Abstract
Differences in gene expression underlie central questions in plant biology extending from gene function to evolutionary mechanisms and quantitative traits. However, resolving expression of closely related genes (e.g. alleles and gene family members) is challenging on a genome-wide scale due to extensive sequence similarity and frequently incomplete genome sequence data. We present a new expression-profiling strategy that utilizes long-read, high-throughput sequencing to capture the information-rich 3′-untranslated region (UTR) of messenger RNAs (mRNAs). Resulting sequences resolve gene-specific transcripts independent of a sequenced genome. Analysis of approximately 229,000 3′-anchored sequences from maize (Zea mays) ovaries identified 14,822 unique transcripts represented by at least two sequence reads. Total RNA from ovaries of drought-stressed wild-type and viviparous-1 mutant plants was used to construct a multiplex cDNA library. Each sample was labeled by incorporating one of 16 unique three-base key codes into the 3′-cDNA fragments, and combined samples were sequenced using a GS 20 454 instrument. Transcript abundance was quantified by frequency of sequences identifying each unique mRNA. At least 202 unique transcripts showed highly significant differences in abundance between wild-type and mutant samples. For a subset of mRNAs, quantitative differences were validated by real-time reverse transcription-polymerase chain reaction. The 3′-UTR profile resolved 12 unique cellulose synthase (CesA) transcripts in maize ovaries and identified previously uncharacterized members of a histone H1 gene family. In addition, this method resolved nearly identical paralogs, as illustrated by two auxin-repressed, dormancy-associated (Arda) transcripts, which showed reciprocal mRNA abundance in wild-type and mutant samples. Our results demonstrate the potential of 3′-UTR profiling for resolving gene- and allele-specific transcripts.
Functional analysis of plant genomes requires methods for resolving differential expression of closely related genes. The ability to distinguish between paralogs (e.g. gene family members) and alleles on a genome-wide scale is key to understanding the genetic basis of quantitative traits in diverse plant populations. Genes with extensive sequence similarity may comprise a significant portion of a given transcriptome. Among maize (Zea mays) inbreds, for example, 90% of the alleles are polymorphic (Wright et al., 2005) and approximately one-third of maize genes are tandemly duplicated (Messing et al., 2004). The extent of these sequence similarities in maize and other complex genomes poses a clear challenge to delineation of gene-specific function.
Differential expression of related, duplicated genes has been linked to functional diversity within species (Gu et al., 2004), and subfunctionalization can provide a basis for genome evolution (Moore and Purugganan, 2005; Emrich et al., 2007b). The impetus for resolving expression among paralogs is further motivated by the extent of polyploidy, which is estimated to affect 50% to 80% of angiosperm species, including maize, wheat (Triticum aestivum), cotton (Gossypium hirsutum), and other important crops (Osborn et al., 2003; Blanc and Wolfe, 2004). Moreover, allele-specific differences in gene expression that contribute to variations in phenotype are widespread in both animal (Cowles et al., 2002; Yan et al., 2002) and plant species (Cong et al., 2002; Guo et al., 2004). Accordingly, transcript profiling has been adapted as a means of appraising quantitative traits (Schadt et al., 2003; Borevitz and Chory, 2004). Association mapping using expression quantitative trait loci has identified candidate genes for important traits in tomato (Solanum lycopersicum; Baxter et al., 2005) and poplar (Populus spp.; Street et al., 2006). In addition, comparing allele-specific expression among inbred parental varieties and F1 hybrids of reciprocal crosses has revealed deviations from dosage dependency and enabled analysis of imprinting in maize endosperm (Guo et al., 2003). The ability to resolve individual transcripts with similar sequences and quantitatively compare their expression is thus central to addressing questions in functional genomics and defining genetic contributions to hybrid vigor (Birchler et al., 2003; Swanson-Wagner et al., 2006; Springer and Stupar, 2007).
Despite rapid advances in expression profiling techniques, the capacity to distinguish among closely related transcripts on a genome-wide scale remains a challenge. In microarray analyses, cross hybridization of similar transcripts to a given oligonucleotide probe may confound expression of individual genes. With sequencing of several genomes complete (e.g. Arabidopsis [Arabidopsis thaliana], rice [Oryza sativa], and poplar), whole-genome tiling arrays have allowed unbiased interrogation of the transcriptome (Yamada et al., 2003; Mockler and Ecker, 2005). These platforms have successfully uncovered discrete polymorphisms and alternate splice sites but depend on fully sequenced genomes and/or are limited to sequence variants present on the array (typically derived from a single reference strain). In addition, quantitative measures of gene expression are limited by probe-specific hybridization efficiencies.
The emergence of high-volume, short-read sequencing technologies has increased resolution for quantitative transcriptome analysis in organisms for which complete genomic sequence is available. Advances in serial analysis of gene expression (SAGE) have opened transcript profiling to unbiased sampling and quantitative analysis of gene expression (Saha et al., 2002; Bao et al., 2005). Although limited in throughput, the sequencing of novel cDNAs following 3′ extension and amplification of short SAGE tags has been successfully utilized for gene discovery (Chen et al., 2002).
Alternatively, genome-wide profiling by massively parallel sequencing (Meyers et al., 2004; Jongeneel et al., 2005) and the more recent Solexa 1-G technology (Barski et al., 2007) has facilitated detection of rare transcripts and comprehensive cataloging of noncoding RNAs (Lu et al., 2005; Nobuta et al., 2007). The capacity of these massively parallel approaches to generate millions of short sequence tags can enable reliable, cost-effective coverage of the transcriptome. However, in genomes where sequence information is fragmentary, the short length of these reads (17–36 bases) provides a limited capability for unambiguous gene assignment. Likewise, near-identical transcripts are difficult to discern with short-sequence reads, even in a fully sequenced genome. Estimates from rice and Arabidopsis massively parallel sequencing libraries indicate that approximately 11% of signature sequences matched multiple target sites in the genome (Nobuta et al., 2007).
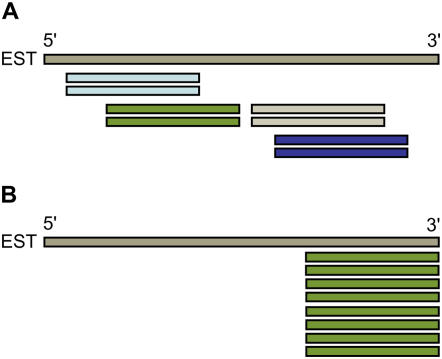
Longer read lengths are achieved with 454-based pyrosequencing, initially described by Margulies et al. (2005) and more recently implemented as a platform for transcript profiling (Emrich et al., 2007a; Weber et al., 2007). A key advantage of the longer reads generated by this technology is greater capability for gene annotation and discovery in both sequenced and nonsequenced genomes. A recent upgrade of the Genome Sequencer FLX system (Harkins and Jarvie, 2007) has extended average read lengths to >200 bases. A tradeoff for obtaining more informative read lengths is a lower depth of sequencing achieved with 454 compared to short-read technologies (e.g. Solexa 1-G). Therefore, one method for improving the efficiency of 454-based transcript profiling is to anchor 454 reads to unique sites near the 3′ ends of expressed sequences to reduce the number of reads necessary to identify individual mRNAs and maximize recovery of gene-specific polymorphisms. The 3′-untranslated region (UTR) is rich in single-feature polymorphisms that distinguish closely related transcripts (Bhattramakki et al., 2002; Vroh Bi et al., 2006). The specificity of 3′-UTR sequence reads thus allows effective annotation of individual mRNAs without assembly of complete cDNAs (Fig. 1).
Figure 1.
Comparison of shotgun versus 3′-anchored approaches for using 454 sequences as a platform for transcript profiling. A, Sequencing of randomly sheared cDNA fragments, followed by contig assembly, can provide coverage of full-length cDNAs. However, the redundancy of sequence reads for a given transcript limits information returned per number of reads and thus statistical analyses of expression. B, Our 3′-UTR profiling method identifies unique ESTs by anchoring each sequence read to a gene-specific region of cDNAs. This increases depth of sequencing and facilitates assembly and analysis.
Here, we present a strategy that harnesses the specificity and information content of the 3′-UTR in a long-read, 454-based sequencing approach to transcript profiling. A key to this method is the use of 3′-anchored sequence reads long enough for unambiguous identification of closely related transcripts. By targeting the 3′-UTR of mRNAs, an unprecedented resolution is achieved for gene- and allele-specific transcripts, even for genomes that are only partially sequenced or lack extensive EST coverage. In addition, detection of haplotypes containing multiple polymorphisms is facilitated by the longer read length. These components of the transcriptome are thus opened to quantitative analysis beyond that currently accessible with short-tag sequencing technologies. In this work, maize provides an ideal system to assess our 3′-anchored strategy, because the genome is rich in genetic complexity from extensive gene duplication (Messing et al., 2004; Messing and Dooner, 2006) and currently is not fully sequenced.
In this study, we introduce a 3′-UTR profiling method that allows quantitative analysis of gene-specific expression on a genome-wide level, here using mutant and wild-type maize ovaries. Concurrent sequencing of multiple mRNA samples was enabled by the use of a multiplexing strategy. Results provided quantitative expression profiles with read output evenly distributed between samples. The frequency of 3′-anchored sequence reads aligning to a given cDNA was used to quantify mRNA abundance and to measure differential gene expression. The long read lengths combined with the specificity of the 3′-UTR were sufficient to distinguish individual members of a previously characterized gene family as well as provide quantitative comparisons of closely related transcripts that matched unique maize ESTs or assembled cDNAs. In addition, insertion deletion (indel) polymorphisms were readily detectable by this method and resolved nearly identical paralogous gene products.
RESULTS
3′-cDNA Library Construction
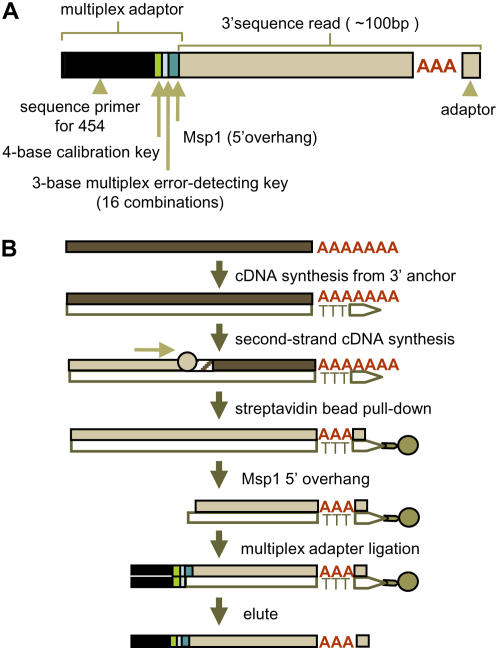
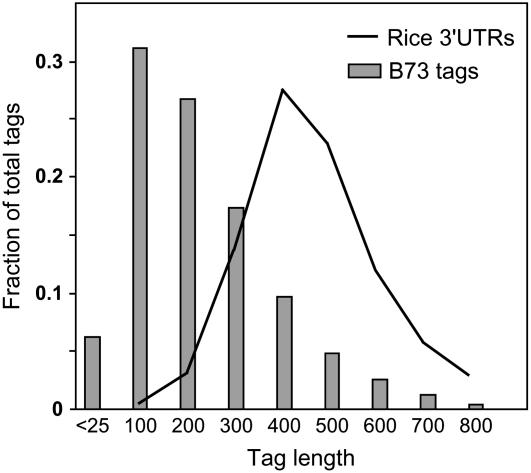
We synthesized 3′-anchored cDNA template libraries to generate gene-specific sequence reads by 454 using the protocol shown in Figure 2. Concurrent sequencing of up to 16 individual sublibraries is enabled by incorporation of a three-base multiplex key in the A-adaptor (Fig. 2A). By using a subset of 16 three-base keys, we could detect single-base errors in the multiplex key. Addition of a fourth base to the multiplex key would enable up to 64 unique combinations with error detection, thus enhancing the number of concurrently sequenced sublibraries. Each 3′-UTR sublibrary was constructed from total RNA (Fig. 2B) using a modified, biotinylated 454-B adaptor that incorporates an oligo(dT) tail for priming cDNA synthesis from poly(A+) RNA. Following second-strand cDNA synthesis, biotinylated cDNAs were bound to Streptavidin beads, purified by magnetic pull down, and digested with MspI to generate 3′-cDNA fragments with 2-base (CG) overhangs. Specific multiplex A-adaptors were then ligated to the purified 3′ fragments. A detailed description is provided in “Materials and Methods.” MspI was selected based on simulated digests of 70,000 3′-orientated ESTs of maize (Fig. 3) from the maize full-length cDNA project (www.maizecdna.org). Predicted tag lengths were used to assess the proportion of 3′ enrichment in comparison to rice 3′-UTRs. While the expected size distribution of MspI-digested cDNA fragments is optimal for the Genome Sequencer 20 read length (approximately 100 bases), the longer reads generated by the 454-FLX instrument (approximately 250 base average) would likely extend through the 3′-UTR of many transcripts. If so, the number of FLX reaction cycles could be configured to optimize average read length (Harkins and Jarvie, 2007). Although a single MspI digest was used in this study, potential increases in coverage of 3′ ends could be achieved by combining digests made with compatible restriction enzymes (e.g. TaqI, MaeII). This would further improve coverage when used in conjunction with the longer read lengths and enhanced read output achieved by 454-FLX technology.
Figure 2.
Schematic strategy for 3′-UTR profiling by 454 sequencing. A, A 3′-anchored cDNA library is restriction digested and tagged via ligation to a multiplex adaptor. Unique combinations of a 4-base multiplex error-detecting key enables sample identification during concurrent sequencing of up to 16 sublibraries. B, Sublibrary construction from total RNA.
Figure 3.
Distribution of tag lengths for a simulated MspI digest of 70,000 3′-oriented ESTs from B73 maize (maize full-length cDNA project [www.maizecdna.org]). A comparison of simulated tag lengths to the distribution of annotated rice 3′-UTRs indicated enrichment of 3′-UTR sequence with an average tag length of 100 to 200 bases. Proportion of short tags is reduced due to the low frequency of MspI sites proximal to the poly(A) tail.
To test the 3′-UTR profiling strategy, we sequenced 3′-anchored cDNA sublibraries prepared from immature ovaries of isogenic viviparous-1 (vp1) mutant and wild-type maize plants in a W22 inbred genetic background. Prior to RNA sampling, plants were subjected to a drought stress treatment (“Materials and Methods”). VP1 is a transcription factor that mediates a subset of responses to the plant stress hormone, abscisic acid (ABA), including maturation and onset of seed desiccation tolerance. The classic vp1 phenotype is that of precocious germination due to reduced ABA sensitivity (McCarty et al., 1991). More recently, however, VP1 has been implicated in stress responses of nonseed tissues (Cao et al., 2007). In addition, preliminary evidence suggests that VP1 may be involved in modulating female reproductive quiescence in maize under drought stress (A.L. Eveland, unpublished data). To resolve differences in expression profiles that would help define roles of the VP1 gene, cDNA sublibraries, each tagged with a unique multiplex key, were prepared from wild-type and vp1-mutant maize ovaries.
Data Assembly and Analysis
A sequencing reaction on the Genome Sequencer 20 instrument (Margulies et al., 2005) yielded 228,595 high-quality reads with an average, trimmed length of 95 bases. Of these, 93% were identified as correctly oriented, 3′-anchored cDNAs with equal representation of wild-type and mutant sublibraries (Table I). The 7% of reads that were excluded from further analysis contained errors in the multiplex key (1.5%) or invalid ligation junctions (5.5%). Assembly of validated, trimmed, high-quality reads using CAP3 (Huang and Madan, 1999) revealed 14,822 nonredundant 3′-anchored consensus sequences, each represented by two to 2,500 reads, and 32,477 singlets. The distribution of consensus sequences per read number is shown in Supplemental Table S1.
Table I.
Summary statistics for a two-sample multiplex 3′-UTR library
| Total high-quality readsa | 228,595 |
| Wild-type sublibrary reads | 105,289 |
| Mutant sublibrary reads | 109,958 |
| Error-detected reads | 13,348 |
| Errors in multiplex key | (1.5%) |
| Invalid ligation junctions | (5.5%) |
| Total unique sequencesb | 47,299 |
| Singletsc | 32,477 |
| Consensus sequences (≥2 reads) | 14,822 |
| cDNA matchesd | 11,559 |
| Genome matchese | 9,740 |
Abundance of reads after filtering for MspI-ligation junctions.
Nonredundant set of sequences representing unique transcripts.
Single-copy transcripts.
Abundance of consensus sequences matching cDNAs in IUC or ZmGI databases.
Abundance of consensus sequences matching available MAGI4.
The capacity of these consensus sequences to identify individual genes based on specificity of their 3′-UTRs was tested by aligning these reads to available maize cDNA databases (The Institute for Genomic Research Zea mays Gene Index [ZmGI] and Industry UniGene [IUC]) using BLASTN (cutoff: E < 10−7). At least 87% of the consensus tags matched cDNAs and 66% aligned with a gene-enriched maize genomic assembly (MAGI; Fu et al., 2005). In addition, BLASTN searches of the The Institute for Genomic Research maize repeat database (http://maize.tigr.org/repeat_db.shtml) indicated that only 1.9% of 3′-anchored consensus sequences contained retrotransposons or other repetitive sequences, whereas another 1.2% were identified as organellar or cytosolic ribosomal RNA (rRNA) contaminants. The latter were most likely due to rare mispriming by the oligo(dT)-B adaptor during cDNA synthesis.
Analysis of 3′-UTR Profile Reveals a Dynamic Range of Expression
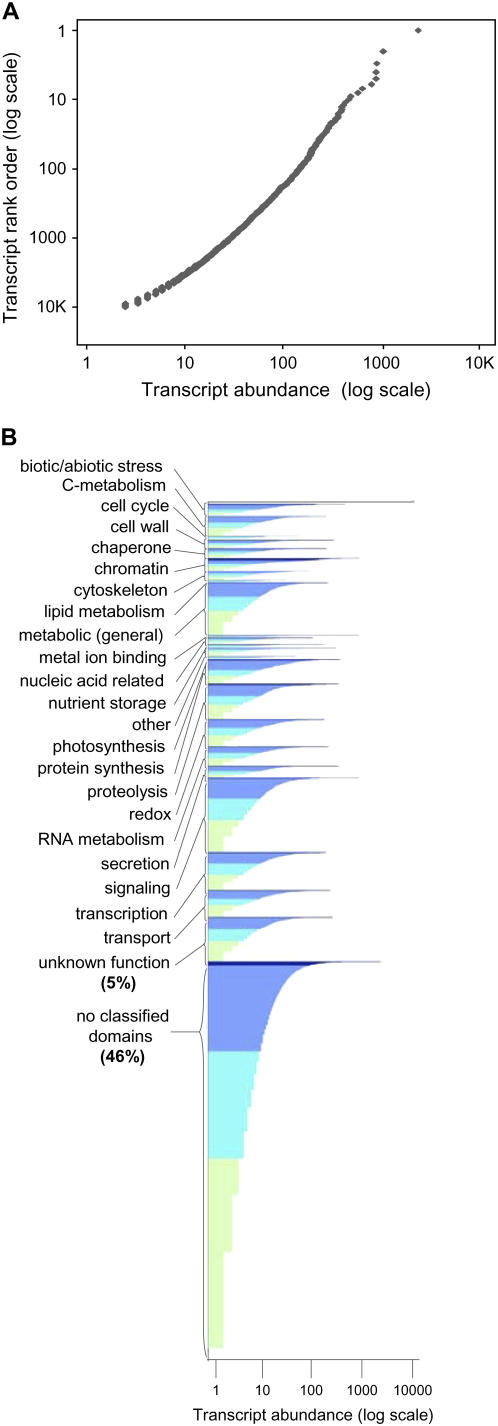
Based on the set of unique consensus sequences obtained from the two-sample library, we developed a graphic display for the quantitative transcriptome profile (Fig. 4, A and B). We quantified gene expression for each of 11,559 consensus sequences that matched unique cDNAs using read frequencies. The results are plotted on a logarithmic scale to capture the full range of expression. The 11,559 3′ sequences profiled were also analyzed based on Gene Ontology functional classifications determined by PFam searches derived from ZmGI and IUC databases. Analysis of respective maize cDNAs revealed 5,202 (45%) that were unclassified and lacked annotation based on sequence similarity. An additional 578 (5%) of consensus sequences matched genes having conserved domains of unknown function.
Figure 4.
A graphic presentation of the quantitative 3′-UTR profile representing 11,559 consensus sequences that matched cDNAs in the two-sample multiplexed library. Read frequency was used as a quantitative measure of mRNA abundance. A, Transcript abundance is plotted on a log-log scale for respective genes in rank order from least- to most-highly expressed. B, Transcripts are grouped into functional classifications based on Gene Ontologies and plotted linearly along the y axis. Read count for each transcript is plotted on the x axis (log scale). Color scale (dark to light) denotes the dynamic range of mRNA abundance. Distribution of consensus sequences according to read count is also shown in Supplemental Table S1.
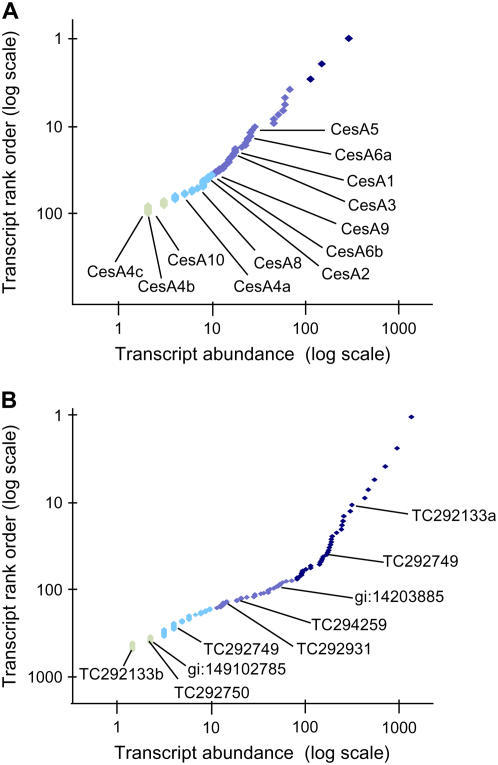
The relationship between abundance of each mRNA and its rank (ordered from least to most prevalent) in the whole dataset approximated a Zipf-power law (ranked slope near −1 on a log-log scale). This distribution was evident among transcripts overall (Fig. 4A) and within individual functional classes (Figs. 4B and 5A). Zipf's power law relationships are observed in a wide range of natural phenomena, including the distribution of gene expression in a variety of organisms (Kuznetsov et al., 2002; Furusawa and Kaneko, 2003). Accordingly, it has been used as a tool for normalization in some SAGE and microarray analyses. Although our results were consistent with this distribution, an interesting exception is shown for the chromatin-related functional class. As shown in Figures 4B and 5B, distribution of expression was skewed for this group of mRNAs by a disproportionate number of highly abundant transcripts.
Figure 5.
Distribution of transcripts (log-log scale) in selected functional classes from Figure 4B and read count for each. Resolution of individual gene family members was enabled by the specificity of the 3′-UTR. A, Quantitative measure of mRNAs for all transcripts classified with cell wall-related functions (Fig. 4B) and resolution of 12 unique mRNAs representing nine previously characterized members of the CesA gene family (including transcript variants for CesA4 and CesA6) in maize ovaries. Read frequencies for CesA gene family members range from two to 27 reads. B, Quantitative measure of mRNAs for all transcripts with chromatin-related functions (Fig. 4B) and identification of H1-like transcripts that matched unique but uncharacterized maize cDNAs (indicated by ZmGI TC nos. or GenBank IDs). Read frequencies for H1 gene family members range from two to 684 reads. TC292133: drought-induced H1; TC29749, TC292749, TC292750, gi:149102785: H1-like HON101; gi:110544310: TC294259; TC292931: HMGI/Y.
Distinguishing Gene Family Members
To evaluate 3′-UTR profiling for resolution of individual gene family members, we analyzed the cellulose synthase (CesA) gene family (Fig. 5A). The assembled 3′-anchored sequences distinguished 12 unique transcripts representing nine annotated CesA gene family members (Supplemental Table S1) that were previously characterized in maize (Holland et al., 2000; Appenzeller et al., 2004). The full-length CesA cDNAs (ZmGI) share up to 94% sequence identity. In some cases, extensive sequence similarity between CesA genes and their proximal mapped locations to each other in the genome are suggestive of paired duplications (e.g. CesA1 and CesA2 on chromosomes 6 and 8 and CesA4 and CesA9 on chromosome 7). The cDNA sequences for CesA4 and CesA9 differ almost exclusively in their 3′-UTRs, thus complicating resolution of these two genes in previous expression studies (Holland et al., 2000). Here, the corresponding 3′-anchored 454 reads for these closely related gene family members aligned with gene-specific regions in the 3′-UTR (Supplemental Fig. S1). Polymorphic variants for CesA4 and CesA6 were also identified. Alignments of consensus tags to a CesA4 cDNA (TC287832) indicated that a novel transcript variant (CesA4c) contained an MspI restriction site polymorphism as well as 35 bp of an unspliced intron (Supplemental Table S2). Although no other ESTs having these features were detected in maize databases, nine reads in our maize-ovary dataset aligned with the CesA4c variant. Consistent with the possibility that these sequences identify a second CesA4 gene, CesA4 has been mapped to two locations (2.06 and 7.01; Holland et al., 2000) corresponding to duplicated chromosome segments (Helentjaris et al., 1988).
In addition, we analyzed a group of closely related histone H1-like transcripts (Fig. 5B). These transcripts matched a unique, nonredundant set of ESTs from various maize cDNA libraries and were annotated based on sequence similarities in other species. Although these H1 genes have not been individually characterized in maize, BLASTN results provided insight for eventual functional analysis. For example, a very highly expressed H1-like transcript (TC292133a) matched a drought- and ABA-induced gene that had been characterized in tomato (Bray et al., 1999). These results indicate that unbiased profiling of closely related transcripts can facilitate studies of functional genomics with or without a fully annotated EST dataset or a completely sequenced genome.
Evaluation of Differential Expression between Multiplexed Sublibraries
The use of 3′-UTR profiling as an effective strategy for detecting quantitative differences in transcript abundance between samples was evaluated based on read frequencies generated from individual sublibraries. Read frequencies representing each expressed gene were determined for wild-type and mutant sublibraries by parsing the CAP3 ace file output. We analyzed 4,147 consensus sequences that were represented by a total of 10 or more reads using a χ2 statistic. Of these, 202 showed significant differences (P < 0.0015) in frequency between the two samples, indicating putative differences in transcript levels. A subset of these consensus sequences with highly significant differences between libraries was annotated by BLASTN to identify best-match ESTs (Table II). Of the 30 sequences listed, three matched to unannotated cDNAs that appeared to be maize specific (TC286704, TC300122 [ZmGI], and 2569799 [National Center for Biotechnology Information {NCBI} UniGene]) and, based on searches of public databases, 10 were found exclusively or highly represented in cDNA libraries from reproductive tissues (2568974/TC285721, 514900, 2566963, 2568212/TC286030, 507904/TC301902 [NCBI UniGene/ZmGI]) or from drought-stressed plants (2564044/TC285867, 2714857/TC286791, 508486/TC29233, 2561245/TC299973, 2567165 [NCBI UniGene/ZmGI]).
Table II.
Best-match cDNAs and associated annotations (BLASTN) for consensus sequences showing highly significant differences in transcript abundance between wild-type and vp1 mutant drought-stressed ovary sublibraries
Read frequency differences for consensus sequences were analyzed using a χ2 statistic. Consensus sequences were aligned to best-match cDNAs using BLASTN in ZmGI and IUC databases. NCBI UniGene IDs are listed for sequences where TC numbers were not available.
| cDNA ID | Percent Match (Length) | BLASTN Annotation (Species) | Read Frequency
|
P Value | |
|---|---|---|---|---|---|
| Wild Type | vp1 | ||||
| TC285867a | 100 (87) | Auxin-repressed dormancy (Robinia pseudoacacia) | 238 | 709 | 10−53 |
| TC285721b | 100 (98) | Gly-rich protein (rice) | 57 | 244 | 10−27 |
| TC286704 | 98 (94) | Unannotated | 25 | 167 | 10−24 |
| TC305930 | 98 (80) | Farnesylated protein 3 (Hordeum vulgare) | 14 | 119 | 10−20 |
| 2569891 | 100 (52) | Xyloglucan endotransglucosylase (H. vulgare) | 234 | 81 | 10−18 |
| TC285789 | 100 (96) | Auxin-repressed dormancy (pea) | 53 | 1 | 10−12 |
| TC292711 | 100 (100) | Nodulin MtN3 family (Arabidopsis) | 0 | 52 | 10−13 |
| 514900b | 97 (116) | At1g74950 (Arabidopsis) | 127 | 37 | 10−12 |
| TC286791a | 100 (86) | Dehydrin RAB-17 protein (maize) | 8 | 69 | 10−12 |
| TC292358 | 100 (96) | Thr-rich extensin (maize) | 1,070 | 1,414 | 10−12 |
| 507881 | 100 (95) | Unnamed protein product (rice) | 93 | 21 | 10−11 |
| TC286485 | 100 (87) | Histone H2A (maize) | 575 | 396 | 10−11 |
| 2566963b | 99 (104) | Unknown protein (rice) | 74 | 12 | 10−11 |
| TC286030b | 100 (111) | Harpin-induced gene 1 (rice) | 106 | 31 | 10−10 |
| TC310545 | 100 (93) | Histone H1 (maize) | 259 | 135 | 10−10 |
| TC294233a | 100 (104) | Putative cystatin cc3 (Saccharum officinarum) | 65 | 158 | 10−10 |
| 2708354 | 100 (97) | Unannotated (rice) | 75 | 16 | 10−10 |
| TC298173 | 95 (93) | Histone 3 (rice) | 151 | 61 | 10−10 |
| TC294050 | 100 (95) | EF-hand Ca2+-binding CCD1 (wheat) | 102 | 32 | 10−9 |
| TC301902b | 100 (87) | AP2 domain, EREBP (rice) | 66 | 13 | 10−9 |
| TC305186 | 100 (78) | Subtilisin-like proteinase (rice) | 113 | 41 | 10−9 |
| 1572511 | 100 (49) | Hypothetical protein (rice) | 31 | 0 | 10−8 |
| TC299973a | 95 (84) | Glycogenin-like (rice) | 31 | 0 | 10−8 |
| 654573 | 100 (45) | Ca+-binding EF hand family (Arabidopsis) | 53 | 10 | 10−8 |
| TC280589 | 100 (98) | Phosphate-induced protein 1 like (Pennisetum ciliare) | 146 | 67 | 10−8 |
| 2567165a | 90 (110) | Heavy-metal associated (rice) | 141 | 64 | 10−8 |
| 2569891 | 100 (52) | Xyloglucan endo-1,4-β-d-glucanase (H. vulgare) | 43 | 6 | 10−7 |
| TC310820 | 100 (91) | CCCH-type zinc finger protein like (rice) | 15 | 61 | 10−7 |
| 2569799 | 100 (74) | Unannotated | 59 | 14 | 10−7 |
| TC300122 | 99 (103) | Unannotated | 127 | 56 | 10−7 |
cDNAs highly represented in GenBank libraries from drought-stressed maize plants.
cDNAs highly represented in GenBank libraries from maize reproductive tissues.
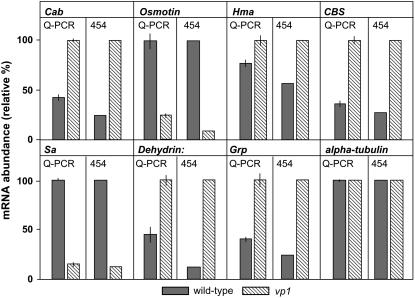
Quantitative differences in levels of specific mRNAs were confirmed for a subset of genes by real-time reverse transcription (RT)-PCR analyses of the wild-type and vp1 mutant samples (Fig. 6). Results showed that differences in transcript abundance between wild-type and mutant RNA samples used in 3′-cDNA sublibrary construction paralleled the 454-based expression profiles.
Figure 6.
Validation of technical accuracy for determining differences in transcript abundance based on read number. Parallel real-time RT-PCR (SYBR green, MyIQ, Bio-Rad) analyses (technical error based on three independent determinations) on identical RNA samples as used in 3′-UTR sublibrary construction validated the significant differences in read frequency (P < 0.0015 except for α-tubulin control) for a subset of genes. Gene-specific primers were designed using the 454 sequence reads and associated EST matches as templates.
Resolution of Near-Identical Transcripts by Polymorphisms
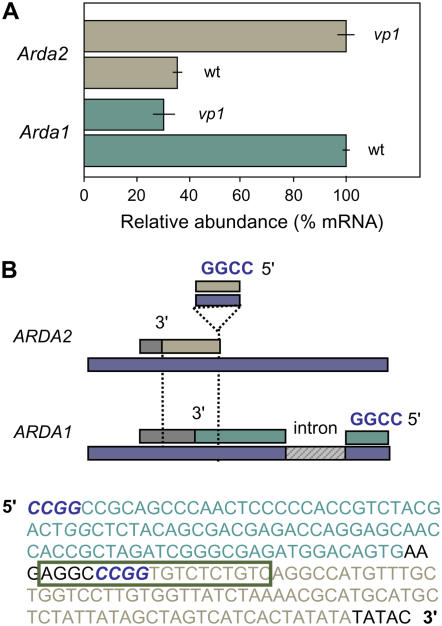
Analyses of the maize genome have revealed a high frequency of nearly identical paralogs with ≥98% identity (Emrich et al., 2007b). In most instances, both gene copies are expressed. Identification of single feature polymorphisms in the 3′ sequences can effectively distinguish a subset of such paralogs. At least one example where 3′-UTR profiling effectively resolved near-identical paralogs was evident for closely related but differentially expressed auxin-repressed, dormancy-associated (Arda) transcripts (we designated these genes as ARDA1 and ARDA2). The ARDA1 and ARDA2 sequences share ≥98% identity (99% in the coding region and 97% in their 3′-UTRs). Two distinct 3′-UTR ARDA consensus sequences detected an 18-bp indel polymorphism that distinguished these two paralogs. Read frequencies showed reciprocal responses in the mutant background by ARDA1 (P < 10−53) and ARDA2 (P < 10−12). Differential profiles for these genes were confirmed by amplifying the region in or around the indel using real-time RT-PCR (Fig. 7A). These reciprocal expression profiles could not be resolved when regions outside of the indel sequence were amplified due to confounding effects of the nearly identical sequences.
Figure 7.
A 3′ sequence polymorphism resolved nearly identical ARDA paralogs with differences in mRNA abundance. A, Q-PCR analyses confirmed the reciprocal responses of near-identical Arda1 and Arda2 (≥98% identity) on wild-type and mutant samples used for sublibrary construction (technical error based on three independent determinations). B, The near-identical ARDA1 (EE188942) and ARDA2 (EE679809) ESTs differ by an 18-bp insertion within ARDA2, which is not present in MAGI4_156527 (region aligning with the ESTs). The 454 sequence reads representing ARDA1 (blue) and ARDA2 (brown) are highlighted within the 3′ end of the ARDA2 EST sequence (top) and within a schematic diagram of the two ESTs (3′ ends) aligned to maize genomic sequence (bottom).
Earlier work identified ARDA1 as a potentially important contributor to stress tolerance in hybrid maize (Guo et al., 2004). The previously undetected ARDA2, resolved in the monoallelic W22 inbred, matched a unique maize EST. Alignment of the two consensus sequences to a region within assembled genomic sequence (MAGI4_156527) verified the presence of two paralogous gene products (Fig. 7B). Both ARDA paralogs appeared to be drought responsive in preliminary analyses. Currently, there is little information for putative roles of ARDA genes. Studies in pea (Pisum sativum) characterized similar genes as markers for dormancy in axillary buds (Stafstrom et al., 1998).
Validation of Single Nucleotide Polymorphisms and Homopolymer-Based Polymorphisms
We conducted a detailed analysis of polymorphisms detected by a preliminary dataset comprised of 1,263 W22 consensus sequences using BLASTN alignment to MAGI4 B73 genomic sequences. We expected that some portion of apparent polymorphisms in consensus sequences ranging from two to 75 reads (56.6%; Supplemental Table S3) was due to sequence errors. To estimate the contribution of sequence errors in the 454 data, we evaluated polymorphisms detected by a subset of 107 cDNA consensus sequences (seven to 75 reads) with respect to B73 MAGI assemblies by independent BLASTN searches of IUC cDNA and public EST databases. We confirmed 93.8% of 146 sequence polymorphisms detected within 52 W22 alleles by identical cDNA matches, indicating that most identify independently documented maize alleles.
Because the pyrosequencing method used by 454 is prone to errors in estimating lengths of long homopolymer runs (Margulies et al., 2005), we investigated the effect this may have on single nucleotide polymorphism (SNP) detection in maize sequences. Overall, 29% of the 1,263 W22 consensus sequences analyzed above contained one or more homopolymer tracts of 5 bp or longer. To assess the impact of homopolymer read errors on SNP detection by 454, we analyzed the polymorphisms detected by a set of 211 W22 consensus sequences (five to 75 reads) in best alignments to the MAGI4 (B73) dataset (examples in Supplemental Fig. S2). Of the total 257 polymorphisms detected (counting indels as one), at least 89.9% were independently confirmed by identical cDNA matches. In addition, only 60 (23%) were potentially attributed to simple or compound (e.g. CCTTT → CCCTT) homopolymer base-calling errors. Moreover, the 60 homopolymer-based polymorphisms were distributed randomly (P > 0.9) between alignments that included long homopolymer tracts of ≥5 bp (21% of consensus sequences analyzed) and alignments that lacked them. Finally, all but seven of 60 homopolymer length polymorphisms were supported by independent EST sequences from W22 or other sources. Hence, these homopolymer-based polymorphisms were not appreciably less reliable (88.3% confirmed) than other substitution and indel polymorphisms (93.8% confirmed by independent cDNA sequences).
DISCUSSION
Our results demonstrate that 3′-UTR profiling is an effective strategy for high-resolution global analysis of gene expression that does not require a complete genome sequence. Using this approach, we were able to identify over 14,000 gene-specific mRNAs and quantify expression based on read frequencies occurring in 3′-anchored consensus sequences. Analysis of the quantitative 3′-UTR profile revealed a dynamic range of gene expression spanning greater than 3 orders of magnitude.
Our strategy of using long-read, 454 sequencing to target gene-specific 3′-UTRs offers several advantages over previous tag-based approaches to global expression profiling. First, depth of sequencing is enhanced by anchoring the 454 reads to unique sites proximal to the 3′ ends of transcripts. This eliminates redundancy associated with shotgun sequencing of cDNA fragments, thus providing more reads per unique transcript and reducing the potential for highly expressed mRNAs to saturate the library (Weber et al., 2007). In this study, the two-sublibrary analysis using the 454 Genome Sequencer 20 instrument identified 47,299 distinct mRNAs (including 14,822 consensus sequences represented by two to 2,500 reads). In comparison, sequencing of nebulized Arabidopsis cDNAs yielded approximately 17,500 unique transcripts after two GS 20 sequencing reactions (Weber et al., 2007). Although we cannot discount the possibility that a portion of the singlets identified in our dataset are due to sequencing errors, deeper sampling with the upgraded FLX technology will provide enhanced statistical support for rare transcripts.
Second, the specificity of these long, 3′-UTR-based sequence reads facilitates unambiguous gene assignment. Our analyses indicated that individual gene family members can be resolved by unique, gene-specific 3′-anchored tags, and the corresponding closely related ESTs can be characterized. Finally, enrichment of 3′-UTR sequences provides a useful source of polymorphic information for studies of natural variation. Identification and analysis of nearly identical paralogous genes is improved on a genome-wide scale by enrichment for polymorphisms in the 3′ sequences. Even in cases where genomic information is very limited, high-throughput sequencing of 3′-UTRs from species' variants allows direct comparison of polymorphic loci. This approach thus provides a tool for genotyping and assessing genetic diversity contributing to quantitative traits without the need for a sequenced genome or extensive EST collections.
Approximately 22% of the unique mRNAs identified in this study by at least two reads did not match ESTs in either ZmGI or IUC databases. A similar percentage of novel sequences (30%) were also observed for a transcript profile from maize shoot apical meristem using 454-based shotgun sequencing of sheared cDNAs (Emrich et al., 2007a). The 8% difference may reflect an increased specificity of our BLASTN results using the IUC cDNA collection and/or more novel transcripts identified in the nondifferentiated shoot apical meristem tissue. Our data also showed that among distinct mRNAs matching cDNAs, approximately 50% either contained domains of unknown function and/or were unclassified based on lack of homology to annotated genes in other species. This percentage demonstrates the potential for gene discovery with unbiased sampling and sequencing of gene-specific 3′-UTRs.
Furthermore, quantitative analyses of closely related transcripts can extend studies of functional genomics to species without completely sequenced genomes and where gene families are largely uncharacterized. We addressed this possibility with an analysis of H1-like transcripts in maize ovaries. Although the individual genes have not been characterized in maize, identification of the corresponding H1 ESTs indicated that these unique, nonredundant transcripts are indeed expressed. One highly represented H1 mRNA in maize, TC292133a, was annotated as a drought- and ABA-inducible H1 gene based on sequence similarities in tomato (Bray et al., 1999). This annotation is consistent with a function for this highly expressed H1 in drought-stressed maize.
For organisms that have limiting cDNA resources, 3′-cDNA tags will be less likely to align with upstream coding sequences, thus constraining functional annotation. Nonetheless, 3′-UTR sequences enable resolution of unique mRNAs and distinguish among closely related transcripts. Quantitative data on transcript abundance is also provided, as well as an open, unbiased sampling of the transcriptome. Where additional cDNA information is available, the 3′-cDNA sequences can be extended by BLASTN alignments. Alternatively, the sequence tags can be used to design primers or probes for screening of cDNA libraries. While the divergence of 3′-UTR sequences facilitates resolution of genes within a genome, it may limit the effectiveness of cross-species comparisons for annotation of transcripts. For example, alignment of maize ovary 3′-cDNA consensus tags to the complete set of rice genes (OsGI) using BLASTN produced matches (expectation score <1e−5) for only 20.6% of the transcripts.
Based on our analysis of SNPs identified within consensus sequences and comparisons with B73 MAGI genomic assemblies (Supplemental Table S1), we confirmed at least 89.9% of polymorphisms independently by identical cDNA matches. These data are consistent with a recent study by Barbazuk et al. (2007) in which 88% of SNPs sampled by two or more 454 reads were validated by Sanger sequencing. Removal of the unconfirmed SNPs from our analysis reduced the estimated polymorphisms in W22 relative to B73 to 43.9%. That estimate is comparable to the 44% polymorphism reported for B73 and Mo17 alleles (Vroh Bi et al., 2006). Due to incomplete coverage of the B73 genome, it is likely that some W22 consensus sequences were aligned to closely related, paralogous MAGI4 sequences rather than alleles (e.g. ARDA paralogs).
In addition, our preliminary results indicate that homopolymer base-calling errors will have a minor impact on the ability to analyze polymorphisms in maize cDNAs. Importantly, even where errors of this type occur, the consistency of base calling in reads derived from independent 454 libraries suggests that nonidentical alleles may still be distinguished if they give rise to different consensus sequences. This level of specificity in gene expression analysis is invaluable to uncovering novel variation in polyploid or paleopolyploid genomes (Osborn et al., 2003). Evidence of ancient tetraploidization in the maize genome can be observed for roughly 60% of genes in duplicated regions (Messing et al., 2004). Conservative estimates indicate that extensive amplification of tandemly duplicated genes may represent approximately one-third (35%) of maize genes (Messing et al., 2004).
Our analysis of expressed CesA gene family members demonstrates the capacity of the approach described to provide quantitative resolution of closely related transcripts. This is achieved by specificity of the 3′-UTRs for individual cDNAs. Cross hybridization of near-identical transcripts often complicates identification of individual gene family members in array-based experiments. Consistent with this, the resolution of three CesA4 transcripts, including a putative splice variant, denotes the complexity within the CesA gene family in maize. Even with the most stringent probe designs, cross hybridization with unknown family members remains a challenge in nonsequenced genomes. With unbiased sampling and sequencing, resolution of tissue and/or temporal-specific transcripts and polymorphic variants will provide functional clues in complex genomes such as maize (Ma et al., 2006). Furthermore, quantitative assessment of transcription among individual members of a gene family can facilitate analyses of functional genomics and address key questions in evolution. Studies in Arabidopsis have identified instances of functional diversification among duplicated genes either in parallel biochemical pathways (Blanc and Wolfe, 2004) or within specific developmental and metabolic processes (Schmid et al., 2005).
Results from the quantitative 3′-UTR expression profile showed that the Zipf power distribution of gene expression observed across the entire dataset overall was not conserved within the chromatin-related functional class. This group of mRNAs showed a skewed distribution of abundance due mainly to a large number of distinct, highly expressed H3 transcripts. Among these, we identified 67 mRNAs having H3 functional domains, and 39% of the consensus sequences were represented by 100 to 1,000 reads. Results may be due to transcriptional responses to the stress treatment or be specific to the reproductive tissues examined.
Validation of differences in transcript abundance for a subset of genes by real-time RT-PCR in RNA samples used for sublibrary construction supports 3′-UTR profiling as a platform for quantitative expression profiling between samples. Furthermore, construction of the 3′-cDNA libraries by this method yielded sequences with very low retrotransposon content and nominal rRNA contamination. In addition, read distribution between multiplexed samples was well balanced. Thus, a multiplexing strategy can be used to concurrently profile multiple samples for increased cost effectiveness. Incorporation of a 4-base error detecting key enables up to 64 unique combinations for individual sample recognition.
Preliminary data generated with the recently upgraded FLX 454 technology (Harkins and Jarvie, 2007) identified approximately 22,920 unique consensus sequences with a much higher depth of sequencing for 12 multiplexed samples in a single reaction (A.L. Eveland, unpublished data). The enhanced sequencing capacity of FLX will therefore provide improved statistical analyses while increasing the number of multiplexed cDNA libraries (e.g. biological replicates and treatments). In this study, a single Genome Sequencer 20 run enabled detection of unique transcripts (two or more reads) with a sensitivity of approximately one in 100,000 mRNA molecules. A similar run on the 454-FLX instrument is expected to increase sensitivity by at least 2-fold. This will be directly applicable to identifying rare transcripts and resolving complex gene families.
Also, the range of gene expression quantified by the 454-based 3′-UTR profile provides higher resolution in global transcript profiling analyses compared to array-based hybridization experiments. Accordingly, our results include detection of many rare mRNAs as well as quantification of highly abundant transcripts. In contrast, this level of resolution was not observed in initial microarray analyses of the same tissues (A.L. Eveland, unpublished data) due to threshold levels of detection and saturation. Likewise, with array-based interpretation of fold-changes, subtle variations in gene expression are often not detected but can have a significant impact on physiology. A quantitative appraisal of all expressed sequences is thus invaluable to studies of quantitative traits such as heterozygosity (Birchler et al., 2003; Stupar and Springer, 2006).
Future Prospects
With 454-based, long-read sequencing of 3′-UTRs, quantitative profiles for allele-specific inheritance patterns can be generated in the absence of a priori data on polymorphisms. Allelic variants are frequently distinguished by single-feature polymorphisms such as those that marked nearly identical paralogs in this study. Identifying allele-specific differences in gene expression and quantifying parental contributions to complex traits in F1 hybrids are key to understanding genetic mechanisms such as imprinting (Guo et al., 2003) and heterosis (Birchler et al., 2003; Springer and Stupar, 2007). In addition, strategies for expression quantitative trait loci analyses (Schadt et al., 2003; Borevitz and Chory, 2004) and genome-wide linkage studies (Cheung et al., 2005) are improved by a high-resolution, nonbiased approach to quantifying allelic imbalances in gene expression (including those resulting from imprinting or X-chromosome inactivation). Furthermore, analysis of natural variation is enhanced by recovery of haplotypes in species where genomic information is limited.
Natural variation can also be assessed with array-based probe sets generated from 3′-anchored sequence reads (Borevitz et al., 2003). For species in which comprehensive microarray platforms are not available, the 3′-UTR sequence reads can serve as blueprints for chip construction with highly specific probe sets representing an unbiased sample of expressed sequences. Alternatively, for genomes with limited EST support, this method can enhance efficiency of cDNA sequencing by prescreening libraries to eliminate redundancy. Fine mapping and marker-assisted breeding can also be facilitated by utilizing indels in the 3′-UTRs as molecular markers (Bhattramakki et al., 2002; Vroh Bi et al., 2006). In addition, anchoring the 454 sequences proximal to the 3′ ends of transcripts enables resolution of 3′-RNA processing variants. Instances of differential polyadenylated transcripts were readily detected in this study (data not shown). Currently, identification and characterization of alternate poly(A) sites is fragmentary for most genes, because the required length of 3′-UTR sequence has been largely outside the range of short-read technologies (Jongeneel et al., 2005).
CONCLUSION
By combining the specificity of 3′-UTRs with long-read, high-throughput sequencing, we are able to distinguish expression of newly identified genes and closely related transcripts on a genome-wide scale. This can also be accomplished without reference to a completely sequenced genome. The approach provides an efficient avenue for gene discovery and elucidation of variations in expression that underlie natural variation and contribute to complex genetics of heterosis and imprinting. In addition, 3′-UTR profiling advances studies of comparative and functional genomics by quantitatively resolving expression of gene families and identifying unknown gene family members.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays) plants were grown in 14-inch, 7-gallon pots under greenhouse conditions (September to November in Gainesville, FL) at 12-h-light/12-h-dark cycles. Sibling wild-type and vp1 mutant plants in a W22 inbred background were derived from a self-pollinated vp1/+ heterozygous ear. A drought-stress treatment was initiated by gradually withholding water beginning 2 weeks prior to tassel emergence. Soil was covered to restrict water loss by evaporation and pots were weighed at the end of each day to determine water loss to transpiration. Water lost to transpiration was added back. One week after ears first appeared, water was withheld completely. Ears were collected right before silk emergence from wild-type and vp1 mutant plants. Immature ovaries (with pedicels) were hand dissected from equivalent sections of each ear (base-to-mid section), weighed to 50 mg fresh weight (15 ovaries per ear), and frozen in liquid N2.
Sublibrary Preparation and Sequencing
Tissue was homogenized in TRIzol reagent (Invitrogen) using a FastPrep lysis system (Q-BIOgene). RNA was extracted using standard methods based on protocols from the University of Arizona (www.maizearray.org). Total RNA (5 μg) from wild-type and vp1 mutant ovaries was used for cDNA synthesis (MessageAmp II, Ambion) and primed with 6 pmol biotinylated (T12) B-adaptor (modified from Margulies et al., 2005) oligo: Biotin, CCTATCCCCTGTGTGCCTTGCCTATCCCTGTTGCGTGTCTCAGTTTTTTTTTTTT[AGC]. Purified cDNA (DNA clear, Ambion) was bound to M-270 Streptavidin beads (Dynal), immobilized on a Magnabot 96 (Promega), and digested with MspI (Promega) to create 2-base CG overhangs for adaptor ligation. A-adaptor oligos (modified from Margulies et al., 2005) included 3-base multiplex keys (wild-type sublibrary top strand, 5′-CCATCTCATCCCTGCGTGTCCCATCTGTTCCCTCCCTGTCTCAGCAT-3′; wild-type sublibrary bottom strand, 5′-CGATGCTGAGACAGGGAGGGAACAGATGGGACACGCAGGGATGA-3′; vp1 mutant sublibrary top strand, 5′-CCATCTCATCCCTGCGTGTCCCATCTGTTCCCTCCCTGTCTCAGACT-3′; vp1 mutant sublibrary bottom strand, 5′-CGAGTCTGAGACAGGGAGGGAACAGATGGGACACGCAGGGATGA-3′).
Adapter pairs were combined and concentrated to 1 pmol/μL in salt buffer (10 mm Tris, 1 mm EDTA, 50 mm NaCl [pH 8]) and annealed by incremental, −1 degree/min decreases (95°C–4°C, with a 30-min hold at 72°C–71°C). Adaptors (5 pmol) with multiplex keys CAT and AGT were ligated to digested wild-type and mutant cDNA samples, respectively. The 3-base key sequences enabled detection of single-base errors in the multiplex key. Unligated adaptors were removed by washing beads twice with 1× B & W buffer (2.5 mm Tris-HCL, pH 7.5, 0.25 mm EDTA, 0.5 m NaCl) and twice with distilled, deionized water. The desired 5′-A-cDNA-B-3′ template strand was eluted with 100 mm NaOH, neutralized, and concentrated on a Qiagen column (Margulies et al., 2005). Sequencing was conducted as per Margulies et al. (2005) using a 454 GS-20 instrument.
The expected yield of approximately 3 × 109 template molecules for the combined libraries was confirmed by a SYBR Green Q-PCR strategy (MyiQ, Bio-Rad). Molecules per microliter of amplified product were calculated from an in vitro transcribed (MAXIscript, Ambion) α-tubulin (maize) standard: α-tubulin forward, 5′-TTGTGCCTGGTGGCGACCTGG-3′ and α-tubulin reverse, 5′-ACCGACCTCCTCGTAGTCCT-3′.
Data Analysis
Quality-trimmed 454 sequences (FASTA format) were filtered for valid key and ligation junction (CGG) sequences at 5′ ends, and poly-A tails were trimmed using custom programs written in Java. Validated, trimmed sequences (93% of total reads) were assembled using CAP3 (http://genome.cs.mtu.edu/sas.html). The nonredundant set of consensus cDNA sequences represented by two or more reads (14,822 total assemblies) were annotated by BLASTN searches of cDNA databases for maize. These included ZmGI and IUC, a collection of cDNAs provided by an industry consortium via a user's agreement (http://www.maizeseq.org). Functional classifications of cDNA matches were based on Gene Ontology terms associated with PFam (http://www.sanger.ac.uk/Software/Pfam/) assignments in IUC. In addition, consensus sequences were aligned by BLASTN to MAGI (version 4.0 [http://magi.plantgenomics.iastate.edu/]).
All 3′-consensus sequence tags were deposited into dbEST (NCBI).
Real-Time PCR Analysis for Validation of 454 Data
Real-time RT-PCR was carried out to validate technical replicates of RNA samples used in sublibrary construction. For real-time PCR analysis, cDNA was synthesized from DNaseI-treated (Ambion) total RNA using an oligo(dT) primer (TaqMan Reverse Transcription Reagents, ABI). Real-time PCR was monitored using the MyiQ Single Color Real-Time PCR Detection system (Bio-Rad). Each reaction contained 10 μL of 2× iQ SYBR Green Supermix (Bio-Rad), 1.0 μL of cDNA sample, and 200 nm gene-specific primer in a final volume of 20 μL. All reactions were performed in triplicate. The relative abundance of transcripts was normalized with 18S rRNA control values using Taqman (Ribosomal RNA Control Reagents, ABI) and to the constitutive expression of an α-tubulin mRNA using SYBR Green on cDNA templates (MyiQ, Bio-Rad). SYBR Green was used to amplify a subset of transcripts with gene-specific primers. Primer pairs were designed using the 454-read and adjacent sequences in best-match ESTs identified by BLASTN as templates.
CBS domain chloride channel ([2562879] CBS forward, 5′-ATGGATGCTGCTGTTCTCATGCTC-3′ and CBS reverse, 5′-ATGGAGTCTCCTGGCGTGCTAC-3′), thaumatin/osmotin ([1321765] Osmotin forward, 5′-TACCGCAGCAGCTGAACAACG-3′ and Osmotin reverse, 5′-ATGTTCCGTCGCAGTCGCTAGG-3′), senescence-associated/tetraspannin ([TC299489] Sa forward, 5′-AACGACGAGGACGACCTCTGC-3′ and Sa reverse, 5′-AGTTTGATTAAGCGTCACCGCCTCG-3′), chlorophyll a/b-binding protein ([TC299127] Cab forward, 5′-TGTACCCTGGCGGCAGCTTC-3′ and Cab reverse, 5′-ATCCACGTACGTACACCCTCTCC-3′), copper transport ATPase/heavy-metal associated ([2562278] Hma forward, 5′-AGCCAAAGCTGACGCCTGATC-3′ and Hma reverse, 5′-TCCTGCAAGGGATGTGTTGTTC-3′), Gly-rich protein ([2923887] Grp forward, 5′-ATCAGGTGAAGGATACGGACAAGGTG-3′and Grp reverse, 5′-ACAGGACAAATTACAAGCCTTGCGGTG-3′), dehydrin DHN1/RAB-17 ([TC286791] dehydrin forward, 5′-ACAGCACTGAGCGGCGCCTATAC-3′ and dehydrin reverse, 5′-ACGTAGCAGCATAAACAGTACACGGACC-3′). Relative expression levels of ARDA1 and ARDA2 were compared by real-time RT-PCR using SYBR Green and gene-specific primers within and around the 18-bp indel sequence (Arda1 forward, 5′-TACAAGCGGGCGCAGTCG-3′, Arda1 reverse, 5′-AGCAAACATGGCCTCTTCACTG-3′; Arda2 forward, 5′-TACAAGCGGGCGCAGTCG-3′, Arda2 reverse 5′-TGGCCTGACAGAGACACCG-3′).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EY950428 through EY965249.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of full-length CesA ESTs and corresponding gene-specific 3′-anchored consensus sequences.
Supplemental Figure S2. Confirmation of SNPs (including homopolymer-based polymorphisms) in W22 3′-anchored sequence reads.
Supplemental Table S1. Distribution of consensus sequences by read number.
Supplemental Table S2. Sequences for gene-specific 3′-anchored 454 reads that resolved individual CesA gene family members.
Supplemental Table S3. Validated polymorphisms in W22 454 consensus sequences compared with B73 genomic sequence.
Supplementary Material
Acknowledgments
We thank William Farmerie and Regina Shaw of the University of Florida Interdisciplinary Center for Biotechnology Research sequencing core for assistance with sequencing. We thank Susan P. Latshaw (Department of Horticultural Sciences, University of Florida) for adaptor oligo design and Wayne T. Avigne (Department of Horticultural Sciences, University of Florida) for lab and greenhouse support.
This work was supported by the National Science Foundation (grant nos. NSF–PGRP–0217552, NSF–PGRP–0077676, and NSF–SGER–0542665).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Karen E. Koch (kekoch@ufl.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Appenzeller L, Doblin M, Barreiro R, Wang H, Niu X, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11 287–299 [Google Scholar]
- Bao JY, Lee S, Chen C, Zhang XQ, Zhang Y, Liu SQ, Clark T, Wang J, Cao ML, Yang HM, et al (2005) Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol 138 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS (2007) SNP discovery via 454 transcriptome sequencing. Plant J 51 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129 823–837 [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Sabar M, Quick WP, Sweetlove LJ (2005) Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J Exp Bot 56 1699–1709 [DOI] [PubMed] [Google Scholar]
- Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC III, Tingey SV, Rafalski A (2002) Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol 48 539–547 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Augar DL, Riddle NC (2003) In search of the molecular basis of heterosis. Plant Cell 15 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Chory J (2004) Genomics tools for QTL analysis and gene discovery. Curr Opin Plant Biol 7 132–136 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA, Shih TY, Moses MS, Cohen A, Imai R, Plant AL (1999) Water-deficit induction of a tomato H1 histone requires abscisic acid. Plant Growth Regul 29 35–46 [Google Scholar]
- Cao X, Costa LM, Biderre-Petit C, Kbhaya B, Dey N, Perez P, McCarty DR, Gutierrez-Marcos JF, Becraft PW (2007) Abscisic acid and stress signals induce Viviparous-1 (Vp1) expression in seed and vegetative tissues of maize. Plant Physiol 143 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Lee S, Zhou G, Rowley JD, Wang SM (2002) Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc Natl Acad Sci USA 99 12257–12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT (2005) Mapping determinants of human gene expression by regional and genome-wide association. Nature 437 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD (2002) Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA 99 13606–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorm JN, Altshuler D, Lander ES (2002) Detection of regulatory variation in mouse genes. Nat Genet 32 432–437 [DOI] [PubMed] [Google Scholar]
- Emrich SJ, Barbazuk B, Schnable PS (2007. a) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res 17 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich SJ, Li L, Wen TJ, Yandeau-Nelson MD, Fu Y, Guo L, Chou HH, Aluru S, Ashlock DA, Schnable PS (2007. b) Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102 12282–12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa C, Kaneko K (2003) Zipf's law in gene expression. Phys Rev Lett 90 088102. [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li WH (2004) Duplicate genes increase gene expression diversity within and between species. Nat Genet 36 577–578 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Danilevskaya ON, Yang X, Hu Z (2003) Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J 36 30–44 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS (2004) Allelic variation of gene expression in maize hybrids. Plant Cell 16 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins T, Jarvie T (2007) Megogenomics analysis using the genome sequencer FLX system. Nat Methods 4 application notes iii–v
- Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenchild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJG, et al (2005) An atlas of human gene expression from massively parallel signature sequences (MPSS). Genome Res 15 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov VA, Knott GD, Bonner RF (2002) General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenchild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569 [DOI] [PubMed] [Google Scholar]
- Ma J, Morrow DJ, Fernandes J, Walbot V (2006) Comparative profiling of sense and antisense transcriptome of maize lines. Genome Biol 7 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905 [DOI] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KFX, et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Dooner HK (2006) Organization and variability of the maize genome. Curr Opin Plant Biol 9 157–163 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S (2004) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Ecker JR (2005) Applications of DNA tiling arrays for whole-genome analysis. Genomics 85 1–15 [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8 122–128 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al (2007) An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25 473–477 [DOI] [PubMed] [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS, Comai L, Madlung A, Doerge RW, Colot V, et al (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19 141–147 [DOI] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE (2002) Using the transcriptome to annotate the genome. Nat Biotechnol 20 508–512 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Collnayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al (2003) Genetics of gene expression surveyed in maize, mouse, and man. Nature 422 297–302 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17 264–275 [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B (1998) Dormancy-associated gene expression in pea axillary buds. Planta 205 547–552 [DOI] [PubMed] [Google Scholar]
- Street NR, Skogstrom O, Sjodin A, Ticker J, Rodriguez-Acosta M, Nilsson P, Jansson S, Taylor G (2006) The genetics and genomics of drought response in Populus. Plant J 48 321–341 [DOI] [PubMed] [Google Scholar]
- Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Joa Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroh Bi I, McMullen MD, Sanchez-Villeda H, Schroeder S, Gardiner J, Polacco M, Soderlund C, Wing R, Fang Z, Coe EH Jr (2006) Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46 12–21 [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol 144 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308 1310–1314 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846 [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297 1143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.