Abstract
Volatile sulfur compounds are key flavor compounds in several cheese types. To better understand the metabolism of sulfur-containing amino acids, which certainly plays a key role in the release of volatile sulfur compounds, we searched the genome database of Lactobacillus casei ATCC 334 for genes encoding putative homologs of enzymes known to degrade cysteine, cystathionine, and methionine. The search revealed that L. casei possesses two genes that putatively encode a cystathionine β-lyase (CBL; EC 4.4.1.8). The enzyme has been implicated in the degradation of not only cystathionine but also cysteine and methionine. Recombinant CBL proteins catalyzed the degradation of l-cystathionine, O-succinyl-l-homoserine, l-cysteine, l-serine, and l-methionine to form α-keto acid, hydrogen sulfide, or methanethiol. The two enzymes showed notable differences in substrate specificity and pH optimum.
Volatile sulfur compounds (VSC) are found in many cheese varieties and are important components of flavor (10). The sulfur compounds methanethiol, methional, and dimethyl trisulfide (DMTS) are key odorants of Swiss Gruyère cheese flavor and are presumably derived from the bacterial metabolism of methionine, which is present in caseins at higher concentrations than cysteine. Swiss Gruyère (place of designated origin) cheese is produced with thermophilic mixed-strain starters which usually contain Streptococcus thermophilus and Lactobacillus delbrueckii subsp. lactis. In addition, depending on the cheese factory, Lactobacillus helveticus can also be present. However, after ripening, Lactobacillus casei and Lactobacillus rhamnosus bacteria are found in high numbers (8). Interestingly, L. casei is also a constituent of the nonstarter lactic acid bacteria of many other cheese varieties (6). Therefore, this species is considered to play an important role during ripening and may be a key player in the formation of flavor compounds.
The catabolic pathway(s) of methionine and cysteine in lactobacilli is not well characterized. Possible pathways involve transaminases, methionine γ-lyases, cystathionine β-lyases (CBL), and cystathionine γ-lyases (CGL) (13). The transamination of methionine by aminotransferases has been studied in lactococci (18, 27, 34). The results of these studies suggested that methionine is converted to 2-oxo-4-methylthiobutyric acid, which is then converted, either enzymatically or chemically, to methanethiol. In Brevibacterium linens, a methionine γ-lyases which catalyzes the deamination of methionine and releases methanethiol has been identified and the corresponding gene has been cloned (2, 14).
CBL, which is encoded by the gene metC in Escherichia coli, is involved in the α,β-elimination of cystathionine to form homocysteine, pyruvate, and ammonia. In comparison, CGL catalyzes the α,γ-elimination reaction of cystathionine, producing cysteine, ammonia, and α-ketobutyrate. CBL and CGL generally show catalytic promiscuity, which means that they catalyze different reactions. Thus, the CBL from Lactococcus lactis subsp. cremoris and L. helveticus have been shown to catalyze α,β- and α,γ-elimination reactions and degrade methionine to methanethiol (1, 17, 21). A CGL has been identified in Lactobacillus fermentum, Lactobacillus reuteri, and L. lactis subsp. cremoris SK11. The purified enzymes degraded methionine to methanethiol and also broke down cysteine, showing that they catalyze α,β-elimination reactions as well (7, 11, 29). CBL and CGL are members of the γ subfamily within the α family of pyridoxal-5′-phosphate-dependent enzymes (24). The malY gene product belongs to a different subfamily within this family but has also been shown to possess cystathionine lyase activity and degrades cysteine and cystathionine in vitro (35). The same activities were found for PatC (L. delbrueckii subsp. bulgaricus), PatB (Bacillus subtilis), Lcd (Streptococcus anginosus), and YtjE (L. lactis), which show similarity to MalY (4, 5, 23, 33). Interestingly, overexpression of malY, patB, and patC in CBL-deficient mutants abolished the methionine requirement, showing that they encode CBL activity in vivo.
Recently, we observed that several L. casei strains isolated from Swiss Gruyère cheese produced VSC in the absence of an amino acceptor (20). Based on this finding, we concluded that the formation of VSC does not necessitate the action of aminotransferases and might be related to lyases. In this study, we studied the ability of L. casei to degrade cystathionine. Furthermore, we cloned two L. casei genes whose product showed homology to CBL and MalY of E. coli and studied the substrate specificity of the gene products.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. casei strains from the Agroscope Liebefeld-Posieux collection were used in this study. They were cultured anaerobically in 10 ml of MRS (12) at 30°C.
Methionine and cysteine auxotrophy.
Strains of interest were assayed for growth in a synthetic medium described by Morishita et al. (25). The medium contained l-methionine (0.1 mg ml−1), l-cysteine (0.1 mg ml−1), both l-methionine and l-cysteine, or neither one. Ten-milliliter volumes of the medium were inoculated with 50-μl cultures grown in MRS. The optical density at 600 nm was determined after 3 days of incubation at 30°C.
Preparation of cell extracts.
Strains were grown for 16 h in 50 ml MRS at 30°C. Bacterial cells were harvested by centrifugation, washed twice with 20 mM sodium phosphate (pH 7.4), and suspended in 500 μl of 20 mM sodium phosphate (pH 7.4). Approximately 0.4 g of glass beads (212 to 300 μm) was added. The cells were then disrupted by violent agitation in a Mini-Beadbeater-8 (Biospec Products, Inc.). After the extract had been cleared by centrifugation, metabolites were removed by using NAP columns (GE HealthCare, Uppsala, Sweden) which had been equilibrated with 20 mM sodium phosphate (pH 7.4) according to the manufacturer's instructions.
Protein determination.
Protein concentrations were determined by Bio-Rad protein assay according to the manufacturer's instructions. Bovine serum albumin was used to obtain a standard curve.
Cystathionine lyase assay.
Cystathionine lyase activity was assayed by measuring the formation of free thiol groups by spontaneous disulfide interchange with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) similar to the method of Uren (32). The reaction buffer contained a 200-μl solution of 50 mM buffer, 2 mM l-cystathionine, 200 μM DTNB, 20 μM pyridoxal-5′-phosphate, and enzyme solution. Depending on the pH, 2-morpholinoethanesulfonic acid, sodium phosphate, potassium phosphate, or Tris was used as a buffer. The reaction was initiated by the addition of enzyme and carried out at 37°C. The yellow coloration that developed as a consequence of enzymatic activity was recorded at 10-min intervals at 412 nm for 90 min. For high-performance liquid chromatography (HPLC) analysis, assays were performed without DTNB and stopped after 1 h of incubation at 37°C by the addition of 2 μl of 380 mM H2SO4 and by heating the mixture at 95°C for 5 min.
HPLC analysis.
Samples were filtered through disposable filter holders (0.45-μm pore size). Separation of the filtrate was carried out on a ion-exchange column (300 by 7.8 mm; HPX-87H Aminex; Bio-Rad) protected with a cation H+ Microguard cartridge. The operating conditions were as follows: a flow rate of 0.6 ml min−1, 65°C, and detection at 210 nm. The mobile phase was 3.8 mM H2SO4. Pyruvate and α-ketobutyrate (both from Sigma-Aldrich) were used as standards.
DNA techniques.
Plasmid preparations, transformation, use of restriction enzymes, and ligation were carried out as described by Sambrook et al. (28) The putative malY and metC genes were amplified from the genomic DNAs of L. casei FAM18149 and FAM18168 by PCR. The primers used for the malY gene were 5′-CGGCAGCGCTAGCATGGAAGAATTGCCAGCTGATG-3′ and 5′-CCTAAGCTTAAAGTTGCGGCTGAGCCAAGT, and those used for the metC gene were 5′-CGGCAGCGCTAGCATGACCCAATTCAATACCAAACTAG-3′ and CCTAAGCTTGTCTTTATTTCTCCGCAGCCGTCAA-3′. Restriction sites are underlined. The PCR products were digested with NheI and HindIII. The pET SUMO vector (Invitrogen) was also digested with NheI and HindIII. Thus, the region encoding the SUMO fusion protein was removed from the plasmid and the PCR fragments could be cloned in the frame directly downstream of the polyhistidine region.
Sequencing was performed with the BigDye Terminator cycle sequencing kit (Applied Biosystems) and analyzed in an ABI Prism 310 genetic analyzer (Applied Biosystems).
Expression and purification of His-tagged proteins.
Recombinant E. coli BL21 was grown in 100 ml of Luria-Bertani broth containing kanamycin at a final concentration of 50 μg ml−1 at 37°C on a shaker. When the optical density of the bacterial cells at 600 nm reached 0.5, induction of genes under the control of isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoters was performed by adding 0.5 mM IPTG and lowering the incubation temperature to 27°C. After 4 h of incubation on a shaker, the bacterial cells were harvested by centrifugation, washed twice with 20 mM sodium phosphate (pH 7.4), and frozen at −20°C.
Cells were suspended in 500 μl binding buffer (20 mM sodium phosphate [pH 7.4], 150 mM NaCl, 20 mM imidazole) and disrupted as described above. The extract was cleared by centrifugation and then applied to a 1-ml HiTrap chelating HP column (GE HealthCare, Uppsala, Sweden) which had been loaded with Ni2+ and equilibrated with binding buffer. After the column had been washed with 8 ml binding buffer, bound proteins were eluted with 20 mM sodium phosphate (pH 7.4)-500 mM NaCl-250 mM imidazole. Afterwards, the imidazole was removed by applying the eluate to NAP columns (GE HealthCare, Uppsala, Sweden) which had been equilibrated with 20 mM sodium phosphate (pH 7.4) and 150 mM NaCl. The eluted protein fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were visualized by colloidal Coomassie staining.
Detection of VSC by gas chromatography (GC)-pulsed flame photometric detection (PFPD).
The enzymatic assays were performed at a 1-ml scale as described in the cystathionine lyase assay section except that, instead of l-cystathionine, either 10 mM l-methionine or 10 mM l-cysteine was used as the substrate and incubated for 6 h.
VSC were detected as described previously (26). Due to the use of a different matrix, the following modifications were applied. The samples (1 ml) were cooled at 4°C and then transferred to a 10-ml vial with a magnetic screw cap, which had been flushed with argon. After preheating the sample (45 min, 60°C), 1 ml of the headspace was injected into the cooled injection system (CIS-4; Gerstel GmbH). The cooled injection system inlet was packed with deactivated wool (Restek GmbH) and set at −100°C. The temperature was programmed to 180°C at a rate of 12°C/s (hold time, 8 min). The temperature was held at 40°C for 1 min before increasing again. Transfer to the capillary column was performed in split mode (10:1). GC was conducted with an SPB-1 sulfur capillary column (30 m by 0.32 mm [inside diameter], 4-μm film thickness; Supelco) with helium as the carrier gas. The oven temperature was programmed from 29°C (hold time, 7 min) to 180°C at a rate of 10°C min−1 (hold time, 10.5 min). Detection was performed with a PFPD model 5380 (Ol Analytical).
Nucleotide sequence accession numbers.
The nucleotide and amino acid sequences reported in this paper appear under NCBI accession numbers EF531299 and EF531300.
RESULTS
Cystathionine lyase activity in L. casei.
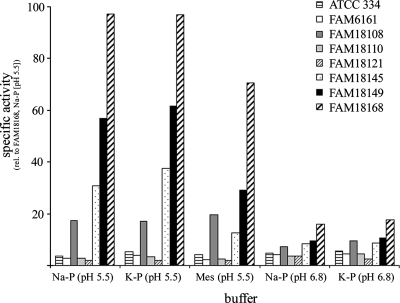
We assayed the cell lysates of 39 L. casei genotypes for cystathionine lyase activity with different buffers and different pHs. First, DTNB was used as a reagent to detect the formation of thiols during the incubation of cell extracts with l-cystathionine. We found that cystathionine lyase activity was dependent on the strain, pH, and buffer used. Only the extracts of 23 genotypes released thiols which reacted with DTNB. Considerable cystathionine lyase activity was only observed at pH 5.5. In 2-morpholinoethanesulfonic acid buffer, the activity of some genotypes was slightly reduced compared to that in phosphate buffer. At pH 6.8, the activity was greatly reduced (Fig. 1), and no activity was detected in Tris (pH 9.0) buffer (data not shown).
FIG. 1.
Cystathionine lyase activities from different L. casei genotypes measured by DTNB reaction with thiols released from l-cystathionine. Since FAM18168 was one of the strains showing the highest activity, it was always included as a reference in the assays. The mean values of three independently performed assays are shown. rel., relative; Na-P, sodium phosphate; K-P, potassium phosphate; Mes, 2-morpholinoethanesulfonic acid.
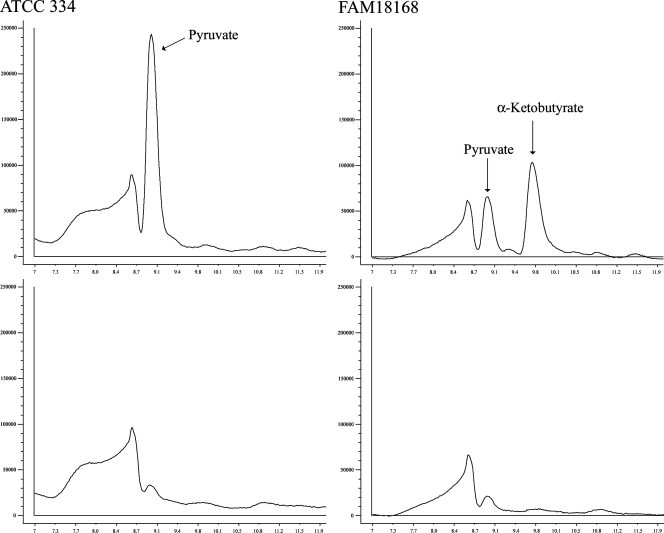
In order to determine whether an α,β- or an α,γ-elimination took place, enzyme reactions were performed in the absence of DTNB and then analyzed by HPLC for the presence of α-keto acids. The analysis showed that the genotypes with cystathionine lyase activity in the assay with DTNB released pyruvate and α-ketobutyrate from cystathionine (Fig. 2). Interestingly, among the apparently inactive genotypes, strains were identified which released pyruvate but not α-ketobutyrate from cystathionine. Reactions carried out at pH 6.8 contained considerably less pyruvate or α-ketobutyrate than those carried out at pH 5.5 (data not shown). Thus, the strains could be classified into three groups, (i) strains with no detectable activity, (ii) strains with low activity producing detectable amounts of pyruvate, and (iii) strains with high activity producing pyruvate and α-ketobutyrate (Table 1). From this observation, we concluded that at least two enzymes are present in L. casei which catalyze the degradation of l-cystathionine.
FIG. 2.
HPLC analysis of cystathionine lyase assays performed at pH 5.5 with cell extracts of L. casei ATCC 334 (left panel) and FAM18168 (right panel) genotypes. The upper panel illustrates reaction products obtained with l-cystathionine, and the lower panel shows the control (assay performed without l-cystathionine). Peaks were identified by retention time.
TABLE 1.
Growth of several L. casei genotypes in synthetic medium with or without methionine and cysteinea
| Strain | Mb | Cc | MCd | HPLC analysise |
|---|---|---|---|---|
| ATCC 334 | − | + | + | Pyr |
| FAM6161 | − | ++ | ++ | |
| FAM8407 | ++ | ++ | ++ | Pyr + AKB |
| FAM17407 | − | ++ | ++ | |
| FAM18108 | ++ | ++ | ++ | Pyr + AKB |
| FAM18110 | − | ++ | ++ | |
| FAM18121 | − | ++ | ++ | Pyr |
| FAM18124 | ++ | ++ | ++ | Pyr + AKB |
| FAM18129 | ++ | ++ | ++ | Pyr + AKB |
| FAM18145 | ++ | ++ | ++ | Pyr + AKB |
| FAM18149 | + | + | + | Pyr + AKB |
| FAM18168 | ++ | + | ++ | Pyr + AKB |
The amount of growth relative to growth in the medium without methionine and cysteine is indicated as follows: −, no growth; +, detectable growth; ++, strong growth.
M, medium contained l-methionine but no l-cysteine.
C, medium contained l-cysteine but no methionine.
MC, medium contained l-methionine and l-cysteine.
The α-keto acids detected by HPLC after incubation of cell-free extract with l-cystathionine for 1 h are presented. Pyr, pyruvate; AKB, α-ketobutyrate.
Methionine auxotrophy.
All of the lactobacilli tested were able to grow in the absence of methionine, but several genotypes showed no growth in the absence of cysteine (Table 1). All strains that were able to grow on methionine as the sole source of sulfur degraded cystathionine to pyruvate and α-ketobutyrate.
Cloning and overexpression of two genes encoding cystathionine lyases.
Two enzymes in E. coli are known to cleave cystathionine by α,β-elimination (16, 35). The proteins are encoded by the genes metC and malY, respectively. We used both amino acid sequences (Swissprot accession no. P06721 and P23256) to search for homologs in the draft genome sequence of L. casei ATCC 334 (genome.jgi-psf.org/mic_home.html). The MetC sequence showed homology to the product of gene 3332 of scaffold 58; MalY was shown to be homologous to the product of gene 1334 of scaffold 16. After the genome sequence had been completed and moved to the NCBI database, it was confirmed that L. casei ATCC 334 contained genes homologous to malY (LSEI_0894) and metC (LSEI_0600) (22).
We cloned both genes from L. casei FAM18149 and FAM18168. The amino acid sequence deduced from LSEI_0894 of FAM18168 was identical to that of FAM18149 and showed 99% identity with the sequence of ATCC 334. Furthermore, the amino acid sequences from LSEI_0600 of FAM18168 and FAM18149 were identical and were 99% identical to that of ATCC 334. A search for protein homology revealed the similarity of LSEI_0894 to PatB (34%) of B. subtilis, PatC (33%) of L. delbrueckii, Lcd (31%) of S. anginosus, YtjE (31%) of L. lactis, and MalY (29%) of E. coli. LSEI_0600 showed similarity to MetC of L. lactis (46%) and L. helveticus (44%), methionine γ-lyase (35%) of B. linens, and MetB (33%) and MetC (25%) of E. coli. For clarity, the gene names malY for LSEI_0894 and metC for LSEI_0600 are used in this report.
In order to produce large amounts of protein, the genes were cloned in frame with a His6 tag coding sequence (pET-SUMO). The region encoding the SUMO protein was omitted since we considered that a large, prolonged N terminus would impair the quaternary structure of the enzymes. Due to the construct, the N terminus of each protein was nevertheless prolonged by 21 amino acid residues (MGSSHHHHHHGSGLVPRGSAS). After purification by affinity chromatography, the apparent molecular mass of the His-tagged MalY protein was 47 kDa (theoretical molecular mass of 46.9 kDa) and that of MetC was 43 kDa (theoretically, 42.7 kDa).
Cystathionine lyase activity was determined at different pHs. We found that the MalY protein showed the highest activity at pH 6.0, whereas the MetC protein had an optimum pH of 9.0. At pH 5.5, a pH usually found in 4-month-old Gruyère cheese, the residual activities of MalY and MetC were 55 and 15%, respectively. To ensure that a cystathionine lyase of E. coli was not copurified by affinity chromatography, the SUMO-chloramphenicol acetyltransferase fusion protein encoded by the unmodified plasmid was isolated under the same conditions. No cystathionine lyase activity was detected in the purified SUMO-chloramphenicol acetyltransferase preparation (data not shown).
After incubation at 56°C for 30 min, the enzyme preparations were no longer active. After several weeks of storage at 4°C or −20°C, both enzyme preparations still showed considerable cystathionine lyase activity. The addition of 4% NaCl completely inhibited the activity of MetC but not that of MalY.
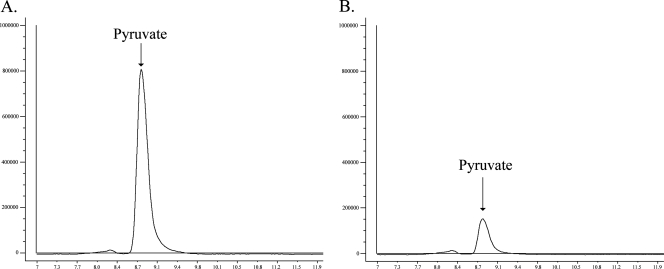
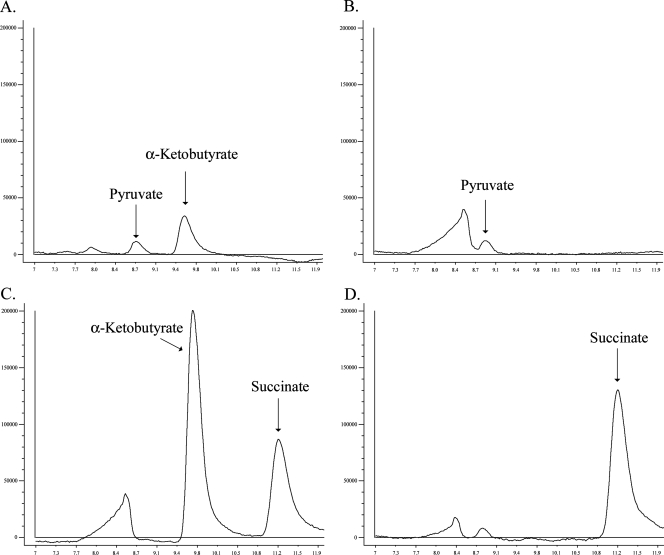
The formation of α-keto acids from l-cystathionine was determined by HPLC (Fig. 3 and 4). The results showed that the product of the malY gene degraded cystathionine to pyruvate, whereas the product of the metC gene released pyruvate and α-ketobutyrate. At pH 9.0, MetC produced more α-ketobutyrate than pyruvate.
FIG. 3.
HPLC analysis of assays carried out at pH 5.5 with purified MalY protein (6 μg) and 2 mM l-cystathionine (A) and 2 mM l-cysteine (B) for 1 h. Peaks were identified by retention time.
FIG. 4.
HPLC analysis of assays carried out with purified MetC (9 μg) and 2 mM l-cystathionine at pH 9.0 for 1 h (A), 2 mM l-cysteine at pH 6.8 for 1 h (B), 2 mM O-succinyl-l-homoserine at pH 6.8 for 1 h (C), and 2 mM O-succinyl-l-homoserine and 2 mM l-cysteine at pH 6.8 for 1 h (D). Peaks were identified by retention time.
To determine substrate specificity, assay mixtures containing l-cysteine, l-methionine, dl-homocysteine, l-serine, O-succinyl-l-homoserine, l-leucine, l-isoleucine, l-valine, l-phenylalanine, l-aspartate, l-glutamate, and l-lysine were prepared. It was found that MalY formed pyruvate from l-cysteine (Fig. 3B) and l-serine at pH 5.5 and pH 6.8. The latter reaction was clearly observed after a prolonged incubation time (data not shown). Reaction vials containing cysteine as a substrate emitted a strong hydrogen sulfide odor after incubation.
The MetC protein formed α-ketobutyrate and succinate from O-succinyl-l-homoserine (Fig. 4C) at pH 5.5, pH 6.8, and pH 9.0. After incubating O-succinyl-l-homoserine and l-cysteine together with MetC, the formation of α-ketobutyrate was no longer observed (Fig. 4D). This indicated that the α-ketobutyrate moiety was used for the synthesis of l-cystathionine. Reaction vials containing MetC and methionine had a cabbage-like odor after a prolonged incubation, which indicated the release of methanethiol. However, the formation of α-ketobutyrate was only detected by HPLC analysis after 20 h of incubation at pH 9.0 but not at pH 6.8. When DTNB was added, the solution turned yellow, indicating the presence of a thiol-containing group. At pH 9.0, a more intense color formation was observed than at pH 6.8. Control experiments with methionine alone or with MalY and methionine did not turn yellow (data not shown). We concluded that MetC was able to liberate methanethiol from methionine.
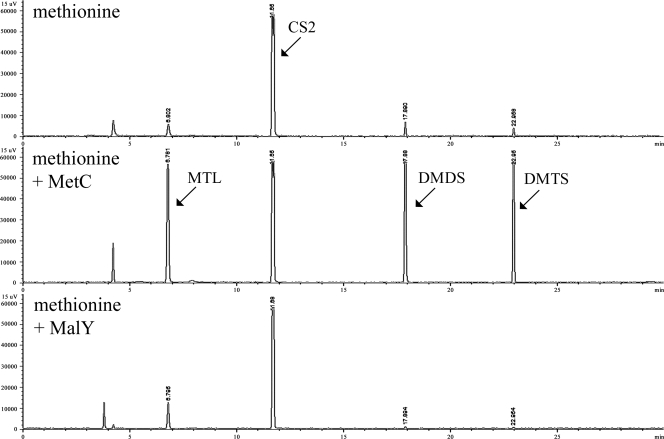
In order to confirm the release of methanethiol from methionine, we used GC coupled to flame photometric detection, which is selective for phosphorus and sulfur. The headspace of an enzyme assay carried out with methionine in the absence of enzyme contained very small amounts of methanethiol, dimethyl disulfide, and DMTS. Whereas the addition of the MalY protein had no effect on the content of VSC, the MetC protein clearly released methanethiol, dimethyl disulfide, and DMTS from methionine (Fig. 5). Furthermore, it was confirmed that MalY released hydrogen sulfide from l-cysteine, as mentioned above (data not shown).
FIG. 5.
Headspace analysis by GC-PFPD of enzymatic assays carried out with methionine at pH 6.8. Fifty micrograms of purified protein was used for the enzymatic reaction. CS2, carbon disulfide; MTL, methanethiol; DMDS, dimethyl disulfide; DMTS, dimethyl trisulfide.
DISCUSSION
In Gruyère and other cheese types, L. casei is the predominant species found after ripening. Therefore, L. casei strains are promising adjuncts and an understanding of sulfur metabolism and the formation of VSC may contribute to the development of cultures with optimized aromatic properties. It is quite well established that the catabolism of most amino acids is initiated by a transamination step in lactic acid bacteria (3, 30). Recently, we found that several L. casei strains degraded methionine to VSC in the absence of amino acceptors such as α-ketoglutaric acid (20), showing that amino acid degradation is not always initiated by transamination.
The availability of genome sequences makes it possible to predict metabolic pathways. However, two items have to be taken into consideration. Firstly, the high genetic variability found in lactic acid bacteria makes it difficult to deduce the pathways of several genotypes from only one genome sequence. Secondly, genes encoding enzymatic activities are identified by homology, which does not always explain their function. Thus, for example, the substrate specificity of enzymes with broad substrate specificity such as cystathionine lyases has to be investigated by biochemical methods.
In this report, we show that L. casei possesses two genes which encode cystathionine lyases and are therefore implicated in the metabolism of sulfur-containing amino acids. On the one hand, the gene product of malY degraded cystathionine to pyruvate and cysteine to pyruvate and hydrogen sulfide, showing that it catalyzes an α,β-elimination reaction. On the other hand, the product of the metC gene released pyruvate and α-ketobutyrate from cystathionine, formed pyruvate from cysteine, and produced methanethiol from methionine. The concomitant formation of dimethyl disulfide and DMTS with methanethiol is probably due to the oxidation of the latter compound (9). Taking the results together, we concluded that MetC catalyzes both an α,β- and an α,γ-elimination reaction. This property has also been reported for the CBL of L. lactis (1, 15).
Furthermore, we found that the MetC protein of L. casei released succinate and α-ketobutyrate from O-succinyl-l-homoserine, which again showed that the enzyme catalyzes an α,γ-elimination. O-Succinyl-l-homoserine is the precursor for cystathionine biosynthesis and on the addition of cysteine, only the production of succinate was observed. This can be explained by the replacement of the succinyl group of O-succinyl-l-homoserine by cysteine. The synthesis of cystathionine could be confirmed by mass spectrometry analysis (data not shown). This reaction is actually catalyzed by cystathionine γ-synthase. Since MetC mainly showed cystathionine lyase activity at a high nonphysiological pH, we think that it mainly functions as a cystathionine γ-synthase within the cell and suggest that the gene be designated metB instead of metC. It may be not unusual for a cystathionine γ-synthase to show lyase activity. Thus, for example, Salmonella cystathionine γ-synthase also degraded cystathionine at high pH (19).
The malY gene of E. coli was initially identified as a regulator of the maltose operon. So it is possible that the malY gene from L. casei also encodes a function as a regulator of gene expression. It has also been shown that overexpression of malY in E. coli abolished the methionine auxotrophy of a metC mutant, demonstrating a possible role in methionine biosynthesis (35). Taking into account the in vitro data presented in this report and the poor homology with MalY, it is possible that MalY from L. casei is involved in methionine biosynthesis. However, only the disruption of the gene in L. casei or its expression in a malY metC double mutant which is unable to grow in the absence of methionine could confirm this hypothesis.
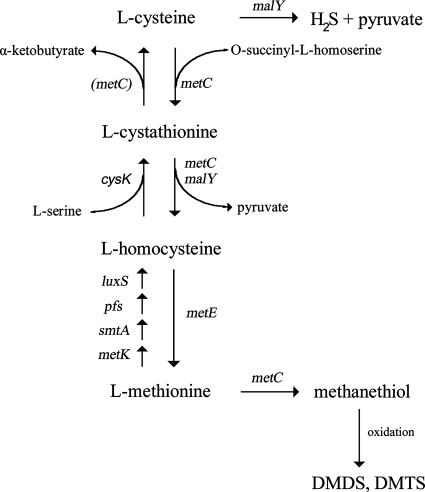
Taking all of the results together, we have put forward the pathways illustrated in Fig. 6. Apart from a gene for cystathionine β-synthase, all of the genes have been identified by homology (22). Cystathionine β-synthase could be encoded by the cysK gene since recent results suggest that the lactococcal cysK gene usually encoding a cysteine synthase may also encode a cystathionine β-synthase (31). Due to their broad substrate specificity, MetC and MalY may be involved in several steps. The release of pyruvate from l-serine by MalY has not been presented. However, it is an interesting observation, since the formation of pyruvate from cysteine and serine indicates that the enzyme may play a role in the generation of energy.
FIG. 6.
Proposed transsulfuration pathway and pathways for the formation of VSC in L. casei. metC and malY have been shown to encode the illustrated activities. The degradation of cystathionine to α-ketobutyrate in several Lactobacillus genotypes could not be explained solely by the activity of metC, which is why it is in parentheses. The functions of other genes have been identified by homology (22). metC, cystathionine γ-synthase (with cystathionine γ- and β-lyase activities); malY, CBL (and probably maltose regulon repressor activities); cysK, cysteine synthase (and putative cystathionine synthase), luxS, S-ribosylhomocysteinase; metE, methionine synthase II (cobalamin independent); metK, S-adenosylmethionine synthetase; smtA, S-adenosylmethionine-dependent methyltransferase; pfs, S-adenosylhomocysteine nucleosidase.
The formation of VSC due to the cystathionine lyase activity of MalY and MetC indicates that the enzymes contribute to the formation of flavor components during cheese ripening. Thus, the observed formation of dimethyl disulfide after bacteria had been incubated with methionine for 1 week may be connected to MetC activity. The reduced activity at low pH might explain why we did not detect production of VSC after 1 day of incubation.
Interestingly, we found a high cystathionine lyase activity with a concomitant release of pyruvate and α-ketobutyrate in cell extracts in several L. casei strains. That this activity was highest at low pH indicates that it is not encoded by metC. Furthermore, the formation of α-ketobutyrate cannot be explained by MalY activity. Therefore, we believe that in these strains a third gene is present which encodes a cystathionine lyase activity. The presence of this gene seems to be strain specific and could explain why several L. casei genotypes were able to grow on methionine as the sole source of sulfur-containing amino acids. That the pathways for the interconversion of methionine and cysteine are present but not always fully functional has also been shown by mutagenesis experiments with L. casei strain S1 (25). Currently, we are interested in identifying the gene that encodes this activity. We think that by understanding the transsulfuration reactions and VSC formation, we will be able to identify and select strains which are the most suitable as flavor-producing adjuncts.
Acknowledgments
We thank Monika Haueter for providing technical assistance and Michael G. Casey for critical reading of the manuscript.
Footnotes
Published ahead of print on 11 September 2007.
REFERENCES
- 1.Alting, A. C., W. J. M. Engels, S. van Schalkwijk, and F. A. Exterkate. 1995. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl. Environ. Microbiol. 61:4037-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarita, F., M. Yvon, M. Nardi, E. Chambellon, J. Delettre, and P. Bonnarme. 2004. Identification and functional analysis of the gene encoding methionine-γ-lyase in Brevibacterium linens. Appl. Environ. Microbiol. 70:7348-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardö, Y. 2006. Flavour formation by amino acid catabolism. Biotechnol. Adv. 24:238-242. [DOI] [PubMed] [Google Scholar]
- 4.Aubel, D., J. E. Germond, C. Gilbert, and D. Atlan. 2002. Isolation of the patC gene encoding the cystathionine β-lyase of Lactobacillus delbrueckii subsp. bulgaricus and molecular analysis of inter-strain variability in enzyme biosynthesis. Microbiology 148:2029-2036. [DOI] [PubMed] [Google Scholar]
- 5.Auger, S., M. P. Gomez, A. Danchin, and I. Martin-Verstraete. 2005. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie 87:231-238. [DOI] [PubMed] [Google Scholar]
- 6.Beresford, T., and A. Williams. 2004. The microbiology of cheese ripening, p. 287-317. In P. F. Fox, P. L. H. McSweeney, T. M. Cogan, and T. P. Guinee (ed.), Cheese: chemistry, physics and microbiology, vol. 1. Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 7.Bruinenberg, P. G., G. de Roo, and G. K. Y. Limsowtin. 1997. Purification and characterization of cystathionine γ-lyase from Lactococcus lactis subsp. cremoris SK11: possible role in flavor compound formation during cheese maturation. Appl. Environ. Microbiol. 63:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey, M. G., J. P. Haeni, J. Gruskovnjak, W. Schaeren, and D. Wechsler. 2006. Characterisation of the non-starter lactic acid bacteria (NSLAB) of Gruyère PDO cheese. Lait 86:407-414. [Google Scholar]
- 9.Chin, H. W., and R. C. Lindsay. 1994. Ascorbate and transition-metal mediation of methanethiol oxidation to dimethyl disulfide and dimethyl trisulfide. Food Chem. 49:387-392. [Google Scholar]
- 10.Curioni, P. M. G., and J. O. Bosset. 2002. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 12:959-984. [Google Scholar]
- 11.de Angelis, M., A. C. Curtin, P. L. H. McSweeney, M. Faccia, and M. Gobbetti. 2002. Lactobacillus reuteri DSM 20016: purification and characterization of a cystathionine γ-lyase and use as adjunct starter in cheesemaking. J. Dairy Res. 69:255-267. [DOI] [PubMed] [Google Scholar]
- 12.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 131:82-91. [Google Scholar]
- 13.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias, B., and B. Weimer. 1998. Purification and characterization of l-methionine γ-lyase from Brevibacterium linens BL2. Appl. Environ. Microbiol. 64:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobric, N., G. K. Y. Limsowtin, A. J. Hillier, N. P. B. Dudman, and B. E. Davidson. 2000. Identification and characterization of a cystathionine β/γ-lyase from Lactococcus lactis ssp. cremoris MG1363. FEMS Microbiol. Lett. 182:249-254. [DOI] [PubMed] [Google Scholar]
- 16.Dwivedi, C. M., R. C. Ragin, and J. R. Uren. 1982. Cloning, purification, and characterization of β-cystathionase from E. coli. Biochemistry 21:3064-3069. [DOI] [PubMed] [Google Scholar]
- 17.Fernández, M., W. van Doesburg, G. A. M. Rutten, J. D. Marugg, A. C. Alting, R. van Kranenburg, and O. P. Kuipers. 2000. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine β-lyase. Appl. Environ. Microbiol. 66:42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, S., E. S. Mooberry, and J. L. Steele. 1998. Use of 13C nuclear magnetic resonance and gas chromatography to examine methionine catabolism by lactococci. Appl. Environ. Microbiol. 64:4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guggenheim, S., and M. Flavin. 1969. Cystathionine γ-synthase. A pyridoxal phosphate enzyme catalyzing rapid exchanges of β and α hydrogen atoms in amino acids. J. Biol. Chem. 244:6217-6227. [PubMed] [Google Scholar]
- 20.Irmler, S., M. L. Heusler, S. Raboud, H. Schlichtherle-Cerny, M. G. Casey, and E. Eugster-Meier. 2006. Rapid volatile metabolite profiling of Lactobacillus casei strains: selection of flavour producing cultures. Aust. J. Dairy Technol. 61:123-127. [Google Scholar]
- 21.Lee, W.-J., D. S. Banavara, J. E. Hughes, J. K. Christiansen, J. L. Steele, J. R. Broadbent, and S. A. Rankin. 2007. Role of cystathionine β-lyase in catabolism of amino acids to sulfur volatiles by genetic variants of Lactobacillus helveticus CNRZ 32. Appl. Environ. Microbiol. 73:3034-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Cuesta, M. C., C. Pelaez, J. Eagles, M. J. Gasson, T. Requena, and S. B. Hanniffy. 2006. YtjE from Lactococcus lactis IL1403 is a C-S lyase with α,γ-elimination activity toward methionine. Appl. Environ. Microbiol. 72:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta, P. K., and P. Christen. 2000. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 74:129-184. [DOI] [PubMed] [Google Scholar]
- 25.Morishita, T., T. Fukada, M. Shirota, and T. Yura. 1974. Genetic basis of nutritional requirements in Lactobacillus casei. J. Bacteriol. 120:1078-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauhut, D., B. Beisert, M. Berres, M. Gawron-Scibek, and H. Kuerbel. 2005. Pulse flame photometric detection: an innovative technique to analyse volatile sulfur compounds in wine and other beverages, p. 363-368. In T. Hofmann, M. Rothe, and P. Schieberle (ed.), State-of-the-art in flavour chemistry and biology. Deutsche Forschungsanstalt für Lebensmittelchemie, Garching, Germany.
- 27.Rijnen, L., S. Bonneau, and M. Yvon. 1999. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 65:4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Smacchi, E., and M. Gobbetti. 1998. Purification and characterization of cystathionine γ-lyase from Lactobacillus fermentum DT41. FEMS Microbiol. Lett. 166:197-202. [DOI] [PubMed] [Google Scholar]
- 30.Smit, G., B. A. Smit, and W. J. M. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 31.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guedon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uren, J. R. 1987. Cystathionine β-lyase from Escherichia coli. Methods Enzymol. 143:483-486. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, Y., Y. Nakano, A. Amano, M. Yoshimura, H. Fukamachi, T. Oho, and T. Koga. 2002. lcd from Streptococcus anginosus encodes a C-S lyase with α,β-elimination activity that degrades l-cysteine. Microbiology 148:3961-3970. [DOI] [PubMed] [Google Scholar]
- 34.Yvon, M., S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zdych, E., R. Peist, J. Reidl, and W. Boos. 1995. MalY of Escherichia coli is an enzyme with the activity of a βC-S lyase (cystathionase). J. Bacteriol. 177:5035-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]