Abstract
The function of small ubiquitin-like modifier (SUMO)-binding proteins is key to understanding how SUMOylation regulates cellular processes. We identified two related Schizosaccharomyces pombe proteins, Rfp1 and Rfp2, each having an N-terminal SUMO-interacting motif (SIM) and a C-terminal RING-finger domain. Genetic analysis shows that Rfp1 and Rfp2 have redundant functions; together, they are essential for cell growth and genome stability. Mammalian RNF4, an active ubiquitin E3 ligase, is an orthologue of Rfp1/Rfp2. Rfp1 and Rfp2 lack E3 activity but recruit Slx8, an active RING-finger ubiquitin ligase, through a RING–RING interaction, to form a functional E3. RNF4 complements the growth and genomic stability defects of rfp1rfp2, slx8, and rfp1rfp2slx8 mutant cells. Both the Rfp-Slx8 complex and RNF4 specifically ubiquitylate artificial SUMO-containing substrates in vitro in a SUMO binding-dependent manner. SUMOylated proteins accumulate in rfp1rfp2 double-null cells, suggesting that Rfp/Slx8 proteins may promote ubiquitin-dependent degradation of SUMOylated targets. Hence, we describe a family of SIM-containing RING-finger proteins that potentially regulates eukaryotic genome stability through linking SUMO-interaction with ubiquitin conjugation.
Keywords: SUMO, SUMO-interacting motif, ubiquitin, RING, E3 ligase
Introduction
Post-translational modification adds layers of complexity to the control of protein function. Ubiquitin and ubiquitin-like proteins, such as the small ubiquitin-like modifier (SUMO), are covalently linked through their C-terminal carboxyl group to an ɛ-amine group of a lysine in the modified protein. Higher eukaryotes express three and possibly four SUMO family members, SUMO1–4, encoded by different genes. Newly synthesized SUMO protein is matured through proteolytic cleavage of a C-terminal peptide to expose the two consecutive glycine residues essential for conjugation. SUMOylation occurs via a biochemical pathway analogous to ubiquitylation: mature SUMO is charged with a high-energy thioester bond by the SUMO-activating enzyme (E1), a heterodimeric protein complex, and transferred to the SUMO-conjugating enzyme (E2), which catalyzes the formation of the isopeptide bond between SUMO and the target protein. SUMO ligases (E3s) provide a platform facilitating the conjugation of SUMO from E2 to the target (Gill, 2004; Johnson, 2004).
SUMO is essential for growth, division, and the maintenance of genome stability in eukaryotic cells. Many SUMOylated proteins are found either in the nucleus or at the nuclear periphery, implying an important role for SUMOylation in biological processes in the nucleus (Johnson, 2004). Mutations in the enzymes of the SUMO conjugation pathway result in sensitivity to genotoxic challenges. In the fission yeast Schizosaccharomyces pombe, mutations in rad31, encoding a SUMO E1 subunit, and hus5, encoding the SUMO E2 (Ubc9 orthologue), render the cells sensitive to DNA damage (al-Khodairy et al, 1995; Shayeghi et al, 1997). Deletion of pmt3, the only SUMO gene in S. pombe, results in slow growth, sensitivity to disruption of mitosis, and increased telomere length (Tanaka et al, 1999). Pli1, the S. pombe orthologue of PIAS family SUMO E3s, is essential for the stability of centromeres and telomeres (Xhemalce et al, 2004). Nse2/Mms21, a component of the Smc5/6 complex, is also a functional SUMO E3 (McDonald et al, 2003; Andrews et al, 2005; Zhao and Blobel, 2005). In budding yeast, Ubc9 and Mms21 are needed to prevent detrimental homologous recombination events caused by stalled replication forks, and function in concert with the RecQ family DNA helicase Sgs1 to maintain genome stability (Branzei et al, 2006).
Like many other forms of post-translational modification, SUMOylation is believed to tag the modified protein for novel protein–protein interactions. Therefore, SUMO-interacting domains would be expected to play a crucial role in regulating SUMOylated proteins, and identifying proteins with SUMO-interacting motifs (SIMs) is important for an understanding of the SUMOylation system (Hannich et al, 2005; Lin et al, 2006; Shen et al, 2006). In contrast to the multiple characterized ubiquitin-binding domains, so far only one SIM (or SUMO-binding motif (SBM)) is known (Minty et al, 2000; Song et al, 2004; Reverter and Lima, 2005). The core of the SIM is composed of three hydrophobic (I, L or V) residues, arranged as V/I-V/I-X-V/I/L or V/I-X-V/I-V/I. These residues form a β-strand and are incorporated into a β-sheet together with SUMO's second β-strand when SIM and SUMO interact; the incorporation of the SIM β-strand can be bidirectional, forming either parallel or antiparallel interactions (Song et al, 2005). The SIM-binding surface on SUMO lies between its second β-strand and its α-helix. A number of hydrophobic residues in SUMO1, including F36 and V38, form a conserved hydrophobic patch to accommodate the hydrophobic side chains of the SIM (Song et al, 2004, 2005; Reverter and Lima, 2005; Hecker et al, 2006). Additional residues surrounding the core SIM residues contribute to its association with SUMO, especially through electrostatic interactions involving acidic SIM residues. The residues lying outside the core may also contribute to the specificity in recognizing different SUMO isoforms (Hecker et al, 2006).
Here, we report the identification of a family of proteins that contain both a SIM and a RING-finger domain. Many RING-finger proteins act as ubiquitin E3 ligases. Ubiquitin E3s can be classified into three subsets: the SCF (Skp1-Cullin-F box) family E3s form multisubunit platforms to promote E2-catalyzed ubiquitylation of target proteins; the HECT (homology to the E6-AP carboxyl terminus) domain family E3s contain their own catalytically active Cys to extend the ubiquitin transfer cascade from E1 and E2 to the substrate. In contrast, RING-finger family E3s possess a substrate-binding site and form a RING-domain-mediated physical interaction with E2, thus facilitating the E2-catalyzed ubiquitin conjugation to the substrate (Joazeiro and Weissman, 2000). Our study indicates that this novel family of SIM/RING-finger proteins can directly couple ubiquitin E3 ligase activity to SUMOylated proteins through their SIM.
Results
Fission yeast proteins Rfp1 and Rfp2 are RING-finger proteins containing a SUMO-interacting motif
Initially, in a two-hybrid screen with Ark1, the fission yeast Aurora kinase that we had characterized previously (Leverson et al, 2000), we identified a small RING-finger protein. Subsequently, we found a second, closely related RING-finger protein encoded by the S. pombe genome (28% identity, Supplementary Figure S1). We named these two proteins Rfp1 and Rfp2, for RING-finger protein 1 and 2, corresponding to the annotated S. pombe proteins SPAC19A8.10 and SPAC343.18, respectively. These two proteins share sequence similarity in two regions: the C-terminal C3HC4-type RING domain and an N-terminal region corresponding to Rfp1 residues 14–37 (Supplementary Figure S1). In a second yeast two-hybrid screen against Rfp1, we identified 17 potential Rfp1-interacting clones (Supplementary Figure S2); the top hit (six clones isolated) was Pmt3, the fission yeast version of SUMO. Among the additional Rfp1-interacting clones identified (Supplementary Figure S2), Rad60 (three hits) contains two tandem SUMO-like motifs (Novatchkova et al, 2005), and Pli1 (1 hit) is a PIAS-family SUMO E3 ligase (Xhemalce et al, 2004), further supporting a link between Rfp1 and the SUMOylation pathway.
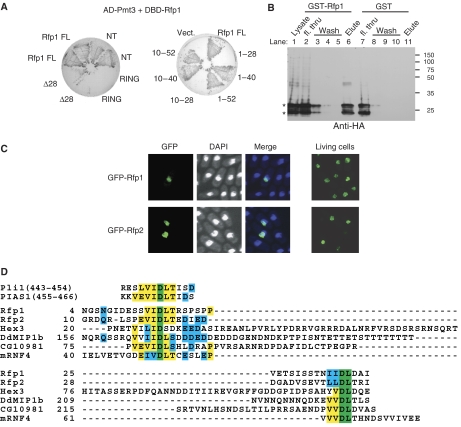
In further yeast two-hybrid interaction analysis, the Pmt3-binding site of Rfp1 was mapped to residues between 10 and 40, congruent with the region near the N-terminus conserved between Rfp1 and Rfp2 (Figure 1A). This region contains two sequences resembling the SIM in PIAS family proteins (Figure 1D), both of which are essential to mediate its interaction with Pmt3 (Figure 1A and D); truncated versions of Rfp1, containing only one or other SIM-like sequence from this region (Δ28, Figure 1A, left panel; 1–28 and 10–28, Figure 1A, right panel), failed to interact with Pmt3. Recombinant GST–Rfp1 protein but not GST alone was able to pull down an epitope-tagged Pmt3 from a lysate of S. pombe cells expressing HA-Pmt3, further confirming the physical recognition of Pmt3 by Rfp1 (Figure 1B, compare lane 6 with lane 11). Consistent with the predominantly nuclear function of SUMO, when green fluorescent protein (GFP)-tagged Rfp1 and Rfp2 were expressed from a mid-strength nmt1 promoter (nmt41), the fluorescent signals were concentrated in the nucleus and formed discrete nuclear foci (Figure 1C). We conclude that Rfp1 and Rfp2 are nuclear proteins possessing an N-terminal SIM and a C-terminal RING domain.
Figure 1.
Identification of SIM in RNF4 family proteins. (A) Yeast two-hybrid interaction between Pmt3 and full-length Rfp1 (Rfp1 FL) and various Rfp1 mutants as indicated. Left panel shows the two-hybrid assay testing interaction between Pmt3 and the full-length (FL), N-terminal region (NT, 1–188), RING domain (RING, 189–254), or N-terminally truncated form (Δ28, deletion of 1–28) of Rfp1 (transformed strains were streaked out in duplicates). Right panel shows further analysis of the interaction between Pmt3 and the N-terminal region of Rfp1. AD, Gal4 activation domain; DBD, Gal4 DNA-binding domain. (B) Physical interaction between Pmt3 and Rfp1. Recombinant GST-Rfp1 or GST alone was incubated with the cell lysate of an S. pombe strain expressing HA-tagged Pmt3, followed by affinity purification on glutathione beads. Co-purified HA-Pmt3 was detected by anti-HA immunoblotting; the doublet of HA-Pmt3 signal (*) was presumably due to partial proteolytic processing of Pmt3 C-terminus. Fractions throughout the purification steps are as indicated. ‘fl. thru', flow through; molecular weight shown in kDa. (C) Episomal expression of N-terminally GFP-tagged Rfp1 and Rfp2 from the nmt41 promoter in the pSLF273 plasmid vector in wild-type cells. Images were taken 24 h after the transformed yeast cells were grown in EMM medium without thiamine. DAPI stained cells were fixed through air-drying; live cells were spread on a thin layer of EMM medium with 2% agarose. (D) Alignment of the SIM of the RNF4 family proteins from different organisms: S. pombe Rfp1 and Rfp2, S. cerevisiae Hex3 (Slx5), Dictyostelium discoideum DdMIP1b (NCBI XP_644602), Drosophila melanogaster CG10981, and mouse mRNF4; the SIMs in Pli1 and PIAS1 are also shown for comparison. Colors indicate completely conserved (green), strongly conserved (yellow), or weakly conserved residues (cyan).
In a BLAST search for Rfp1 or Rfp2 orthologues in other organisms, the top hit against Rfp2 was a mammalian protein called RNF4 (RING-finger protein 4, also known as SNURF, small nuclear RING-finger protein), based largely on the similarity in their RING domains (Supplementary Figure S1). Moreover, close inspection of the RNF4 sequence revealed there is also a high degree of similarity to both Rfp1 and Rfp2 in the N-terminal SIM (Figure 1D). In addition, we noticed conservation at the C-termini: all three proteins end with a triplet of hydrophobic residues immediately adjacent to their RING-finger domains (Supplementary Figure S1). Related proteins are found in lower eukaryotic species, indicating that this is a conserved family of proteins (Figure 1D and Supplementary Figure S3). Notably, Hex3/Slx5 in Saccharomyces cerevisiae, MIP1 (MEK1-interacting protein 1) in Dictyostelium discoideum, and the mammalian RNF4, have all been implicated in SUMOylation pathways, but the existence of their SIMs has not previously been reported (Sobko et al, 2002; Hazbun et al, 2003; Hakli et al, 2005; Hannich et al, 2005; Wang et al, 2006).
Rfp1 and Rfp2 together are required for cell growth and genome stability
To determine the function of Rfp1 and Rfp2, we deleted the rfp1+ and rfp2+ loci separately by targeted disruption. Loss of either the rfp1+ or the rfp2+ gene did not cause any visible phenotype. We then carried out experiments to determine the phenotype of Δrfp1Δrfp2 cells. First, we crossed the haploid Δrfp1 and Δrfp2 strains and carried out both random spore analysis and tetrad dissection. We found that among the viable spores, only 4.6% had both Δrfp1 and Δrfp2 disruption makers, in contrast to the percentage of wild type (33.4%), Δrfp1 (34%), and Δrfp2 (28%) (Figure 2A). Poor viability/reduced growth of the Δrfp1Δrfp2 cells is also apparent in the tetrad analysis (Figure 2B). Next, using a double-null Δrfp1Δrfp2 mutant strain generated from the viable spores and a single rfp2-null strain, we constructed a diploid strain carrying a single Δrfp1 allele and two Δrfp2 alleles. Dissection of the tetrads formed by this strain showed a 2:2 separation; half of the spores either formed colonies that are much smaller, or failed to form visible colonies at all (Figure 2C). Microscopic colonies could be seen in both tetrad analyses shown in Figure 2B and C, however, indicating that the spores could germinate but only survive for a limited number of cell divisions (data not shown). The surviving cells grew much slower and appeared significantly elongated compared to the wild-type (Figure 2D, left panel). These experiments suggest that the rfp1+ and rfp2+ genes compensate for each other and that simultaneous deletion of both rfp1+ and rfp2+ results in a severe growth defect. When we examined the nuclear morphology of the Δrfp1Δrfp2 cells, we noticed fragmented chromosomes, elongated nuclei, and asymmetric positioning of the nuclei (Figure 2D, right panels). These phenotypes indicate that the deletion of both rfp+ genes results in loss of genome integrity, which is manifested in the form of defects in chromosomal segregation and cell division, compromised viability of the mutant cells, and a low efficiency in germination of the mutant spores. These observations are also consistent with the results of our yeast two-hybrid screen (Supplementary Figure S2), where the Rfp1-interacting proteins Pmt3, Rad60, Pli1, and Rhp18 proteins have all been implicated in pathways for genome stability and DNA-damage repair (Tanaka et al, 1999; Verkade et al, 2001; Xhemalce et al, 2004; Raffa et al, 2006).
Figure 2.
Rfp1 and Rfp2 are required for genome stability. (A) Scores of a random spore analysis. The diploids were obtained by crossing Δrfp1 and Δrfp2 strains of opposite mating type and sporulated directly. (B) Tetrads from zygotic asci of the diploids as described in (A). Genotypes: wt, wild type; 1, Δrfp1; 2, Δrfp2; 12, Δrfp1Δrfp2; (12), Δrfp1Δrfp2 (inferred). (C) Tetrad analysis of a diploid strain generated through crossing h−ade6- M216Δrfp1Δrfp2 with h+ade6-M210Δrfp2 followed by selection on adenine-deficient medium. The diploid cells were sporulated to form azygotic asci for dissection. (D) Morphology of cells lacking both the rfp1 and rfp2 genes. A panel with cell morphology observed by differential interference contrast microscopy (DIC), and three panels of fields of cells with DAPI-stained nuclei are shown as indicated.
RNF4 is a functional orthologue of Rfp1 and Rfp2
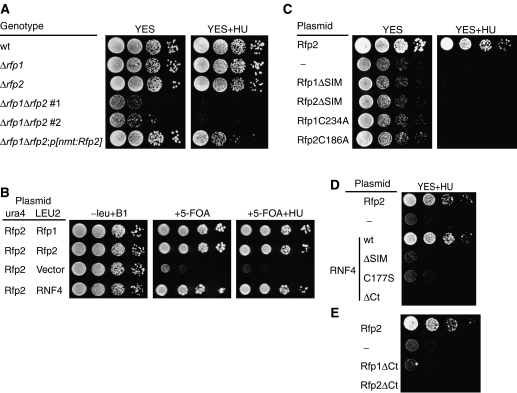
To confirm that the growth defect of Δrfp1Δrfp2 cells was caused by the loss of rfp1+ and rfp2+ genes, we transformed the viable mutant cells with plasmids containing either Rfp1 or Rfp2 cDNA under the wild-type nmt promoter (nmt1). We then compared the growth of these cells in complete (YES) medium (nmt1 promoter activity is thereby kept at minimum level). In contrast to Δrfp1Δrfp2 cells, the growth of rfp1+ or rfp2-transformed cells was similar to both the wild type and the cells with a single deletion (Figure 3A, left panel). A rescue of the growth defect was also obtained with plasmids containing the genomic fragments of rfp1+ or rfp2+ expressed under the control of their own promoters (Supplementary Figure S4). Observing the transformed cells under the microscope, we noticed a reduction in elongated cells that were predominant among the culture of Δrfp1Δrfp2 cells (data not shown). In a parallel comparison, we found that the Δrfp1Δrfp2 cells were very sensitive to 10 mM hydroxyurea (HU), which causes replicative stress. Ectopically expressed Rfp2 partially suppressed HU sensitivity as compared to the strains with single deletions, which grew as well as wild type in the presence of 10 mM HU (Figure 3A, right panel). We conclude that the ectopic expression of Rfp2 can significantly suppress the slow growth phenotype of Δrfp1Δrfp2 cells, although this does not fully mimic the activity of the endogenous rfp+ loci, which are presumably subject to precise expression control.
Figure 3.
Rescue of the growth defect of Δrfp1Δrfp2 cells. (A) The growth of various S. pombe strains (as indicated) were compared by spot assay on complete medium (YES) in the absence (left) or presence (right) of 10 mM HU. Two independently isolated Δrfp1Δrfp2 strains (1 and 2) are shown. (B) Rescue of Δrfp1Δrfp2 by plasmid shuffle. A Δrfp1Δrfp2p[nmt:Rfp2 ura4+] strain was constructed by crossing Δrfp1 with Δrfp2p[nmt:Rfp2], and transformed with a LEU2 plasmid containing Rfp1, Rfp2, RNF4, or empty vector. The growth assay was conducted on various EMM media as indicated. (C) Activity of Rfp1 or Rfp2 mutants either missing the SIM or carrying a mutation in an essential RING finger Cys in Δrfp1Δrfp2 cells. (D) Rescue of Δrfp1Δrfp2 cells by RNF4 requires the SIM, an intact RING finger and the hydrophobic C-terminus. (E) The hydrophobic C-termini in Rfp1 and Rfp2 conserved in RNF4 family proteins are required to rescue Δrfp1Δrfp2 cells. ‘—', blank vector.
To determine whether RNF4 is a functional orthologue of Rfp1 and Rfp2, we examined whether mouse RNF4 could compensate for the loss of both Rfp1 and Rfp2. We transformed expression plasmids for the wild-type forms of Rfp1, Rfp2, and RNF4 into a Δrfp1Δrfp2 strain covered with an Rfp2 plasmid with a ura4+ gene, and compared growth of the transformed strains in media containing 5-fluoroorotic acid (5-FOA) (Figure 3B). The RNF4-transformed cells grew as well as Rfp1- or Rfp2-transformed Δrfp1Δrfp2 cells, suggesting that RNF4 can functionally replace Rfp1 or Rfp2 in vivo.
Next, we examined whether the conserved motifs shared in this protein family were also essential for function in yeast. We first tested the in vivo activity of Rfp mutants designed to be defective in either SIM or RING function. Both the deletion of the SIM and mutation of a critical Cys in the RING finger resulted in the loss-of-function of the two Rfp proteins, that is, the mutants failed to rescue growth of Δrfp1Δrfp2 cells (Figure 3C). Hence, the SIM and the RING domain are essential for the molecular function of Rfp1 and Rfp2 in vivo. We then generated mutant forms of RNF4 with either a SIM deletion (RNF4ΔSIM) or a point mutation in its RING domain (RNF4C177S); both mutants failed to support the growth of Δrfp1Δrfp2 cells in HU-containing medium (Figure 3D). Because of the apparent sequence similarity in the C-termini of RNF4, Rfp1, and Rfp2, we also tested the activity of an RNF4 mutant missing the C-terminal four residues (RNF4ΔCt) in the same assay. The RNF4ΔCt mutant also acted as a loss-of-function mutant. Similar C-terminal deletions in both Rfp1 and Rfp2 also abolished their function (Figure 3D). Our observation is reminiscent of the studies on Bre1 family proteins, each containing a C-terminal RING domain with conserved sequence at the C-terminus (Hwang et al, 2003). Moreover, addition of extra residues to the Bre1 C-terminus also abolished its function (Wood et al, 2003). Thus, these conserved termini appear to be integral parts of the C-terminally positioned RING-finger domains (Supplementary Figure S3). Taken together, all three motifs conserved between RNF4 and Rfp1/2 prove to be essential for RNF4 to replace Rfp1 and Rfp2 functionally. We conclude that mammalian RNF4 protein is a true orthologue of the fission yeast Rfp1 and Rfp2.
RNF4 can replace both Rfp1/Rfp2, and Slx8
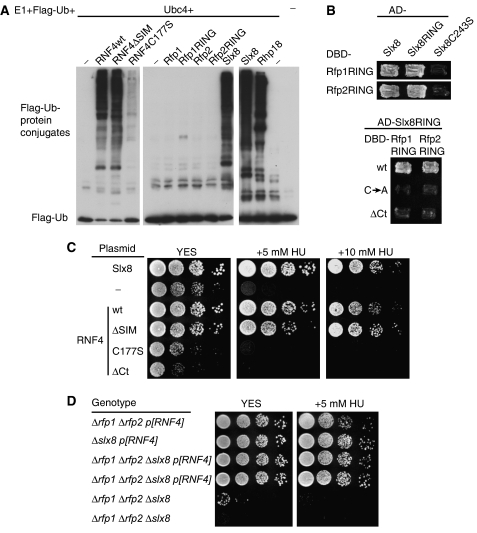
To determine if the RING fingers in Rfp and RNF4 have any E3 ligase activity, we carried out both SUMO and ubiquitin ligase assays. For the SUMO ligase assay, we used S. pombe versions of E1 (the Rad31-Fub2 dimer), E2 (Hus5/Ubc9), and Pmt3 (SUMO). In contrast to Pli1, a known SUMO E3 in S. pombe, Rfp1, Rfp2, and RNF4 all failed to exhibit detectable SUMO E3 ligase activity; neither was any SUMO ligase activity detected with RNF4 in the SUMO ligase assay using mammalian components (data not shown).
In the ubiquitin ligase assay, RNF4 showed robust E3 ligase activity comparable to the activity of Rhp18, a known ubiquitin E3 (Figure 4A). This is consistent with prior studies on RNF4 by Palvimo and co-workers (Hakli et al, 2004). Moreover, whereas the C177S mutation in the RING domain of RNF4 diminished its ubiquitin ligase activity, the SIM in RNF4 was not essential for the activity in vitro (Figure 4A). In contrast to RNF4, we failed to detect any ubiquitin E3 activity with either full-length Rfp1 or Rfp2, or their RING-finger-alone deletion derivatives (Figure 4A; the minor higher molecular weight species associated with the Rfp1 RING is likely the result of the conjugating activity of E2). Therefore, although Rfp1 and Rfp2 can be replaced by RNF4 in vivo, they do not possess the same biochemical activity as RNF4 in vitro. This may be explained by the unique structure of the RING domains of Rfp1 and Rfp2 that is not shared by genuine ubiquitin E3 ligases: first, the spacing between the C-terminal two Cys is unusual, with four residues instead of two. Second, the bulky hydrophobic residue (mostly Ile) that usually precedes the second Cys in other ubiquitin E3s is not conserved (Supplementary Figure S3). The S. cerevisiae protein Hex3/Slx5, the budding yeast orthologue of Rfp1/2, shows further sequence diversity with extra residues between the second and third Cys, and between the fifth and sixth Cys, in its RING domain. Thus the RING domain of Rfp1, Rfp2, or Hex3 alone may not be able to couple a ubiquitin-conjugated E2 to substrate.
Figure 4.
Functional equivalence between RNF4 and Rfp–Slx8 complex. (A) A comparison of ubiquitin ligase activity among RNF4, Rfp1, Rfp2, Slx8, Rhp18, and various mutants as indicated. See Methods for detailed descriptions of the mutant forms and for assay conditions. Flag-Ub was detected by immunoblotting with anti-Flag antibodies. (B) Interaction between the RING domains of Rfp1/2 and that of Slx8 in yeast two-hybrid tests. Top panel: Gal4 DBD fusion of Rfp1 RING (aa 174–254) or Rfp2 RING (aa 134–205) was tested against Gal4 AD fusion of full-length Slx8, Slx8 RING (aa 197–269), or Slx8 C243S mutant. Bottom panel: AD fusion of Slx8 RING was tested against the DBD fusion constructs of Rfp1 and Rfp2 RING domains, in the forms of wild type (wt), Cys mutants (CA, Rfp1RING C234A or Rfp2RING C186A), or C-terminal deletion mutants (ΔCt). (C) Suppression of the growth defect of Δslx8 cells by episomal expression of Slx8, RNF4, and RNF4ΔSIM. The growth was compared by spot assay on complete medium (YES) or YES containing 5 mM or 10 mM HU. (D) Suppression of the growth defect of Δrfp1Δrfp2Δslx8 cells by RNF4. Δrfp1Δrfp2Δslx8p[RNF4] strain was constructed by crossing p[RNF4]-covered single mutants to avoid propagating strains with growth defects. Different strains as indicated were compared for growth in both YES medium and YES containing 5 mM HU; duplicate strains are independently isolated clones.
In S. cerevisiae, the Hex3/Slx5 protein associates with another RING-finger protein, Slx8, both genetically and physically (Uetz et al, 2000; Mullen et al, 2001; Wang et al, 2006; Yang et al, 2006). In S. pombe, an Slx8 homologue can be identified (SPBC3D6.11c). Immunoprecipitation of Rfp2 was able to bring down the fission yeast Slx8 when both proteins were overexpressed in 293T cells (data not shown). In contrast to Rfp1 and Rfp2, we found that S. pombe Slx8 had robust E3 ubiquitin ligase activity (Figure 4A). The stable association of the budding yeast Hex3 and Slx8 is reminiscent of two other heterodimeric ubiquitin E3 complexes, that of BRCA1 and BARD1, and of RING1b and Bmi1, both of which are formed through RING–RING dimeric interaction (Brzovic et al, 2001; Xia et al, 2003; Buchwald et al, 2006; Li et al, 2006). We therefore examined whether the interaction between Rfp1/2 and Slx8 was specifically mediated by their RING domains. We conducted pairwise yeast two-hybrid tests with the RING domains of Rfp1 and Rfp2. Both Rfp1 and Rfp2 RINGs interacted with full-length Slx8 and the RING domain of Slx8, but not with an Slx8 mutant (C243S) that presumably disrupts its RING structure (Figure 4B, top panel). Moreover, the Slx8 RING domain interacted specifically with both the Rfp1 and Rfp2 RING domains, but not with their mutant forms (Figure 4B, bottom panel), consistent with our observation that the same set of mutations also abolished the function of Rfp1 and Rfp2 in vivo (Figure 3). Hence, the physical interaction between Hex3 and Slx8 orthologues is conserved between the two yeast species; this represents a third example of an E3 ligase that functions as a RING–RING heterodimer. We hypothesize that a functional ubiquitin E3 ligase complex can be formed by heterodimeric association of the active Slx8 and the inactive Rfp1 or Rfp2.
This hypothesis suggests that the mammalian RNF4 might act as a functional fusion of Slx8 and Rfp1/Rfp2/Hex3 proteins, comprising both a SIM (as in Rfp1/Rfp2/Hex3) and an E3-active RING finger (as in Slx8). We first attempted to verify this prediction by testing whether RNF4 could also replace Slx8 in yeast. We deleted the slx8 gene in S. pombe and observed a phenotype similar to that of Δrfp1Δrfp2 cells, for example, elongated cell bodies, defects in chromosomal segregation, and significantly reduced viability (Supplementary Figure S5 and data not shown). We then assayed for the rescue of the Δslx8 growth defect by transforming Slx8 or RNF4 cDNAs into Δslx8 cells. As expected, Slx8 rescued the Δslx8 growth defect, and RNF4 also almost completely suppressed the Δslx8 phenotype (Figure 4C). Significantly, the RNF4 mutant lacking the SIM was as effective as the wild type in rescuing Δslx8 growth and reducing sensitivity to HU (Figure 4B). In contrast, the RNF4C177S and the RNF4ΔCt mutants were inactive in Δslx8 cells just as in Δrfp1Δrfp2 cells (Figures 3D and 4B). Therefore, in the presence of endogenous Rfp1/2, an intact RING finger is essential for RNF4 to compensate for the loss of Slx8, whereas SUMO interaction is not, possibly because the RING domain of RNF4 can interact with Rfp1/2, which then provide SIM function. Our results are consistent with the recent findings by Brill and co-workers, showing that the RING domain alone was sufficient for Slx8's physiological function in S. cerevisiae in the presence of Hex3 (Yang et al, 2006).
We next tested whether RNF4 could compensate for the simultaneous loss of both Rfp1 and Rfp2 and Slx8, by comparing the growth of Δrfp1Δrfp2, Δslx8, and Δrfp1Δrfp2Δslx8, all carrying RNF4 cDNA (pSLF173-RNF4 or p[RNF4]), in parallel (Figure 4D). We found that all three strains grew equally well on YES medium, although Δrfp1Δrfp2p[RNF4] appeared to be slightly more sensitive in the presence of HU. In contrast, the triple mutant failed to propagate in the absence of RNF4 expression. Therefore, the Rfp1/2 pair and Slx8, both individually and together, can be functionally replaced by RNF4. These data strongly support the hypothesis that RNF4 is a functional chimera between Slx8 and Rfps.
SUMO-directed ubiquitylation by RNF4 family proteins
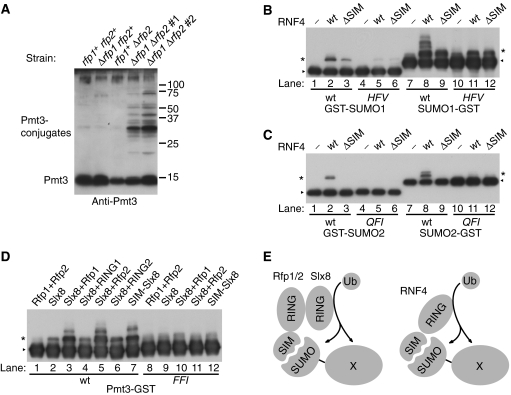
The molecular structure and biochemical activity of RNF4 and its complementing activity in S. pombe revealed an intriguing link between SUMOylation and ubiquitylation. Given that a major role of ubiquitylation is to mark proteins for degradation, we speculated whether the RNF4 family proteins function to destabilize at least a subset of SUMOylated proteins in the cell through ubiquitylation. We therefore examined the level of SUMOylated proteins in both wild type and Δrfp1Δrfp2 mutant cells, using an anti-Pmt3 antiserum to probe for SUMOylated proteins. Cells lacking both rfp1+ and rfp2+ contained a significantly higher level of SUMOylated proteins than wild type or the single Δrfp1 or Δrfp2 mutants (Figure 5A). A similar accumulation of SUMOylated proteins was detected in Δslx8 cells (data not shown). SUMOylated proteins have also been found to accumulate in the Δhex3 or Δslx8 background in S. cerevisiae (Wang et al, 2006). Therefore, the conserved function of the RNF4 family proteins may be to target SUMOylated proteins for ubiquitin-mediated degradation.
Figure 5.
SUMO-directed ubiquitylation by RNF4 family proteins. (A) Immunoblotting detection of Pmt3 and Pmt3-conjugated proteins in vivo in various strains as indicated. (B) Ubiquitylation of GST-SUMO1 (lanes 1–6) or SUMO1–GST (lanes 7–12) by wild-type or SIM-deficient RNF4. HFV, SUMO1 mutant (H35A/F36A/V38A) with disrupted SIM interaction site (lanes 4–6 and 10–12). (C) Ubiquitylation of GST-SUMO2 (lanes 1–6) or SUMO2–GST (lanes 7–12). QFI, SUMO2 mutant (Q30A/F31A/I33A) with disrupted SIM-binding surface (lanes 4–6 and 10–12). The amount of SUMO2-fusion proteins used was equivalent to 1/10 of the SUMO1-fusion proteins in (C). (D) Ubiquitylation of Pmt3-tagged GST. Combinations of Rfp and Slx8 constructs are as indicated. FFI, Pmt3 mutant (F49A/F50A/I52A) with disrupted putative SIM-binding site corresponding to SUMO1 H35F36V38 (lanes 8–12). All versions of SUMO constructs in (B–D) were N-terminally Flag-tagged and were detected (together with their ubiquitin conjugated forms) with anti-Flag immunoblotting; triangles indicate the position of the original SUMO substrates, and asterisks indicate mono-ubiquitylated species. (E) A diagram illustrating the different models of SUMO-directed ubiquitylation of a SUMOylated target protein (X) by RNF4 family proteins in yeast (left) and mammalian (right) cells.
Prompted by the accumulation of SUMOylated proteins in Δrfp1Δrfp2 mutant cells and the unique presence of the SIM in RNF4 family members, we tested the hypothesis that RNF4 family ubiquitin ligase activity might be directed towards SUMOylated proteins. For this purpose, we carried out in vitro ubiquitin ligase assays using different SUMO–GST fusion proteins as artificial substrates to mimic a SUMOylated protein; an epitope tag (Flag) was included at the N terminus of SUMO in the fusion proteins to enable the detection of specific ubiquitin-conjugated species. We detected significant ubiquitylation of both GST-SUMO and SUMO–GST by RNF4 for both the SUMO1 and SUMO2 fusion proteins (Figure 5B and C, lanes 2 and 8). Based on size, the majority of the modification appears to be mono-ubiquitylation (marked by *), although multi-ubiquitylation is apparent with SUMO1–GST as substrate (Figure 5B, lane 8). The amount of ubiquitylated SUMO-tagged GST was significantly reduced when RNF4ΔSIM was used (Figure 5B and C, lanes 3 and 9), although the nonspecific E3 ligase activity of RNF4 and RNF4ΔSIM was indistinguishable (Figure 4A). The fact that SUMO fused either to the N-terminus or the C-terminus of GST was ubiquitylated by RNF4 is consistent with the idea that RNF4 would be able to recognize conjugated SUMO independent of its linkage position in a target SUMOylated protein. We conclude that the SIM in RNF4 is therefore critical for promoting specific ubiquitin ligase activity towards a SUMO-tagged protein in vitro.
As a control experiment, we generated mutant forms of SUMO in all the SUMO-fusion substrates. A conserved β-strand on SUMO is key to mediating its interaction with the SIM (see Introduction), with its residues being involved in van der Waal's contacts with the hydrophobic residues in SIM (Song et al, 2004, 2005; Hecker et al, 2006). On this β-strand, we generated combined alanine substitutions of three residues corresponding to H35, F36, and V38 in SUMO1 (labeled HFV; Figure 5B, lanes 4–6 and 10–12), and Q31, F32, and I34 in SUMO2 (labeled QFI; Figure 5C, lanes 4–6 and 10–12). These SUMO mutants formed SUMO conjugates comparably to wild-type SUMO in an in vitro SUMOylation assay (data not shown). We then tested the mutant forms of both GST-SUMO and SUMO–GST as ubiquitylation substrates for RNF4. In contrast to their wild-type counterparts, ubiquitylation of the mutated SUMO2 fusion proteins by RNF4 was abolished (Figure 5C, lanes 5 and 11), and was significantly reduced to exclusive mono-ubiquitylation on SUMO1 mutants (Figure 5B, lanes 5 and 11); in fact, these SUMO1 mutants were only mono-ubiquitylated to a similar level by either RNF4 or RNF4ΔSIM (Figure 5B). These results demonstrate that the physical contacts between SIM and SUMO are critical for RNF4 to act as a SUMO-targeted ubiquitin ligase.
We next tested whether the Rfp–Slx8 complex recapitulates the SIM-dependent ubiquitin ligase activity of RNF4. For this purpose, the fission yeast SUMO, Pmt3, was used in the form of Pmt3-GST in the ubiquitin ligase assay (Figure 5D). Consistent with the results in the nonspecific ligase assay (Figure 4A), Rfp1 or Rfp2 was not capable of ubiquitylating the Pmt3-tagged substrate; Slx8 alone, while a potent ligase, could only mono-ubiquitylate Pmt3-GST to a limited degree (Figure 5D, lanes 1 and 2). On the contrary, Slx8 together with either Rfp1 or Rfp2 showed greater Pmt3-specific E3 ligase activity (Figure 5D, lanes 3 and 5), generating multi-ubiquitylated forms. This observation clearly demonstrates that Rfp1 or Rfp2 is capable of targeting Slx8 to SUMOylated proteins, presumably through the physical interaction between Rfp and Slx8. When either Rfp was replaced by its RING-alone mutant form, Pmt3-specific ubiquitylation was lost, indicating the essential role of the Rfp SIM for targeting the Rfp–Slx8 complex to Pmt3-GST (Figure 5D, lanes 4 and 6). This role was further clarified with a chimeric protein in which the SIM of Rfp2 was fused with Slx8. This SIM-tagged Slx8 alone ubiquitylated Pmt3-GST to the same level as an Rfp–Slx8 complex did (Figure 5D, lane 7). As was the case with the mammalian versions, the combined mutations in Pmt3 disrupting its interaction with SIM (labeled FFI) also abolished all the SIM-mediated specific ubiquitylation (Figure 5D, lanes 8–12). We conclude that Slx8 can be targeted to SUMO-tagged substrates by the SIM in Rfp1 or Rfp2. Our studies demonstrate that RNF4 mimics the activity of the heterodimeric Slx8-Rfp1 or Slx8–Rfp2 complex both in vivo and in vitro (Figure 5E).
Discussion
We have characterized a family of SUMO-interacting RING-finger proteins, which in principle have the ability to direct the ubiquitylation machinery to SUMOylated proteins, thereby forming an intriguing link between the two post-translational modification pathways. Many ubiquitin E3 ligases exist with targeting motifs that recognize post-translational modifications (Seet et al, 2006), for example, the SH2 domain in c-Cbl for phosphorylated tyrosine (Joazeiro et al, 1999), and the Fbx2 subunit in an SCF complex for N-linked glycosylation (Yoshida et al, 2002). The RNF4-like proteins thus represent a novel class of ubiquitin E3 ligases specifically recognizing SUMOylation.
RNF4 associates with a number of transcription factors (Moilanen et al, 1998; Kaiser et al, 2003; Wu et al, 2004). However, the nature of RNF4's activity as a transcriptional co-regulator was not clear. RNF4 was found to be a potential DNA-binding protein (Hakli et al, 2001). The DNA-binding activity of the budding yeast Hex3/Slx5–Slx8 complex lies in the non-RING region of Slx8 and is not essential for Slx8's function in vivo (Yang et al, 2006). Thus, DNA binding may not be a conserved function of these proteins. On the other hand, the SIM-mediated interaction with SUMO apparently controls the cellular localization of RNF4, such as its association with the PML nuclear bodies (Hakli et al, 2005). We have also noticed that the Rfp2ΔSIM mutant showed a different pattern of nuclear localization than wild-type Rfp2 (Sun and Hunter, unpublished observations). We suggest that RNF4 exerts transcriptional regulatory activity through directly interacting with SUMOylated transcription factors. Consistently, its SIM coincides with the identified androgen receptor-interaction region (Moilanen et al, 1998).
Hex3/Slx5 and Slx8 are linked to the SUMOylation pathway both biochemically and genetically (Hazbun et al, 2003; Hannich et al, 2005; Wang et al, 2006; Burgess et al, 2007). Hex3/Slx5 was found to interact with SUMO in yeast two-hybrid screens (Uetz et al, 2000; Hannich et al, 2005). Loss-of-function mutations in Hex3/Slx5 and Slx8 are able to suppress the temperature-sensitive mutation of mot1 (mot1–301), which encodes an inhibitor of TATA-binding protein in S. cerevisiae (Wang et al, 2006). Significantly, this unbiased approach identified almost exclusively components of SUMOylation pathway, including both subunits of E1, E2, and two SUMO proteases, together with Hex3 and Slx8. The same report, as well as our studies and those of Kosoy et al (2007) in S. pombe, shows the accumulation of SUMOylated species in cells lacking these RING-finger proteins. In Dictyostelium, DdMIP1, the RNF4 homologue, was found to interact with MEK1 through its SIM in yeast two-hybrid assays. Interestingly, DdMIP1 can promote the ubiquitylation of activated MEK1 in a RING-finger-dependent manner, and the loss of DdMIP1 resulted in accumulation of SUMOylated MEK1 during cAMP-induced chemotaxis (Sobko et al, 2002). Therefore, a conserved function of the RNF4 family proteins could be to destabilize SUMOylated transcription factors and activated protein kinases via RING-finger-mediated ubiquitylation.
Although there are other explanations, the accumulation of SUMOylated proteins in Δrfp1Δrfp2 and Δslx8 mutant cells is consistent with these proteins normally being targeted for degradation following SUMO-dependent ubiquitylation. In addition, we cannot rule out other possible fates for the SUMOylated proteins that are ubiquitylated by RNF4 family proteins. In our in vitro assays with artificial substrates, RNF4 catalyzed predominantly mono-ubiquitylation, and mono-ubiquitylation of target proteins could play a distinct role, perhaps in subcellular localization through interaction with mono-ubiquitin-binding proteins. In addition, our current observations are not yet able to differentiate between degradation and de-SUMOylation (Figure 5A). SUMOylation and de-SUMOylation are known to be critical in regulating the partitioning of proteins between nucleus and cytoplasm (Gill, 2004; Johnson, 2004), and in Dictyostelium MEK1 is subject to both DdMIP1-mediated ubiquitylation and SUMOylation-regulated nuclear export (Sobko et al, 2002). Nevertheless, current findings suggest RNF4 family proteins participate in processes that dramatically change the cellular activity of their targets.
What other in vivo targets might there be for RNF4 family proteins? RNF4 family proteins are crucial for maintaining eukaryotic genome integrity in both budding and fission yeast (Zhang et al, 2006; Burgess et al, 2007; Kosoy et al, 2007). RNF4 is expressed in proliferating tissues and tumors, indicating an important role in cell-cycle regulation (Galili et al, 2000; Cavallo et al, 2005). Rfp1 and Rfp2 are essential for cell proliferation in fission yeast and their mutation resulted in a plethora of phenotypes due to loss of genome integrity. In S. cerevisiae, Hex3/Slx5 and Slx8 are essential for DNA damage repair during cell-cycle progression; they interact genetically with Sgs1, a RecQ family DNA helicase; Δslx5 and Δslx8 are both synthetically lethal with Δsgs1 (Mullen et al, 2001; Zhang et al, 2006). They also genetically interact with the telomerase gene (tlc1); Δslx5 and Δslx8 enhance the senescence phenotypes of the tlc mutant (Azam et al, 2006). Both Sgs1 and one of its mammalian orthologues, the Bloom's syndrome protein (BLM), are SUMOylated (Eladad et al, 2005; Branzei et al, 2006). In mammalian cells, SUMOylation of BLM changes its nuclear localization and may be essential for its role in DNA repair. In budding yeast, Sgs1 may function together with Ubc9, the SUMO pathway E2, and Mms21/Nse2, an Smc5/6 complex-associated SUMO E3 ligase, to prevent formation of the toxic DNA recombination intermediates resulting from stalled replication forks (Branzei et al, 2006). Interestingly, Hex3 was found to associate with components of the Smc5/6 complex in S. cerevisiae (Hazbun et al, 2003). In addition, Rad60, which associates with Smc5/6 has two functional C-terminal SUMO-like domains (Boddy et al, 2003; Raffa et al, 2006), was identified in our yeast two-hybrid screens for Rfp1/2-binding proteins (Supplementary Figure S2), and Boddy and co-workers have shown that Rad60 can be ubiquitylated by Rfp/Slx8 complexes in vitro, suggesting that Rad60 may be a physiological target (Prudden et al, 2007). Therefore, we suggest that SUMOylated subpopulations of Sgs1 and other DNA repair proteins are likely targets for RNF4/Rfp/Hex3-mediated ubiquitylation in vivo. Degradation of SUMOylated forms of these proteins may be necessary to balance their activity and to prevent excessive processing of certain DNA structures generated by the DNA repair machinery.
In summary, we suggest that SUMOylation-dependent ubiquitylation is a new regulatory principle that is likely to play an important role in cellular processes that are mediated by SUMOylation. We do not know how SUMOylated proteins are recognized by RNF4 family proteins for ubiquitylation, but we suspect that only certain SUMOylated proteins are targeted, and that this will represent only a subpopulation of each target. Target SUMOylated proteins could be selected because the conjugated SUMO lies in a particular sequence context, because a branched SUMO chain is needed, or because multiple SUMO adducts on a single protein molecule are required (this could explain why all RNF4 family members have a tandem SIM motif). Our studies also establish another type of post-translational modification that can be used to target proteins for ubiquitylation in a regulated fashion. Nevertheless, whereas protein degradation is obviously a candidate means of regulating the metabolism of SUMOylated proteins, we do not rule out other possible mechanisms, such as the regulation of SUMOylation pathway per se (i.e., conjugation or de-SUMOylation). As indicated in the studies by Wang et al (2006), RNF4 family proteins may be integral members of the SUMOylation machinery that act to balance the overall SUMOylation activity in eukaryotic cells.
Materials and methods
cDNAs and expression vectors
Mouse RNF4 cDNA was amplified from mouse testis total RNA with RT–PCR. The cDNAs of Rfp1, Rfp2, Slx8, Rhp18, and Pli1 were directly PCR-amplified from S. pombe genomic DNA; 5′ introns in Rfp2 and Pli1 genes were removed with a second PCR. Ubiquitin E1 and E2 (Ubc4) have been described previously (Joazeiro et al, 1999; Leverson et al, 2000; Xia et al, 2003). Bacterial expression plasmids for fission yeast for SUMO E1 subunits (Rad31 and Fub2), E2 (Hus5), and Pmt3 were gifts from Felicity Watts (Ho et al, 2001). Mammalian cDNAs for SUMO E1 (Sae1 and Sae2), E2 (Ubc9), SUMO1, and SUMO2 were gifts from Ronald Hay (Desterro et al, 1999; Tatham et al, 2001). For gene expression in S. pombe, all ORFs were inserted into the nmt1 vector pSLF173 designed to express proteins with an N-terminal HA tag, except for the GFP-fusion constructs, in which the EGFP (Clontech) coding sequence was fused in-frame to 5′ of the ORFs of Rfp1 and Rfp2 in the nmt41 vector pSLF273. Both vectors were provided by Susan Forsburg (Siam et al, 2004). For gene expression in E. coli, the ORFs of RNF4, Rfp1, Rfp2, and Slx8 were inserted in-frame in the GST-fusion vector pGEX4T-1 (Pharmacia); the Rhp18 and Pli ORFs were inserted in His tag vector pHis8. PCR or PCR-based site-directed mutagenesis (QuikChange, Stratagene) was used to generate the following mutations used in this study: RNF4ΔSIM (deleting residues 50–66); RNF4C177S, RNF4ΔCt (deleting C-terminal 4 residues); Rfp1ΔSIM (deleting residues 14–38); Rfp1C234A, Rfp1ΔCt (deleting C-terminal 3 residues); Rfp1RING (RING1, deleting Rfp1 N-terminal 173 residues); Rfp2ΔSIM (deleting residues 19–42); Rfp2C186A, Rfp2ΔCt (deleting C-terminal 4 residues); Rfp2RING (RING2, deleting Rfp2 N-terminal 133 residues); Slx8RING (deleting Slx8 N-terminal 196 residues); and Slx8C243S. Rfp2 residues 1–48 were fused to the N-terminus of Slx8 to form the SIM-tagged Slx8 (SIM-Slx8). To generate the bacterial expression plasmids for GST-fusions of SUMO1, SUMO2, and Pmt3, the coding sequences of Flag tag and SUMO (ending with GlyGly) were PCR fused, and either inserted in pGEX4T-1 with a stop codon (for GST-SUMO1/SUMO2), or ligated to the 5′ end of GST ORF and inserted in pHis8 (for SUMO1/SUMO2/Pmt3-GST; the extra His tag from pHis8 thus contributing to the higher molecular weight is shown in Figure 5B and C). Purification of GST- and His-tagged proteins was carried out as described previously (Leverson et al, 2000; Xia et al, 2003).
Fission yeast strains and methods
The genetic and biochemical manipulation of S. pombe was conducted as described by Moreno et al (1991) and by Forsburg and Rhind (2006). Targeted disruption of rfp1+, rfp2+, and slx8+ genes in S. pombe was carried out according to Wang et al (2004). All strains used in this study are ade6-M210/M216 his3-D1 leu1–32 ura4-D18. Specific strains are: HSY6, Δrfp1∷his3+; HSY53, Δrfp2∷leu1+; HSY213 and HSY215, Δrfp1∷his3+Δrfp2∷leu1+ (1 and 2, respectively); HSY337 Δslx8∷leu1+; HSY443, Δrfp1∷his3+Δrfp2∷kanMX pSLF173-Rfp2; HSY475, Δrfp1∷his3+Δrfp2∷kanMX Δslx8∷leu1+pSLF173-mRNF4; HSY481, Δrfp1∷his3+Δrfp2∷kanMX Δslx8∷leu1+. Diploid and other episomal plasmid-transformed strains are as described in the Results. All rescue assays were carried out in YES, presumably containing thiamine, such that the nmt1 promoter is kept at minimal activity; expression of the proteins was verified with anti-HA immunoblots. For biochemical analysis, all S. pombe strains were freshly recovered from frozen stock, amplified in YES medium to late log phase. The S. pombe cells were then collected with centrifugation and washed once with water and frozen by liquid nitrogen. To make S. pombe cell lysates, frozen cells were thawed, resuspended in 250 μl of breaking buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris–HCl (pH 8.0), and 1 mM EDTA, pH 8.0), and cell suspension was mixed with ∼250 μl of acid-washed glass beads (Sigma, 425–600 μm) on a vortex for 5 min at 4°C. Cell lysate was cleared by centrifugation and analyzed in immunoblotting. Goat anti-Pmt3 antiserum, a gift from Felicity Watts (Ho et al, 2001), was used for detection of endogenous Pmt3 and its conjugates in S. pombe. GST-pull-down experiments were performed as described previously (Joazeiro et al, 1999; Leverson et al, 2000).
Two-hybrid assay
The yeast two-hybrid tests were performed essentially as described before (Leverson et al, 2002). Budding yeast two-hybrid strain AH109 harboring the HIS3, ADE2, and lacZ reporters downstream of heterologous GAL4-responsive promoter elements was transformed with Gal4 DNA-binding domain (DBD) fusion and activation domain (AD) fusion plasmids as indicated (Matchmaker, Clontech, Palo Alto, CA). The transformed cells were grown on Trp−Leu−His−Ade− plates for 3 days to assay for interaction. All constructs used were tested to be negative in self-activation by co-transformation with blank vectors.
Protein conjugation assay
The in vitro ubiquitin ligase assay has been described before (Joazeiro et al, 1999; Leverson et al, 2000; Xia et al, 2003). For nonspecific ubiquitin ligase assays, a bacterially expressed, Flag-tagged ubiquitin was used for detection of ubiquitin conjugates. For assays with SUMO-based substrates, free ubiquitin (Sigma) was used. All purified proteins were dialyzed and frozen in the buffer containing 50 mM Tris–HCl, pH 7.5–8.0, 400 mM NaCl, 1 mM DTT, and 10% glycerol. The ubiquitin ligase assay was carried out by incubating 1 μg ubiquitin, ∼2 μg His-E1, ∼2 μg His-Ubc4, and 1 μg SUMO–GST substrate (when appropriate) in 25 μl of reaction buffer (50 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT, and 5 mM ATP) for 1 h at 30°C. The reaction was terminated by addition of SDS–PAGE sample buffer and was analyzed by anti-Flag (M2, Sigma) immunoblotting. The SUMO ligase assay was performed using SUMO E1/E2/E3 essentially in the same reaction buffer and conditions as the ubiquitin assay.
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Susan Forsburg for help in initiating this project and Michael N Boddy for communication of the results from his group before publication. We thank Susan Forsburg, Felicity Watts, and Ronald Hay for reagents, and Steven Miller for carrying out the GST-Rfp1 pull-down assays. HS thanks Julie Bailis, Beth Baber, Han-Kuei Huang, and Hua Gao for assistance with yeast genetics, Zhongsheng You for helpful discussions, and the Lundblad laboratory for sharing equipment. This study was supported by NIH Grants CA80100 and CA82683 to TH; HS was an Abbott Fellow of the Life Sciences Research Foundation (2003–2006); JDL was an American Cancer Society Postdoctoral Fellow; TH is a Frank and Else Schilling American Cancer Society Research Professor.
References
- al-Khodairy F, Enoch T, Hagan IM, Carr AM (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci 108: 475–486 [DOI] [PubMed] [Google Scholar]
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ (2005) Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 25: 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB (2006) Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res 34: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Shanahan P, McDonald WH, Lopez-Girona A, Noguchi E, Yates IJ, Russell P (2003) Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol Cell Biol 23: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M (2006) Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (2001) Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 8: 833–837 [DOI] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25: 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X (2007) The Slx5/8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol (doi:10.1128/MCB.00787-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo F, Astolfi A, Iezzi M, Cordero F, Lollini PL, Forni G, Calogero R (2005) An integrated approach of immunogenomics and bioinformatics to identify new tumor associated antigens (TAA) for mammary cancer immunological prevention. BMC Bioinformatics 6: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT (1999) Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem 274: 10618–10624 [DOI] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA (2005) Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 14: 1351–1365 [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Nayak S, Epstein JA, Buck CA (2000) Rnf4, a RING protein expressed in the developing nervous and reproductive systems, interacts with Gscl, a gene within the DiGeorge critical region. Dev Dyn 218: 102–111 [DOI] [PubMed] [Google Scholar]
- Gill G (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ (2001) The RING finger protein SNURF is a bifunctional protein possessing DNA binding activity. J Biol Chem 276: 23653–23660 [DOI] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ (2005) SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res 304: 224–233 [DOI] [PubMed] [Google Scholar]
- Hakli M, Lorick KL, Weissman AM, Janne OA, Palvimo JJ (2004) Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett 560: 56–62 [DOI] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110 [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, Snydsman BE, Bradley P, Muller EG, Fields S, Baker D, Yates JR III, Davis TN (2003) Assigning function to yeast proteins by integration of technologies. Mol Cell 12: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Ho JC, Warr NJ, Shimizu H, Watts FZ (2001) SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res 29: 4179–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kaiser FJ, Moroy T, Chang GT, Horsthemke B, Ludecke HJ (2003) The RING finger protein RNF4, a co-regulator of transcription, interacts with the TRPS1 transcription factor. J Biol Chem 278: 38780–38785 [DOI] [PubMed] [Google Scholar]
- Kosoy A, Calonge TM, Outwin EA, O'Connell MJ (2007) Fission yeast RNF4 homologs are required for DNA repair. J Biol Chem 282: 20388–20394 [DOI] [PubMed] [Google Scholar]
- Leverson JD, Huang HK, Forsburg SL, Hunter T (2002) The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol Biol Cell 13: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson JD, Joazeiro CA, Page AM, Huang H, Hieter P, Hunter T (2000) The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell 11: 2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu R-M (2006) Structure of a Bmi-1-Ring1B polycomb group Ubiquitin ligase complex. J Biol Chem 281: 20643–20649 [DOI] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24: 341–354 [DOI] [PubMed] [Google Scholar]
- McDonald WH, Pavlova Y, Yates JR III, Boddy MN (2003) Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem 278: 45460–45467 [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275: 36316–36323 [DOI] [PubMed] [Google Scholar]
- Moilanen AM, Poukka H, Karvonen U, Hakli M, Janne OA, Palvimo JJ (1998) Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol 18: 5128–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F (2005) Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJP, Tainer JA, McGowan CH, Boddy MN (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J [E-pub ahead of print: advance online publication, 30 August 2007; doi:10.1038/sj.emboj.7601838] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Wohlschlegel J, Yates JR III, Boddy MN (2006) Sumo-binding motifs mediate the RAD60-dependent response to replicative stress and self association. J Biol Chem 281: 27973–27981 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T (2006) Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 7: 473–483 [DOI] [PubMed] [Google Scholar]
- Shayeghi M, Doe CL, Tavassoli M, Watts FZ (1997) Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res 25: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP (2006) The mechanisms of PML-nuclear body formation. Mol Cell 24: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R, Dolan WP, Forsburg SL (2004) Choosing and using Schizosaccharomyces pombe plasmids. Methods 33: 189–198 [DOI] [PubMed] [Google Scholar]
- Sobko A, Ma H, Firtel RA (2002) Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev Cell 2: 745–756 [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem 280: 40122–40129 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y (1999) Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19: 8660–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276: 35368–35374 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Verkade HM, Teli T, Laursen LV, Murray JM, O'Connell MJ (2001) A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol Genet Genomics 265: 993–1003 [DOI] [PubMed] [Google Scholar]
- Wang L, Kao R, Ivey FD, Hoffman CS (2004) Strategies for gene disruptions and plasmid constructions in fission yeast. Methods 33: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jones GM, Prelich G (2006) Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 172: 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Wu SM, Kuo WC, Hwu WL, Hwa KY, Mantovani R, Lee YM (2004) RNF4 is a coactivator for nuclear factor Y on GTP cyclohydrolase I proximal promoter. Mol Pharmacol 66: 1317–1324 [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B (2004) Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J 23: 3844–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Pao GM, Chen HW, Verma IM, Hunter T (2003) Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem 278: 5255–5263 [DOI] [PubMed] [Google Scholar]
- Yang L, Mullen JR, Brill SJ (2006) Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res 34: 5541–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, Tai T (2002) E3 ubiquitin ligase that recognizes sugar chains. Nature 418: 438–442 [DOI] [PubMed] [Google Scholar]
- Zhang C, Roberts TM, Yang J, Desai R, Brown GW (2006) Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 5: 336–346 [DOI] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures