding motifs are concerned.
Here, we examine the impact of changes in TF-binding sequences on the associated gene expression on a genome-wide scale using comparative expression data sets of related organisms (Ranz et al, 2003; Rifkin et al, 2003; Su et al, 2004; Khaitovich et al, 2005; Gilad et al, 2006; Tirosh et al, 2006) combined with comparative data sets of predicted TF-binding sites. We find that most predicted changes in TF-binding sequences, in both yeast and mammals, have only little effect on gene expression. To examine the connection between changes in TF-binding sequences and ED in a more controlled setting, we measured the transcription response of three closely related yeast species to mating pheromone. Analysis of this response allows us to assess the relative contribution of specific cis-regulatory elements and of general promoter structure to the divergence of gene expression.
Results
Reported changes in TF-binding sequences have only little detectable impact on gene expression
A recent study has characterized thousands of matches to binding site motifs that were conserved in the promoters of both chimpanzee and mouse (and are thus likely to be functional in both organisms) but are mutated, and do not match the binding site motif, in human (Donaldson and Gottgens, 2006). To examine the impact of these mutations on gene expression, we assembled two data sets comparing gene expression between human and either chimpanzee or mouse. The first data set compares the expression levels of human and chimpanzee genes across five tissues (Khaitovich et al, 2005) and the second data set compares the expression patterns of human and mouse genes across 30 orthologous tissues (Su et al, 2004). In both cases, we found that the ED of genes with diverged sequence motifs in their proximal promoters (1 kb) was indistinguishable from the ED of genes with conserved motifs (Supplementary Figure 1).
Analysis of mammalian regulatory divergence is hindered by several limitations, including the inherent complexity of mammalian promoters, the presence of multiple (often far away) enhancers, and the poor knowledge of TFs and their binding specificities. To try and circumvent these problems, we turned to yeast, whose promoters are significantly shorter (∼600 bp) and well defined. Doniger and Fay (2007) have recently analyzed the conservation of sequence motifs in promoters of closely related yeast species and predicted the loss of TF-binding sites. We examined the impact of predicted changes in TF-binding sites on ED using a comparative data set that we have recently reported, where we examined the genome-wide expression programs of the same species to several environmental stresses (Tirosh et al, 2006).
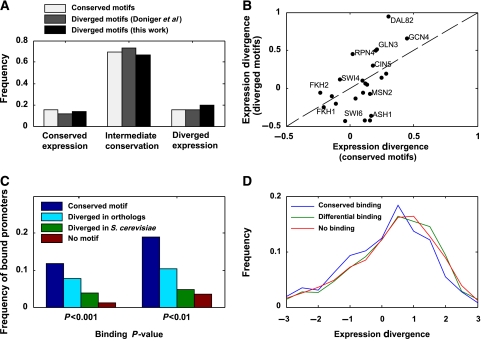
Also here, genes predicted to loose TF-binding sites (i.e. whose promoters contained diverged sequence motifs) had the same level of ED as genes with conserved sequence motifs (Figure 1A). To further verify these results, we have also analyzed the promoters of these species and generated another set of predicted changes in TF-binding sites (see Materials and methods). Notably, both our analysis and that of Doniger and Fay (2007) predicted the loss of a TF-binding site only if there were no other matches to that sequence motif at the same promoter, thus effectively removing cases of ‘binding site turnover' (Dermitzakis and Clark, 2002). However, also for these predictions we observed similar levels of ED (Figure 1A; see Supplementary Figure 2 for results with different parameters). Since the expression data are taken from stress conditions, it could be expected that only changes in sequence motifs for TFs that participate in the stress response will have an impact in this data set. We thus separately analyzed the impact of changes in sequence motifs for each TF (Figure 1B). Although changes in sequence motifs for stress-related TFs were, in general, associated with higher ED than other TFs, none of these TFs were significantly associated with high ED.
Figure 1.
Expression divergence of yeast genes with diverged TF sequence motifs. (A) The percentage of genes with conserved, intermediate or diverged expression among those with conserved or diverged motifs as predicted by Doniger and Fay (2007) and by a similar analysis (see Materials and methods and Supplementary Figure 2). The difference between any pair of the three sets is not statistically significant (P>0.05). (B) Average expression divergence for genes with conserved or diverged motifs for various TFs. Some stress-related TFs (e.g. GCN4, DAL82) have relatively high ED of genes with diverged motifs, but in none of these cases it is significantly higher than the respective ED of genes with conserved motifs. (C) Percentage of S. cerevisiae-bound promoters at two different binding P-values (Harbison et al, 2004) among promoters with different patterns of motif conservation and divergence. The difference between each pair of different patterns is significant (P<0.05). (D) Expression divergence between human and mouse liver cells of genes with conserved, diverged or no binding by four liver-related TFs.
One possible explanation for the lack of correlation between the divergence of sequence motifs and that of gene expression is that the reported sequence variations do not affect the binding of the respective TFs to promoters. To explore this possibility, we examined the binding of multiple TFs, with conserved or diverged motifs, to S. cerevisiae promoters (Harbison et al, 2004) (Figure 1C). We found that S. cerevisiae promoters with diverged motifs are bound by the respective TF less often than promoters with conserved motifs but more often than promoters without sequence motifs for that TF in any of the species (Figure 1C). Thus, in certain cases, TFs retain their binding to promoters despite divergence of the respective sequence motifs, although this may also represent the differences between the promoter regions examined by sequence analysis and those experimentally tested. However, in other cases, promoters with diverged motifs are not bound by the respective TF, suggesting that TF binding has also diverged. Notably, also for these promoters, the percentage of genes with diverged expression is not higher than average (Supplementary Figure 2). Thus, despite the apparent loss of TF binding, gene expression remained conserved, perhaps through compensation by other regulatory elements.
Since divergence of sequence motifs corresponds only partially to divergence of TF binding, interspecies differences in TF binding should be experimentally determined. The binding of four TFs (FOXA2, HNF1A, HNF4A and HNF6) to 4000 orthologous gene pairs in human and mouse liver cells was recently analyzed by chromatin immunoprecipitation (Odom et al, 2007) and extensive differences were identified between the binding of these TFs to orthologous genes. To examine the impact of these differences, we compared the expression levels of human and mouse liver cells (Xing et al, 2007). However, we found no connection between differences in TF binding and ED (Figure 1D).
Genome-wide analysis of the mating response in three yeast species
The difficulty to predict expression changes from analysis of sequence motifs could stem from the coordinated activity of multiple TFs that affect gene expression through combinatorial regulation. The binding of multiple TFs to multiple promoter elements may conceal the influence of specific differences, but at the same time provide more raw material for regulatory changes and is, in general, correlated with an increased ED (Tirosh et al, 2006; Landry et al, 2007). We thus sought to analyze a simpler situation where gene expression is defined primarily by a single TF. The yeast mating response appears suitable. This response is relatively isolated and is induced by the activation of the STE12 TF whose consensus binding motif, as well as target promoters, is well defined (Roberts et al, 2000; Zeitlinger et al, 2003; Bardwell, 2005).
Mating in yeast occurs when two haploid cells of the opposite mating types (a and α) fuse and form a single diploid cell. Each cell secrets a unique pheromone (a-factor or α-factor, respectively) that is sensed by the other cell, triggering it to initiate the mating response. Mating involves extensive changes in the gene expression program (Roberts et al, 2000), with more than 200 upregulated genes and additional genes downregulated. Many of the upregulated genes have specific roles in mating and are regulated by STE12, while the downregulated ones are associated with cell-cycle arrest.
The α-factor peptide secreted by S. cerevisiae has been isolated and synthesized in vitro. Currently, most studies of the mating response are performed by subjecting haploid yeast cells of the a-type to this synthetic α-factor (e.g. Roberts et al, 2000). Since the gene coding for the α-factor peptide is highly conserved within the sensu–stricto complex, we used the synthetic α-factor from S. cerevisiae to elicit the mating response in three closely related species: S. cerevisiae, S. paradoxus and S. mikatae. As expected, all species responded to this α-factor by arresting their cell cycle and growing a visible shmoo (Supplementary Figure 3).
We measured the gene expression response to α-factor using microarrays containing complete-ORF probes for the ∼6000 S. cerevisiae genes. To control for technical variations, we performed biological repeats (three in S. cerevisiae and S. paradoxus and four in S. mikatae), with all experiments (for all species and repeats) executed in parallel. The sequence of S. paradoxus and S. mikatae genes is highly similar to those of S. cerevisiae (∼90 and ∼85% on average, respectively), and accordingly produced significant and reproducible hybridization. Notably, while absolute hybridization intensities are affected by sequence mismatches, our analysis is based solely on the ratios of hybridization intensities in samples taken with and without pheromone. Indeed, this cross-species hybridization platform was validated in both yeast and other organisms by us and others (Sartor et al, 2006; Tirosh et al, 2006; Oshlack et al, 2007).
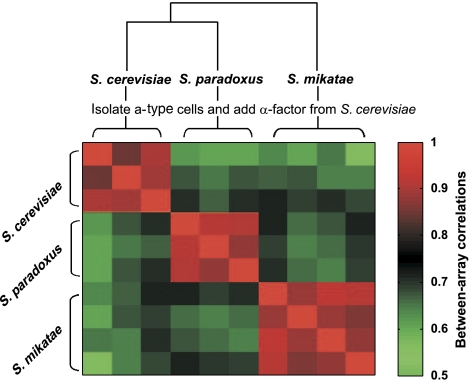
In each of the three species, α-factor induced significant (P<0.05) expression changes of more than 1000 genes. As expected, about 100 genes were upregulated by at least two-fold, and these genes were enriched with previously known mating-related genes (P<10−5 in each species). The response of each species was highly reproducible, with a genome-wide correlation of ∼0.9 among biological repeats (Figure 2). The correlations between the responses of the different species were significantly lower (r∼0.6–0.7), although clearly far from being random. Thus, the overall transcriptional response is conserved but also includes substantial species-specific differences. As additional controls, we compared our results with those of a previous study of the mating response in S. cerevisiae (Roberts et al, 2000) and with expression measurements of S. cerevisiae cells undergoing natural mating. As expected, both data sets had high correlations with the response of the three species to α-factor and especially with the response of S. cerevisiae (Supplementary Figure 4).
Figure 2.
Correlations between the mating expression program in different species. We isolated a-type cells from S. cerevisiae, S. paradoxus and S. mikatae, subjected them to S. cerevisiae α-factor and measured their genome-wide expression profiles using S. cerevisiae arrays. Each species was measured with three or four biological repeats. The correlations among these genomic responses were calculated over 3248 genes with a significant response in at least one experiment.
We identified 408 genes that are differentially expressed between at least one pair of yeast species (see Materials and methods and Supplementary Table 1). Interestingly, these diverged genes had high ED also in the stress-related comparative data (Tirosh et al, 2006) (P=5 × 10−26), and were enriched with TATA-containing genes (35% compared with 22%, P=1.5 × 10−8). This suggests that genes vary in their tendency for ED, such that the same genes diverge in expression at different processes. Furthermore, this tendency may be related to the presence of TATA boxes, as we and others have previously suggested (Tirosh et al, 2006; Landry et al, 2007). In contrast, these diverged genes do not have higher then average protein sequence divergence (Wall et al, 2005) (P=0.46), suggesting a decoupling between evolution of protein sequence and expression.
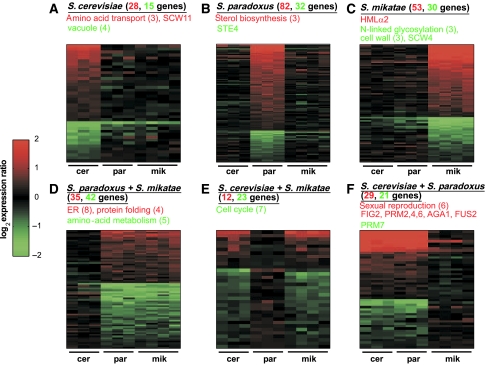
To better understand the pattern of differential expression, we classified the differentially expressed genes into 12 classes and separately analyzed the genes within each class (Figure 3). Each class corresponds to genes that are up- or downregulated only in a specific subset of the three species (Materials and methods). Interestingly, the largest class corresponds to 82 genes with an S. paradoxus-specific upregulation (Figure 3B), and the smallest class corresponds to 12 genes with the opposite pattern (Figure 3E; no upregulation only in S. paradoxus).
Figure 3.
Differential expression pattern in the mating response. Differentially expressed genes were classified into 12 patterns of up- or downregulation in a subset of the species (see Materials and methods). Each subfigure shows the log2 expression ratios of genes from two classes corresponding to up- and downregulation in a specific subset of species. (A) S. cerevisiae, (B) S. paradoxus, (C) S. mikatae, (D) S. paradoxus+S. mikatae, (E) S. cerevisiae+S. mikatae, (F) S. cerevisiae+S. paradoxus. Cer, par and mik indicate the columns corresponding to expression of S. cerevisiae, S. paradoxus and S. mikatae, respectively. The corresponding subset of species, number of genes within each class, enriched GO annotations and selected genes are indicated at the top of each subfigure. Red and green correspond to up- and downregulation, respectively.
We analyzed the enrichment of gene ontology (GO) annotations and the presence of mating-related genes within each class (Figure 3). The most notable class was composed of genes that were upregulated in S. cerevisiae and S. paradoxus but not in S. mikatae (Figure 3F). This class is comprised of 29 genes and includes six mating-related genes (FIG2, AGA1 and PRM2,4,6 and FUS2). This class, as well as the entire set of differentially expressed genes, is also enriched with cell wall genes (P<10−4 for both gene sets), which are important for various aspects of the mating response such as shmoo formation, cell adhesion and fusion. Additional studies are needed to elucidate the possible impact of these expression differences on the mating behavior of these yeast species.
TF-binding sequences versus ED in the mating response
The mating response is orchestrated by a single TF, STE12, whose binding site motif (TGAAACA) is well characterized. This binding consensus, as well as the coding sequence of STE12, and specifically its binding domain are well conserved between the species analyzed (Supplementary Figure 5). As mentioned above, these properties enable a more focused analysis of evolution of TF-binding sequences that minimizes trans-acting and combinatorial effects. We thus searched for genes with a conserved STE12 sequence motif that has diverged in at least one of the three yeast species for which expression was measured (Materials and methods). This analysis identified 64 diverged sequence motifs. In each species, we classified diverged STE12 sequence motifs as a ‘conserved motif' if the sequence motif was conserved in that species but lost in another species, and ‘lost motif' if the sequence motif was conserved in two other species but lost in that species.
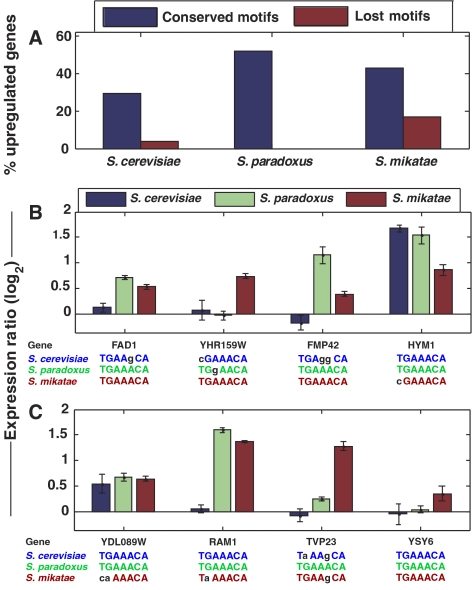
Divergence of STE12 sequence motifs was highly correlated with ED in the mating response. In each of the three species, genes with ‘conserved motif' were significantly more likely to be upregulated in response to α-factor than genes with ‘lost motif' (Figure 4A). For example, more than half of the conserved motifs in S. paradoxus were associated with upregulated S. paradoxus genes (24 out of 46), while none of the 17 lost motifs in S. paradoxus were associated with upregulated S. paradoxus genes. Thus, genes with an STE12-binding site that is conserved in two species but lost in the third species tend to respond to α-factor only in the species in which the site is conserved (Figure 4B). For example, the promoter of FAD1 (flavin adenine dinucleotide synthetase) has a perfect match to the STE12 sequence motif in S. paradoxus and S. mikatae, but not in S. cerevisiae. Accordingly, FAD1 was upregulated only in S. paradoxus and S. mikatae.
Figure 4.
Mating response of genes with conserved or diverged STE12 sequence motifs. (A) The percentage of upregulated genes in response to α-factor is significantly higher among genes whose STE12 sequence motif is conserved (blue) than those whose sequence motifs has diverged (red) in each species (P=1 × 10−3, 4 × 10−6, 1 × 10−4 for S. cerevisiae, S. paradoxus and S. mikatae, respectively). Only sequence motifs which are conserved in two species and diverged in another were considered, and these were defined as ‘conserved' and ‘lost' in the respective species (including 27, 17 and 52 genes lost in S. cerevisiae, S. paradoxus and S. mikatae, respectively). Upregulation was defined by log2 ratio>0.5, yet other thresholds gave similar results. (B) Four examples of genes in which divergence of STE12 sequence motifs was associated with reduced response to α-factor. Conserved and mutated STE12 sequence motifs are shown beneath each gene; mutated positions are indicated by black and lowercase. (C) Four examples of genes in which the presence of STE12 sequence motifs is not correlated with the response to α-factor.
A significant number of exceptions were still observed, however (Figure 4C). For example, RAM1 (β subunit of the farnesyltransferase complex which prenylates the a-factor) was upregulated in S. mikatae despite a mutation in the STE12 sequence motif, and was not upregulated in S. cerevisiae despite the conservation of its STE12 sequence motif. In yet other cases, differential expression was found despite the presence of conserved sequence motifs (e.g. YSY6).
We next asked how much of the observed interspecies differential expression can be accounted for by differences in STE12 sequence motifs (Figure 5). Since STE12 controls the upregulation, but not downregulation, in response to α-factor, we examined the presence and divergence of STE12 sequence motifs in promoters of genes that are upregulated only in a subset of the three yeast species. We found that only 11% of this differential expression can be accounted for by divergence of STE12-binding sequences. If we restrict this analysis to genes with STE12 sequence motifs in at least one species, which are thus more likely to be directly regulated by STE12, divergence of STE12-binding sequences is consistent with approximately one-third of the expression differences.
Figure 5.
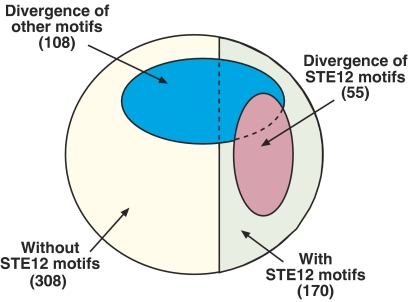
Differential upregulation classified by the presence and divergence of TF-binding sites. Venn diagram showing all the pairwise species differences in upregulation, classified by the presence of STE12 sequence motifs (yellow—no motif, green—motif in at least one of the species), the divergence of STE12 sequence motifs (red—expression differences are consistent with divergence of STE12 sequence motifs) and the divergence of additional TFs which are significantly associated with differences in the mating response (blue—expression differences are consistent with divergence of TF sequence motifs). Numbers in parentheses indicate the number of genes in each class.
While the mating response is predominately activated by STE12, other TFs may also impact the observed gene expression. We thus examined whether divergence of sequence motifs for other TFs is correlated with expression differences in the mating response. Divergence of sequence motifs for seven additional TFs were found to be significantly (P<0.05) associated with differential upregulation (MBP1, TEC1, SKO1, RDS1, SWI6, HAP2 and HSP1). For example, out of 16 pairs of orthologs which are differentially upregulated and also have a diverged MBP1 sequence motif, divergence of the motif is correlated with the loss of upregulation in 15 gene pairs (P<0.001), consistent with the predicted cooperative binding of MBP1 and STE12 (Banerjee and Zhang, 2003; Das et al, 2004). Conversely, loss of TEC1 sequence motifs is correlated with the gain of upregulation, consistent with the role of TEC1 in shifting the binding of STE12 from mating-related genes to pseudohyphal-related genes (Zeitlinger et al, 2003). Moreover, in four of these genes, the differential appearance of the TEC1 motif in S. cerevisiae and S. mikatae is also consistent with their differential binding by TEC1 in pseudohyphal conditions (Borneman et al, 2007), while none show the opposite pattern. Altogether, divergence of sequence motifs for STE12 and these seven additional TFs, could account for 32% of the observed differential upregulation and up to 49% of the differential upregulation of genes with an STE12 sequence motif (Figure 5). In contrast, divergence of these motifs is inconsistent with differential upregulation (e.g. loss of STE12 motifs coincides with gain of upregulation) in 17% of all genes and 27% of STE12-dependent genes.
Flanking promoter sequences and their impact on chromatin structure
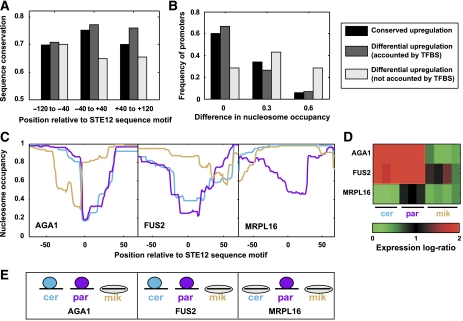
Taken together, changes in TF sequence motifs can account for approximately half of the ED of STE12-dependent genes. To understand the genetic basis of the remaining expression differences, we examined the divergence of promoter sequences that flank the STE12 sequence motifs (Figure 6A). Interestingly, we found that these flanking sequences are less conserved in genes with unexplained differential upregulation than either in genes with conserved upregulation or in those with differential upregulation that could be accounted by diverged binding sequences. This difference did not extend to the entire promoter and was only significant at the region surrounding the STE12-binding sequences (−40 to +40). Moreover, this region was less conserved than the remaining promoter among genes with unexplained differential upregulation but not among other genes (Figure 6A). These results suggest that sequences flanking STE12-binding sites are important for STE12 binding and that divergence of these sequences is responsible for some of the observed differential upregulation.
Figure 6.
Divergence of flanking promoter sequences and chromatin structure. (A) The average conservation of promoter sequences flanking conserved STE12 sequence motifs is shown as a function of the distance from the sequence motif (x-axis) and the conservation of the expression response (color). The difference in conservation between genes with unexplained differential expression and the other two gene sets is significant only at the −40 to +40 interval (P<0.05). (B) Maximal interspecies differences in the predicted nucleosome occupancy at the location of conserved STE12-binding sites. (C) Predicted nucleosome occupancy at the regions of conserved STE12-binding sites for the three genes with the largest differences in nucleosome occupancy among those with unexplained differential upregulation (interspecies differences larger than 0.5). The three species are indicated by different colors (blue, purple and orange correspond to cerevisiae, paradoxus and mikatae, respectively). (D) Expression response (log ratio) of the three genes shown in (C). (E) Predicted configuration of STE12 binding (circles) and nucleosomes (ovals) at the STE12-binding sites of the three genes.
Flanking sequences could influence gene expression if they contain unrecognized sites that are bound by either STE12 or by related TFs. However, we find no enrichment of matches (or weak matches) to TF sequence motifs at these regions. Alternatively, these sequences could influence gene expression through other mechanisms, most notably by modulating chromatin structure. To examine this possibility, we applied a computational model to predict the nucleosome occupancy in promoters of the different yeast species (Segal et al, 2006). We found that divergence of the sequences flanking STE12 sequence motifs indeed affects the predicted nucleosome occupancy at the location of the sequence motifs, and that this effect is larger among genes with unexplained differential upregulation compared with other genes (Figure 6B).
For most genes, the predicted differences in nucleosome occupancy are quite small and do not appear to influence gene expression. However, among the three genes with the largest predicted changes in nucleosome occupancy (differences larger than 0.5 at the conserved STE12 sequence motifs), these changes were exactly consistent with the observed differential upregulation (Figure 6C–E). For example, the mating-related gene FUS2 is upregulated in S. cerevisiae and S. paradoxus but not in S. mikatae, and the predicted nucleosome occupancy of the conserved STE12-binding site at its promoter is significantly higher in S. mikatae. These results suggest that divergence of the sequences flanking conserved TF-binding sites have influenced nucleosome positioning and therefore also the accessibility of promoters to TFs. To further examine this possibility, we analyzed the predicted nucleosome occupancy among genes with interspecies differences in the binding of TEC1 and STE12 at pseudohyphal conditions (Borneman et al, 2007). Similarly, we found that the largest predicted changes in nucleosome occupancy are consistent with differences in TF binding (Supplementary Figure 6); predicted changes in nucleosome occupancy could account for six out of seven differences in TF binding among the genes with the largest (>0.5) variations (P=0.03).
Discussion
Our goal in this study was to examine systematically the impact of changes in cis-regulatory elements on the divergence of gene expression. To this end, we first analyzed existing data comparing the gene expression profiles of related species for which promoter analyses predicted the gain and loss of TF-binding sites. Both in mammalian and in yeast data sets, we observed only a poor correlation between the divergence of sequence motifs and gene expression. Furthermore, analysis of experimentally mapped differences in human and mouse TF binding led to similar conclusions.
Although surprising, these results are consistent with various previous studies, as described above. Several reasons may explain the stabilized expression we and others have observed despite the divergence of TF sequence motifs. First, the identification of diverged motifs may not be optimal and may not correspond to divergence in TF binding. We tried to overcome this difficulty by using different data sets and restricting our analysis to the most significant cases of sequence divergence. Second, it might be that divergence of sequence motifs does impact gene expression, but only under conditions which do not appear in our gene expression data sets. However, also for the stress-related TFs in yeast, divergence of sequence motifs could not predict divergence of gene expression during environmental stress. Third, although we tried to control for binding sites turnover, a compensatory motif may have appeared in more distant regions not considered in our analysis. Again, this is an important limitation concerning the mammalian system where transcription is controlled by multiple enhancers, but is less of a problem in the yeast system where the relevant controlling region is believed to be rather limited.
Finally, gene expression might be compensated through other mechanisms, such as chromatin structure or interactions among TFs. For example, loss of binding sites may be buffered by the presence of other binding sites for additional factors that are involved in the same process or that bind cooperatively with the factor whose binding site has diverged. Indeed, both the mammalian tissue-specific expression and the response of yeast to stressful conditions involve a coordinated function of multiple TFs. Thus, the observed variability in TF-binding sequences among related species may reflect the complexity of the underlying regulatory network and the possibility to encode the same expression pattern with many different promoter designs perhaps even more than it reflects the actual divergence of gene expression.
To overcome the above limitations, we extended the analysis to a more controlled setting and generated a new comparative data set describing the transcriptional response of three closely related yeast species to mating pheromone. This response is predominantly orchestrated by a single TF, STE12, whose binding specificity is well characterized. While the response was largely conserved, the expression of ∼400 genes significantly differed between the species. In this case, divergence of STE12 sequence motifs was correlated with the changes in gene expression, and together with other TF sequence motifs could account for approximately one-third of the observed differential upregulation and up to half of that of STE12-regulated genes. Importantly, predictions based on sequence analysis are only partially consistent with the actual binding of TFs. In fact, a number of studies over the past few years indicate that binding does occur at highly diverged and non-consensus sites, reflecting additional regulatory aspects, which are still poorly understood. For example, approximately half of the genes that are bound by STE12 in S. cerevisiae contain an exact match to the STE12-binding motif, and only one-third of the genes with conserved matches to the STE12-binding motif are bound by STE12 in S. cerevisiae (Harbison et al, 2004). Accordingly, the impact of binding site divergence could be actually larger than that inferred from our analysis of binding site sequences alone.
Chromatin structure has a central role in controlling the accessibility of promoters to TFs, and the mechanisms by which it is regulated are widely studied (Li et al, 2007). The influence of DNA sequence on nucleosome positions has been extensively analyzed and several models have been developed to predict nucleosome positioning based on DNA sequences alone (Ioshikhes et al, 2006; Segal et al, 2006). We applied the model developed by Segal et al to examine the effect of sequence divergence in regions surrounding STE12-binding sites and found that sequence divergence may indeed affect nucleosome positions and that, at least in certain cases, this could account for the observed differential expression. Similarly, we found that some of the interspecies differences in the binding of TEC1 and STE12 at pseudohyphal conditions are correlated with changes in nucleosome occupancy. Thus, the location of nucleosomes can evolve among closely related species and influence gene ED. Notably, our analysis may capture only a small proportion of the actual influence of chromatin structure, as it is based on computational predictions of the intrinsic preference of nucleosomes for certain DNA sequences, but ignores additional regulatory mechanisms and particularly the dynamic nature of chromatin structure. It would be interesting, although significantly more complicated, to directly measure the location of nucleosomes among multiple species at similar conditions, such as during mating, and examine their influence on expression differences.
Our results provide a detailed account of the impact of promoter sequence variations on gene expression within the mating system. Such a detailed analysis is enabled by the relative simplicity of this system and would be much more difficult in typical regulatory systems, such as the yeast stress response or the mammalian liver. Similar analysis of additional regulatory systems may indicate the generality of the results shown here and begin to uncover the relative importance of different mechanisms in generating interspecies differences in gene expression.
Finally, we note that while our study focused on the genetic basis of gene expression evolution, a complementary challenge is to describe the contribution of such changes to phenotypic diversity. In this aspect, the data set generated here might be beneficial, as it provides insights into the modulation of the mating response. First, we have found that several mating-related genes are not upregulated in S. mikatae as in S. cerevisiae and S. paradoxus (Figure 3F). Second, we identified a large class of genes that were only upregulated in S. paradoxus (Figure 3B). Third, studies of protein sequence evolution have shown that proteins involved in mating and fertility are among the most rapidly evolving genes (Swanson and Vacquier, 2002). In the future, as more data accumulate, it would be interesting to examine our data set from this respect and to see whether the same is also true for the evolution of gene expression and to examine the possible contribution to phenotypic adaptation and speciation.
Materials and methods
Prediction of changes in human TF-binding sequences
We downloaded the mammalian promoters multiple alignments from the UCSC genome browser. We focused on proximal promoters (1 kb) of human, chimpanzee, mouse and rat and searched them for the presence of exact matches to the mammalian promoter motifs defined by Xie et al (2005). This initial search was performed after excluding the alignment gaps. We restricted this analysis to the promoter motifs with the highest confidence: those which matched the binding sites of known TFs and the 30 promoter motifs with the highest conservation significance.
We searched for promoters that contain an exact match to a specific motif in several species but not in another. Because the human sequences are more accurate than those of the other species, and because our expression data sets compare human with either chimpanzee or mouse, we focused only on matches that are conserved in chimpanzee and mouse (or rat) but mutated in human. Thus, we demanded that the chimpanzee and mouse promoters contain exact matches to the motif and that these matches are aligned (at the same position in the multiple alignments). The entire human proximal promoter was required to have no exact matches. The number of functional mutations was estimated by the number of differences from the motif in the sequence, which was aligned to the matches in the other species. This analysis resulted in prediction of 695 human promoters with at least two mutations in a TF-binding site, which is conserved in chimpanzee and mouse (or rat).
Prediction of changes in yeast TF-binding sequences
Promoters (600 bp) of S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii and S. bayanus were downloaded from SGD and aligned by clustelw. We searched these promoters for the presence of TF sequence motifs as defined by MacIsaac et al (2006). For each TF, we calculated the position weight matrix scores across all promoters and defined binding sites as matches with scores greater than a predefined threshold (t1). Diverged promoters were defined as those with a conserved and aligned match exceeding t1 in at least three of the five species, but no match in another species that exceeds a second threshold (t2). Thus, promoters with multiple binding sequences for the same TF would be recognized as diverged only if all these binding sequences have diverged in one of the species. The thresholds (t1 and t2) were chosen based on their specificity in predicting the binding of TFs (Harbison et al, 2004); the specificity of a given threshold is the fraction of bound promoters among those with a conserved match exceeding that threshold. We chose t1 as the minimal threshold with specificity greater than 0.5, and t2 as the maximal threshold with specificity lower than 0.1. Other thresholds and parameters were also examined and led to similar conclusions (see Supplementary Figure 2).
Expression divergence
Yeast ED was calculated from the response of four yeast species to various environmental stresses (Tirosh et al, 2006). Tissue-specific ED between human and chimpanzee was taken from Khaitovich et al (2005), log2-transformed, centered and normalized by its s.d. Expression patterns of human and mouse across 30 orthologous tissues were taken from Su et al (2004); ED was calculated as in Liao and Zhang (2006), centered and normalized by its s.d. ED between human and mouse liver cells was calculated as the log2 of the absolute difference in log-transformed expression levels, as measured with exon arrays (Xing et al, 2007). Diverged genes were defined in each data set as those with ED higher than mean+s.d.
Analysis of mating and response to α-factor in three yeast species
Haploids isolation
S. paradoxus (hoΔ∷KANr) and S. mikatae (hoΔ∷KANr) cells were grown in liquid YPD to saturation, then transferred to sporulation plates (1% K-acetate, 0.1% yeast extract, 0.05% glucose and 2% agar) and incubated at 25°C for 6–7 days. Haploid cells were isolated by random spore analysis: Asci were collected from the sporulation plates, resuspended in buffer A (10 mM DTT, 100 mM Tris-SO4 pH 9.4) and incubated for 1 h at 30°C with gentle agitation. Next, the asci were resuspended in buffer B (1.1 M Sorbito, 10 mM K-phosphate buffer pH 7.2), treated with yeast lytic enzyme (ICN) (0.5 mg/5 × 108 cells) for 1 h at 30°C and transferred to 0.5% Triton X-100. Asci were disrupted by sonication (three rounds of 30 s) and then spread on YPD plates. Haploids mating type was determined using PCR (Huxley et al, 1990).
α-Factor treatment
S. cerevisiae, S. paradoxus and S. mikatea a-type cells were grown to log phase in YPD at 30°C. Cells (3 × 106/ml) were treated with 3 μM α-factor for 90 min. Samples were collected at the indicated time points. Cell counting was performed in a standard hemocytometer.
Microarray analysis
α-Factor experiments: S. cerevisiae, S. paradoxus and S. mikatae a-type cells (5 × 106/ml of each) were grown on YPD at 30°C to early log phase. Cells were treated with 6 μM α-factor for 60 min. As reference we used the same cells before addition of α-factor. Three to four biological repeats (different colonies) were processed in parallel for each species.
Natural mating experiments: S. cerevisiae a- and α-type cells (5 × 106 of each) were mixed and applied on to 0.22-μm filter. The filters were placed on YPD agar plates at 30°C for 90 min and then snap-frozen with liquid nitrogen. For reference, S. cerevisiae a- and α-type cells were placed on separate filters and incubated on YPD agar plates at 30°C for 90 min. The cells were mixed only prior to mRNA extraction.
Sample labeling, hybridization and scanning: cDNA of experiment and reference samples was synthesized from total RNA using M-MLV Reverse Transcriptase RNase H Minus (Promega) and labeled with Cy3 and Cy5, respectively, by the indirect amino-allyl method. Hybridization and scanning were carried out as described in Tirosh et al (2006).
Expression data were log2-transformed and normalized by intensity-dependent correction (subtracting a lowess regression) and by position-dependent correction (subtracting the median of each subarray from all the genes in it). To account for differences in the overall signal in different arrays, we calculated the linear regression between the first S. cerevisiae array and each subsequent array, and divided the log2 expression ratios of that array by the regression coefficient.
Raw and normalized expression data are available at the GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/) database with the series accession GSE7525.
Differential expression in the mating response
To identify genes that are differentially expressed among the three species, we performed a two-sampled t-test for each gene and for each pairwise species comparison. A false discovery rate of 0.05 was used to generate a list of differentially expressed genes. We noted that many of the genes identified by the t-test as differentially expressed are highly regulated and display the same general trend in all species (up- or downregulated). These differences may arise from technical artifacts such as inflated log ratios due to low signals and different sensitivity of the different arrays. Furthermore, these differences are less biologically meaningful than differences between up/downregulation in one species and no regulation (or opposite regulation) in another species. We thus employed another test for differential expression, which takes into account not only the difference in log ratios but also the magnitude of overall up- or downregulation. We used the formula from Tirosh et al (2006) to estimate the ED of each gene, which can be rewritten as
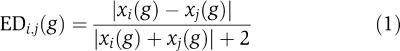 |
where EDi,j(g) is the expression divergence of gene g between species i and j, and xi(g) is the normalized log2 expression ratio of gene g in species i.
The 20% genes with the highest ED were considered to be differentially expressed. These two tests agreed on the differential expression of 408 genes, which we further analyze.
We classified the differentially expressed genes into 12 patterns of differential expression. Each pattern corresponds to higher expression in a subset of the species compared with the remaining species. We first defined characteristic expression vectors for six patterns with values of 1 in a subset of the species and −1 in the remaining species, and classified genes according to the vector with which their expression pattern had the highest correlation. Next, we divided each class into two subclasses. The first subclass corresponds to genes that can be estimated as upregulated in one subset of the species and not regulated in the other species, and the second corresponds to genes that are better estimated as downregulated in one subset of the species and not regulated in the other species. This was done by comparing the absolute values of the highest and lowest response of those genes; if the maximum magnitude of upregulation in any of the species was higher than the maximum magnitude of downregulation in any of the species, then we classified the gene as upregulated, and in the opposite case as downregulated. Each class (and subclass) was analyzed by searching for enrichment of GO annotations and presence of mating-annotated genes.
Impact of changes in TF sequence motifs on mating ED
For each pair of orthologs that are differentially upregulated, we examined whether there are non-conserved matches to TF sequence motifs (MacIsaac et al, 2006). Divergence in sequence motifs for STE12 and five other TFs were significantly (P<0.05) associated with loss of upregulation and divergence in sequence motifs for two other TFs were significantly associated with gain of upregulation. Thus, the differential upregulation of each pair of orthologs with diverged matches to any of these eight motifs in which the difference in expression was consistent with the difference in the sequence motif (e.g. the gene with the STE12-binding site is upregulated and the gene without this binding site is not upregulated) was considered to be accounted by changes in TF sequence motifs.
Nucleosome occupancy
Nucleosome occupancy was estimated across the promoters of the three yeast species using the working version of the yeast model from the Segal lab (http://genie.weizmann.ac.il/pubs/nucleosomes06/index.html). We examined the differences in predicted nucleosome occupancy at the positions of STE12-binding sites. Few promoters had large differences (0.5 or higher) and the predicted changes in nucleosome occupancy among these promoters were correlated with differential upregulation.
Supplementary Material
Supplementary Figures
Supplementary Table 1
Acknowledgments
This study was supported by the Helen and Martin Kimmel Award for Innovative Investigation and grants from the Pasteur–Weizmann joint research program, the Kahn fund for Systems Biology at the Weizmann Institute of Science, the Tauber Fund and the Israeli Ministry of Science.
References
- Banerjee N, Zhang MQ (2003) Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res 31: 7024–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L (2005) A walk-through of the yeast mating pheromone response pathway. Peptides 26: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M (2007) Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Das D, Banerjee N, Zhang MQ (2004) Interacting models of cooperative gene regulation. Proc Natl Acad Sci USA 101: 16234–16239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG (2002) Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Donaldson IJ, Gottgens B (2006) Evolution of candidate transcriptional regulatory motifs since the human–chimpanzee divergence. Genome Biol 7: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Fay JC (2007) Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol 3: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS (2006) Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312: 276–279 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I (1990) Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6: 236. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nat Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S (2005) Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL (2007) Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J (2006) Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol 23: 530–540 [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M (1998) Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125: 949–958 [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E (2006) An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H (2005) Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development 132: 1663–1674 [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, Macisaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E (2007) Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Chabot AE, Smyth GK, Gilad Y (2007) Using DNA microarrays to study gene expression in closely related species. Bioinformatics 23: 1235–1242 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ (2006) An integrated view of protein evolution. Nat Rev Genet 7: 337–348 [DOI] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL (2003) Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, Tyers M, Boone C, Friend SH (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873–880 [DOI] [PubMed] [Google Scholar]
- Romano LA, Wray GA (2003) Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 130: 4187–4199 [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Ruvkun G (2003) Functional tests of enhancer conservation between distantly related species. Development 130: 5133–5142 [DOI] [PubMed] [Google Scholar]
- Sartor MA, Zorn AM, Schwanekamp JA, Halbleib D, Karyala S, Howell ML, Dean GE, Medvedovic M, Tomlinson CR (2006) A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res 34: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J (2006) A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3: 137–144 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Mitani Y, Satoh G, Satoh N (1999) Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126: 3725–3734 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N (2006) A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Wall DP, Hirsh AE, Fraser HB, Kumm J, Giaever G, Eisen MB, Feldman MW (2005) Functional genomic analysis of the rates of protein evolution. Proc Natl Acad Sci USA 102: 5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QF, Prabhakar S, Chanan S, Cheng JF, Boffelli D, Rubin EM (2007) Detection of weakly conserved ancestral mammalian regulatory sequences by primate comparisons. Genome Biol 8: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Ouyang Z, Kapur K, Scott MP, Wong WH (2007) Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol Biol Evol 24: 1283–1285 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gu J, Gu X (2004) How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet 20: 403–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table 1