Abstract
The differentiation of CD4+ CD8+ double positive (DP) thymocytes requires the irreversible choice between two alternative lineages, distinguished by the mutually exclusive expression of either CD4 or CD8. Differentiating DP cells transiently down-regulate both CD4 and CD8, and this has complicated the debate whether the mechanism of CD4/CD8 lineage choice is instructive, stochastic/selective, or more complex in nature. Using fluorescence in situ hybridization, we show that the stable silencing of coreceptor loci, and ultimately lineage choice, is predicted by the spatial repositioning of coreceptor alleles to centromeric heterochromatin domains. These data provide evidence that lineage-specific developmental programs are established early during the transition from the DP to the single positive stage.
Keywords: lineage commitment, FISH
Introduction
As cells make developmental choices, their genomes are modified to reflect their previous developmental history as well as their developmental potential (1). This occurs at an epigenetic level, rather than at the level of primary nucleotide sequence. Epigenetic modifications include DNA (CpG) methylation (2), the packaging of DNA in chromatin, the posttranslational modification of chromatin proteins (3), and the spatial partitioning of gene loci into transcriptionally permissive and repressive domains within the nucleus (4, 5). Silent, but not active, gene loci are often found in close proximity to centromeric heterochromatin in activated B cells (4, 6). In double positive (DP) thymocytes, the spatial repositioning of RAG and Tdt loci to pericentric heterochromatin marks their stable silencing in response to TCR engagement (6) and is associated with the spreading of repressive histone modifications over an extended region of the Tdt locus (7). Centromeric repositioning does not occur during transient RAG and Tdt silencing in the DP cell line VL3-3M2 (6), where repressive histone modifications remain confined to the Tdt promoter region (7). These data suggest that for the Tdt locus at least, centromeric repositioning is part of a concerted program of epigenetic events that stabilize gene silencing. Proximity to centromeres in cis (as a result of chromosomal translocations or the centromeric integration of transgenes) can result in gene silencing (8), and the recruitment of chromatin domains to centromeric heterochromatin in trans has been mechanistically linked to gene silencing by the requirement for polycomb proteins, histone methyltransferases, and heterochromatin binding proteins in Drosophila (9, 10).
The development of T cells in the thymus is understood in considerable detail and provides a useful model for cellular commitment and differentiation in metazoans. Commitment and differentiation of CD4+ CD8+ DP thymocytes to the CD4 or the CD8 lineage is triggered by TCR engagement and proceeds via a series of intermediate (DPlo and CD4+ CD8lo) stages (11, 12). Transiently reduced expression of CD4 and CD8 RNA and protein occurs in thymocytes en route to either lineage and is not predictive of lineage fate (12–19). This has added complexity to the debate as to whether thymocyte lineage choice operates instructive or stochastic/selective mechanisms, or results from cellular computations of signal strength and duration (11, 12).
Here, a functional appraisal of thymocyte lineage commitment and developmental potential is combined with a detailed analysis of CD4 and CD8 locus position (4) in transitional thymocyte subsets. We show that lineage choice is anticipated by the repositioning of coreceptor alleles to centromeric heterochromatin domains. Centromeric repositioning is progressive, developmentally regulated, and selectively affects coreceptor loci that are subject to stable silencing. Our results provide evidence that lineage-specific developmental programs are implemented at an early stage of DP to single positive (SP) differentiation.
Materials and Methods
Mouse Strains, Cell Sorting, and Cell and Organ Culture.
OT-I (20) and AND (21) TCR transgenic mice on a RAG-1o/o (22) background were used as sources of MHC class I– (Kb) or II– (Ek/Ab) selected thymocytes, respectively. Aβo/o (23), β2mo/o (24), or Tap1o/o (25) thymi were used as sources of thymocytes with a wild-type TCR repertoire selected by MHC class I or II, respectively. MHCo/o (Aβo/o β2mo/o) thymocytes were used as the source of MHC-naive thymocytes. All procedures involving animals were approved by the Home Office, UK. Thymocytes were stained with CD4-PE/Cy5, CD8a-PE (Caltag Laboratories), and in some experiments CD69-FITC (BD Biosciences), and sorted on a FACS DIVA (Becton Dickinson) and subjected to fluorescence in situ hybridization (FISH; see below), or reaggregated with MHCo/o or C57BL/10 (H-2b) thymic stroma as described previously (26). Where indicated, CD3/CD4- or CD3/CD3-bispecific antibodies (27) were added to the cultures and thymocyte suspensions were stained with CD4-PE/Cy5 or -APC, CD8-PE or -FITC, and analyzed on a FACSCalibur (Becton Dickinson). Thymocytes were stained with CD4-PE/Cy5, CD5-PE (Caltag Laboratories), and sorted for FISH analysis after 18 h.
FISH.
Three-dimensional (3-D) FISH was performed as described previously (4). For FISH involving heat denaturation, cells were fixed to coverslips in 4% paraformaldehyde/PBS for 10 min, permeabilized in 0.2% Triton X-100/PBS for 12 min, heated to 95°C for 8 min in the presence of probes in hybridization mix, and placed on ice for 2 min. Hybridization and probe detection were performed as described previously (4). Probes for γ satellite repeats, CD4 (p7′.3.1), CD8 (CD8α-1), and TCRα (32.1w7) were provided by N. Dillon (Imperial College, London, UK), A. Rahemtulla (Imperial College, London, UK), D. Kioussis (National Institute for Medical Research, London, UK), and M. Malissen (Centre d'Immunologie de Marseille-Luminy, Marseille, France). Samples were counterstained with DAPI and analyzed on a Leica TCS-4D or SP2 confocal microscope. Distance measurements were taken with Volocity software (http://www.improvision.com).
Online Supplemental Material.
A description of distance measurements (Fig. S1) and data obtained in the coreceptor reexpression assay (Fig. 2) are available at http://www.jem.org/cgi/content/full/jem.20041127/DC1.
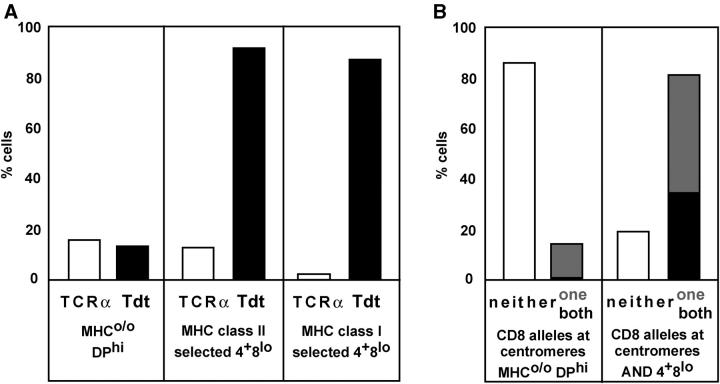
Figure 2.
Nuclear position of control loci and the impact of heat denaturation on the detection of CD8 alleles during thymocyte development. (A) MHCo/o DPhi, MHC class II–selected AND CD4+ CD8lo, and MHC class I–selected CD4+ CD8lo cells were isolated and subjected to 3-D FISH using probes for TCRα, which is expressed throughout the DP to SP transition, and Tdt, which is silenced in the DP to SP transition. (B) MHCo/o DPhi and MHC class II–selected AND CD4+ CD8lo cells were isolated and subjected to a modified FISH protocol including heat denaturation (refer to Materials and Methods). This increased the detection of centromere-associated CD8 alleles in AND CD4+ CD8lo cells compared with 3-D FISH data shown in B.
Results
Coreceptor Locus Repositioning during Lineage Commitment and Differentiation.
To ask if nuclear organization is predictive of lineage choice, we sorted thymocyte populations in transit from the DP to the SP stage from mice where thymocyte differentiation is driven either by MHC class I or II and the ultimate lineage outcome is known. DPhi CD69−, DPlo CD69+, CD4+ CD8lo, and CD4 SP thymocyte populations from MHC class I–deficient mice (Tap1o/o or β2mo/o) and AND TCR (RAG-1o/o H2b) transgenic mice were subjected to 3-D FISH, which preserves the spatial relationship between proteins and DNA in the nucleus (4). The position of CD4 and CD8 alleles was determined relative to pericentric heterochromatin domains, as defined by γ satellite DNA (4). Alleles were classified either as not associated with γ satellite foci, as associated, or as constrained to the dark area surrounding the γ satellite domains, which appeared to exclude background hybridization (Fig. 1 A). Based on previous work using γ satellite probes together with antibodies against nuclear proteins, this area would correspond to pericentromeric foci of Ikaros proteins (4, 6, 28). Distance measurements from the center of locus-specific FISH signals to the nearest γ satellite domain substantiated this classification (>0.8 μm for “nonassociated,” 0.55–0.8 μm for “constrained,” and <0.55 μm for “associated” alleles; Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20041127/DC1).
Figure 1.
Developmental regulation of coreceptor locus position. (A) The position of CD8 alleles (green) relative to γ satellite repeats (red) was analyzed by 3-D FISH and defined as associated, constrained, or not associated as detailed in Fig. S1. Bar, 2 μm. (B) The percentage of cells with centromerically positioned CD8 loci in thymocyte subsets isolated from MHCo/o, class I–deficient, AND TCR transgenic, and OT-I TCR transgenic mice. For MHCo/o, AND, and OT-I thymocyte subsets, the position of CD4 alleles was also evaluated.
CD8 alleles were not spatially associated with centromeric heterochromatin in the great majority (>90%) of MHC-naive (MHCo/o) DP thymocytes (n = 284), or DPhi cells from class I–deficient and AND mice (83%, n = 100 and 91%, n = 200, respectively). In contrast, centromere-associated or constrained CD8 alleles were recorded in 46% of class I–deficient CD4 SP (n = 101) and in 57% of AND CD4 SP cells (n = 111). Critically, centromeric repositioning was already apparent in class II–selected transitional thymocyte subsets: 38% of class I–deficient CD4+ CD8lo (n = 95) cells had constrained or γ satellite–associated CD8 alleles. In AND thymocytes, repositioning of CD8 alleles was seen in DPlo cells (42%, n = 201) as well as in CD4+ CD8lo cells (48%, n = 115; Fig. 1 B).
In contrast to CD8 alleles, CD4 alleles did not undergo centromeric repositioning in developing AND thymocytes (3% of DPhi cells, n = 82; 6% of DPlo cells, n = 101; 7% of CD4+ CD8lo cells, n = 151; 6% of CD4 SP cells, n = 150; Fig. 1 B), indicating that repositioning was selective for CD8 during class II–driven thymocyte differentiation.
Centromeric repositioning of CD8 did not occur during the development of thymocytes transgenic for the class I–restricted OT-I TCR (7% of OT-I DPhi, n = 101; 7% of CD4+ CD8lo, n = 114; 6% of DPlo, n = 94; 10% of CD8 SP, n = 115; Fig. 1 B), which we examined as an example of MHC class I–driven thymocyte differentiation. Instead, the frequency of cells with constrained or γ satellite–associated CD4 alleles, which was constant throughout MHC class II–driven thymocyte differentiation, increased during OT-I differentiation from 6% in DPhi cells (n = 150) to 20% in DPlo (n = 100), and 21% in CD4+ CD8lo cells (n = 150) to 37% in the CD8 SP (n = 150; Fig. 1 B). Hence, repositioning of CD4 in class I–selected OT-I thymocytes appeared less extensive than that of CD8 in class II–selected thymocytes, but was initiated in transitional thymocyte subsets.
To further evaluate the selectivity of coreceptor locus repositioning during MHC class I– and II–driven thymocyte differentiation, we examined the nuclear position of control loci. TCRα expression continues during the DP to SP transition, and TCRα loci remained noncentromeric during both MHC class I– and II–driven thymocyte differentiation (we detected constrained or centromere-associated TCRα loci in 17% of MHCo/o DPhi, n = 103; 13% of class II–selected [AND] CD4+ CD8lo, n = 109; and 2% of class I–selected (Aβo/o) CD4+ CD8lo, n = 102; Fig. 2 A). In contrast to TCRα, expression of Tdt is terminated by TCR signals at the DP stage (6, 7). Tdt was found in noncentromeric positions in MHC-naive DPhi cells (15% of MHCo/o DPhi cells had constrained or centromere-associated Tdt loci, n = 106; Fig. 2 A) and was repositioned to centromeric heterochromatin domains during both MHC class I– and II–driven thymocyte differentiation (87% of Aβo/o CD4+ CD8lo, n = 102 and 92% of AND CD4+ CD8lo, n = 60; Fig. 2 A).
In addition to centromeric repositioning, the shutdown of coreceptor loci during thymocyte differentiation was accompanied by a progressive reduction in locus accessibility to FISH probes under 3-D FISH conditions (not depicted). This was reflected in nonequivalent probe hybridization, which was not due to technical failure because our probes worked with >90% efficiency in DPhi cells (not depicted). To address whether changes in locus accessibility resulted in an underestimate of the actual extent to which silenced coreceptor loci were repositioned, we modified our FISH protocol to include heat denaturation. This compromised the preservation of nuclear structure (not depicted), but improved the detection of CD8 alleles in AND CD4+ CD8lo cells. Under these conditions, we found centromere-associated CD8 alleles in 81% of AND CD4+ CD8lo cells, compared with 48% by 3-D FISH (Fig. 2 B). Hence, denaturation conditions that disrupt protein–DNA interactions revealed previously undetectable CD8 alleles in centromeric positions.
Developmental Potential of Transitional Thymocyte Subsets.
In parallel to evaluating the position of CD4 and CD8 coreceptor loci in the nucleus, we analyzed lineage commitment and developmental potential of sorted thymocyte subsets. Initial experiments suggested that coreceptor reexpression (14) was not necessarily indicative of lineage commitment (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20041127/DC1) and we opted for reaggregate cultures with H2b thymic stroma (to allow for sustained TCR–pMHC contact) or MHCo/o stroma (26). The latter monitors the cells' competence to differentiate without pMHC contact. When combined with bispecific CD3/CD4 or CD3/CD3 antibodies (27), which drive the differentiation of uncommitted thymocytes to the CD4 or the CD8 lineage, respectively (29), the system becomes a powerful tool to probe thymocyte lineage potential.
DPhi CD69−, DPlo CD69+, CD4+ CD8lo, and CD4 SP thymocyte populations were sorted from MHC class I–deficient mice (Fig. 3 A) and subjected to reaggregate culture. The CD4+ CD8lo population readily generated CD4 SP progeny in MHCo/o stroma (67 ± 4% at 96 h, n = 3; Fig. 3 B) as well as a minor CD8 SP population (11 ± 2%, n = 3). To further evaluate residual CD8 lineage potential, we exposed CD4+ CD8lo reaggregate cultures to bispecific CD3/CD3 antibodies, which drive uncommitted thymocytes to the CD8 lineage (29). The CD4 bias of the CD4+ CD8lo population was not reversed by anti-CD3/CD3 (13 ± 8% CD8 SP at 96 h, n = 3), whereas DPhi cells readily generated CD8 SP progeny under these conditions (33 ± 4% CD8 SP at 96 h, n = 3; Fig. 3 C). For comparison, DPhi, DPlo, CD4+ CD8lo, and CD4 SP thymocyte populations were sorted from AND TCR (RAG-1o/o H2b) transgenic mice and placed in MHCo/o reaggregates (Fig. 3 E). AND CD4+ CD8lo differentiated to the CD4 SP stage with similar efficiency as class I–deficient CD4+ CD8lo cells, without generating CD8 SP progeny (Fig. 3, compare B with E). In this experimental system, the CD4+ CD8lo phenotype therefore marks a stage during MHC class II–driven thymocyte differentiation at which cells appear committed to the CD4 lineage and are able to progress to the CD4 SP stage without further pMHC contact. As demonstrated above for AND thymocytes, CD8 but not CD4 alleles are repositioned to centromeric heterochromatin at this stage (Fig. 1 B).
Figure 3.
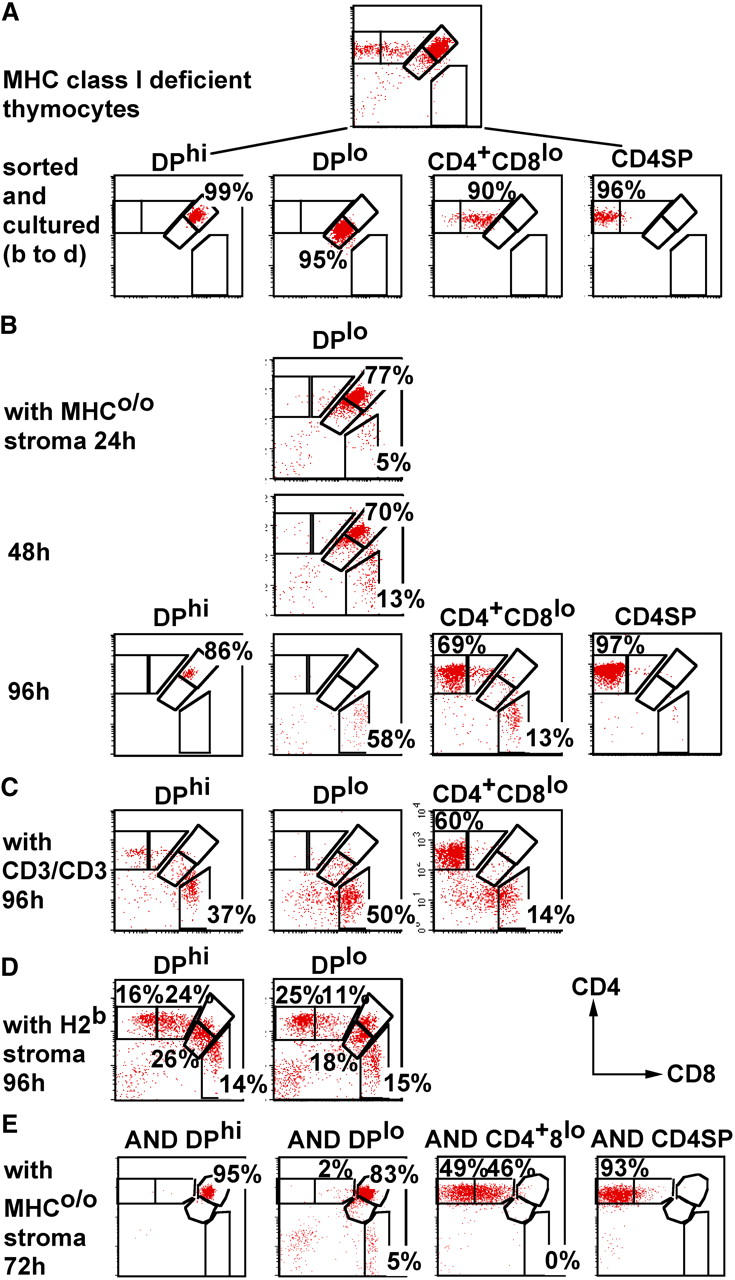
Lineage commitment and loss of CD8 lineage potential during MHC class II–driven thymocyte differentiation. (A) DPhi, DPlo, CD4+ CD8lo, and CD4 SP thymocyte subsets were isolated from MHC class I–deficient (Tap1o/o) mice. (B) Isolated MHC class I–deficient thymocyte subsets were cultured in reaggregates with MHCo/o thymic stroma for the indicated number of hours and then analyzed for CD4 and CD8 expression. (C) Isolated MHC class I–deficient thymocyte subsets were cultured in reaggregates with MHCo/o thymic stroma in the presence of anti-CD3/CD3 for 96 h. (D) Isolated MHC class I–deficient thymocyte subsets were cultured in reaggregates with H2b stroma for 96 h. (E) DPhi, DPlo, CD4+ CD8lo, and CD4 SP thymocyte subsets were isolated from AND TCR transgenic mice and cultured in reaggregates with MHCo/o thymic stroma for 72 h.
In contrast to CD4+ CD8lo cells, the class II–selected DPlo population failed to progress to the CD4 SP stage when deprived of pMHC. Most (77%) returned to a DPhi phenotype within 24 h, whereas others were diverted to a CD8 SP phenotype (5% at 24 h, 13% at 48 h, and 58% at 96 h; Fig. 3 B). CD8 differentiation of DPlo cells could not be ascribed to residual MHC class I expression by class I– (β2m or Tap1) deficient cells because a fraction of AND TCR transgenic DPlo cells also became CD8 SP (5%; Fig. 3 E). The yields of CD8 SP progeny generated by class I–deficient DPlo cells increased moderately (4.5- ± 2.1-fold, n = 4) in the presence of bispecific anti-CD3/CD3 (Fig. 3 C), and they generated CD8 SP (15%) as well as CD4 SP (25%) progeny in reaggregates with H2b stroma (Fig. 3 D). By these criteria and in this experimental system, class II–selected DPlo cells are not uniformly or irreversibly CD4 committed. Nevertheless, CD8 alleles were selectively repositioned to centromeric heterochromatin in AND DPlo cells (Fig. 1 B).
Next, we examined DPhi, DPlo, CD4+8lo, and CD8 SP subsets from OT-I TCR transgenic (RAG-1o/o H2b) mice as an example for MHC class I–selected thymocytes. DPhi cells generated relatively few CD8 SP progeny in MHCo/o stroma (15 ± 12% at 72–96 h, n = 3), but efficiently differentiated to the CD8 SP stage in H2b stroma (Fig. 4 A). We were surprised by the behavior of DPlo cells, which we expected would up-regulate CD4 to become CD4+ CD8lo (12, 16). Only a minority of DPlo cells became CD4+ CD8lo (6% in MHCo/o, Fig. 3 A; 4% in H2b stroma, not depicted) or DPhi (14% in MHCo/o stroma, Fig. 4 A; 4% in H2b stroma, not depicted). Instead, they quickly progressed to the CD8 SP stage (45% at 24 h, 70% at 48 h, not depicted; 97% at 72 h, Fig. 4 A). Conversely, many CD4+ CD8lo cells became DPlo (44% at 24 h) and then progressed to the CD8 SP stage (14% at 24 and 80% at 72 h, Fig. 4 A). Consistent with residual CD4 potential (see below), CD4+ CD8lo but not DPlo cells reproducibly generated a minor CD4 SP population (13 ± 3%, n = 7; Fig. 4 A).
Figure 4.
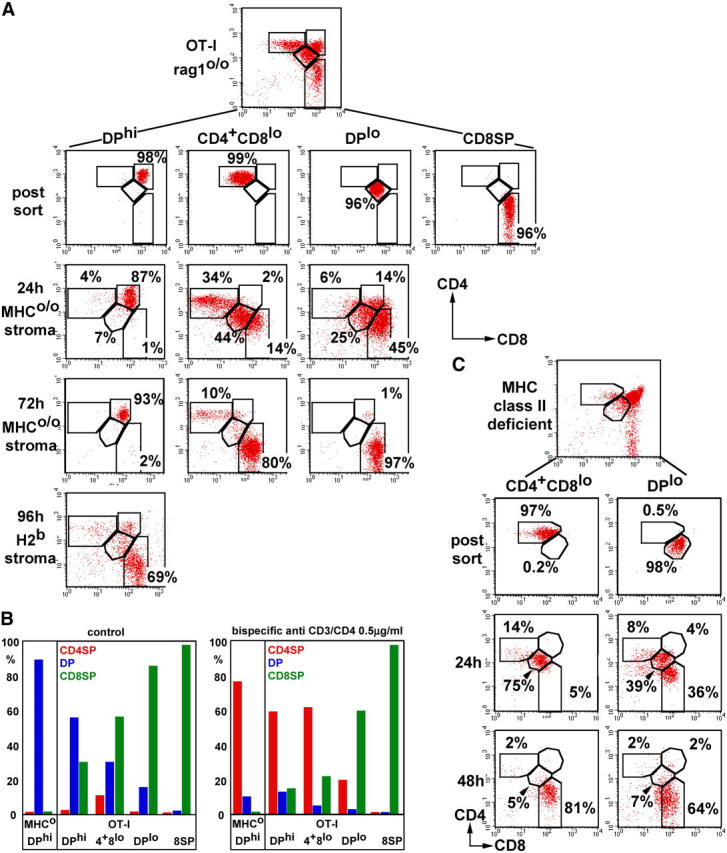
Lineage commitment and loss of CD4 lineage potential during MHC class I–driven thymocyte differentiation. (A) OT-I TCR transgenic RAG-1o/o H2b thymocytes were isolated and analyzed as described in Fig. 2 A. (B) The generation of CD4 SP (red), DP (blue), and CD8 SP (green) progeny by MHCo/o and OT-I thymocyte subsets is plotted for control reaggregates with MHCo/o stroma (left). Bispecific CD3/CD4 antibodies were added to drive the differentiation of uncommitted thymocytes to the CD4 lineage (right). Cell cycle analysis showed that the generation of CD4 SP progeny was not due to the expansion of preexisting cells (CD4+ CD8lo in MHCo/o stroma: G1 = 84.3%, S = 2.3%, G2/M = 3.3%; CD4+ CD8lo with bispecific CD3/CD4: G1 = 85.6%, S = 2.8%, G2/M = 2.4%, not depicted). (C) CD4+ CD8lo and DPlo subsets were isolated from class II–deficient (Aβo/o) mice and analyzed 24 and 48 h after reaggregation with MHCo/o stroma.
These results tentatively placed DPlo downstream of CD4+ CD8lo cells, suggesting that the developmental sequence in the OT-I thymus was reversed relative to that of MHC class II–selected thymocytes (Fig. 3 A). Therefore, we examined the developmental potential of OT-I thymocyte populations in reaggregates exposed to bispecific CD3/CD4 antibodies, which drive uncommitted thymocytes to the CD4 lineage (reference 29 and Fig. 4 B). The CD4+ CD8lo population generated a much higher percentage of CD4 SP progeny (61%) than the DPlo population (20%; cell cycle analysis confirmed that the increase of CD4 SP was due to differentiation, not expansion of preexisting cells; see legend to Fig. 4 B for details). Hence, in contrast to class II–selected CD4+ CD8lo cells (Fig. 3 A), the OT-I CD4+ CD8lo subset appears flexible in its lineage choice. Progressive CD8 lineage commitment and the loss of developmental potential for the CD4 lineage place OT-I thymocyte populations in the following order: DPhi > CD4+ CD8lo > DPlo > CD8 SP. To test whether this applies to MHC class I–selected thymocytes in general, we isolated CD4+ CD8lo and DPlo subsets from MHC class II–deficient (Aβo/o) mice. DPlo cells differentiated to the CD8 SP stage within 24 (36%) to 48 h (64%) and generated few CD4+ CD8lo or DPhi progeny (Fig. 4 C). CD4+ CD8lo cells became DPlo (75% at 24 h) en route to the CD8 SP stage (81% at 48 h; Fig. 4 C). These data confirm that in our experimental system, the majority of class I–selected DPlo cells behave as progeny, not precursors of CD4+ CD8lo cells.
Discussion
Coreceptor Locus Repositioning Is Predictive of CD4/CD8 Lineage Choice.
3-D FISH analysis revealed marked centromeric association of CD8 loci increased in MHC class II–selected CD4+ CD8lo populations, coincident with the cells' ability to differentiate to the CD4 SP stage without continued pMHC contact, and with their reduced potential to differentiate toward the CD8 lineage in response to anti-CD3/CD3. In AND TCR transgenic thymocytes, substantial centromeric association of CD8 occurred at the earlier DPlo stage, suggesting that for this TCR specificity at least, CD8 repositioning precedes the ability to differentiate to the CD4 lineage without further pMHC signals. Analysis of OT-I TCR transgenic cells as an example of class I–selected thymocytes indicates that the CD4 locus is repositioned during the differentiation to the CD8 (not the CD4) lineage, if to a lesser extent than CD8 in CD4-committed cells. Robust repositioning of CD4 during CD8 T cell differentiation has been seen in other models of CD8 T cell differentiation (Robey, E., personal communication). Hence, in contrast to CD4/CD8 protein and RNA expression, locus repositioning appears to be predictive of lineage choice before differentiation to the SP stage.
Functional Nonequivalence of MHC Class I– and II–selected Transitional Thymocyte Populations.
Coreceptor locus repositioning and differential accessibility demonstrates that transitional thymocyte subsets selected by MHC class I and II are not equivalent, a conclusion that is substantiated by the functional analysis of their developmental potential. First, in contrast to class I–selected DPlo cells, which reach the CD4 SP stage independently of stroma pMHC, class II–selected DPlo cells need pMHC to become CD4 SP in our experimental system. They are not uniformly CD4 committed and can generate CD8 SP progeny when deprived of pMHC or exposed to bispecific anti-CD3/CD3. Second, class II–selected CD4+ CD8lo become CD4 SP without pMHC and appear largely CD4 committed because they are not readily redirected to the CD8 lineage by anti-CD3/CD3. Class I–selected CD4+ CD8lo cells also differentiate to the SP stage in MHCo/o stroma, but in contrast to their class II–selected counterparts, they are not uniformly or irreversibly lineage committed and can be redirected to the CD4 lineage by bi-specific antibodies. Because it takes 24 h for MHC-naive OT-I cells to become CD4+ CD8lo when placed in a selecting environment (not depicted), CD8 lineage commitment appears to be a protracted process, which can be overridden for many hours after initial pMHC contact. Finally, MHC class I– and II–selected transitional thymocytes appear to differ with respect to developmental sequence. Our analysis agrees with models that place DPlo cells between the DPhi and the CD4+ CD8lo population (12–16) in MHC class II–driven differentiation. However, three lines of evidence place the majority of class I–selected DPlo cells firmly downstream of CD4+ CD8lo: (a) class I–selected DPlo cells transit to the CD8 SP stage faster than CD4+ CD8lo cells; (b) class I–selected CD4+ CD8lo cells transit through a DPlo stage en route to the CD8 SP stage, but not vice versa; and (c) class I–selected DPlo cells lack CD4 lineage potential in our assay, in contrast to CD4+ CD8lo cells. This does not exclude that some MHC class I–selected thymocytes transit through a DPlo stage before they become CD4+ CD8lo (16).
Implications for Models of CD4/CD8 Lineage Commitment.
We find that CD8 locus repositioning is indicative of the fate of CD4-committed CD4+ CD8lo thymocytes before their differentiation to the SP stage and may precede commitment and/or the competence of MHC class II–signaled DPlo cells to differentiate to the CD4 lineage without further pMHC contact. These data provide novel markers (30–33) of thymocyte lineage choice before overt differentiation and contribute to a growing body of evidence against purely stochastic/selective models of CD4/CD8 lineage commitment (for review see reference 12). Our functional data indicate that MHC class I–selected CD4+ CD8lo thymocytes are competent to progress toward the CD8 lineage, yet retain CD4 lineage potential. This calls into question the conceptual distinction between instructive and stochastic/selective models of lineage choice. It adds to evidence (for review see reference 12) that the CD4/CD8 cell fate decision can be iterative and is not necessarily made “on the spot” as had been implied in the original models. Taken together with data that the specification of CD4 lineage fate is initially labile (as predicted by kinetic signaling models; references 12, 33, and 34), but becomes firmly specified within hours (34), and evidence that CD4 lineage specification is promoted by signal strength/lck activity (12, 29, 35), one could liken thymocyte lineage choice to an auction where thymocytes will firmly accept a “CD4 bid” within a few hours, whereas cells receiving “CD8 bids” may remain open to higher offers (and a CD4 lineage fate) for an extended period. At a mechanistic level, a cascade of chromatin-based events, including repositioning of coreceptors and other silenced loci (7), would ultimately result in lineage progression. The lineage-specific reorganization of the thymocyte nucleus opens new avenues for investigating the molecular basis of lineage choice.
Acknowledgments
We thank Steve Smale and B.J. Fowlkes for discussions, Ellen Robey for communicating unpublished results, J.Y. Tso (Protein Design Labs, Inc.) for CD3/CD4- and CD3/CD3-bispecific antibodies, Katy Smith for help with cell sorting, and Niall Dillon, Steve Smale, Dimitris Kioussis, and Marie Malissen for probes.
This work was supported by the Medical Research Council, UK.
The authors have no conflicting financial interests.
Abbreviations used in this paper: 3-D, three-dimensional; DP, double positive; FISH, fluorescence in situ hybridization; SP, single positive.
References
- 1.Fisher, A.G. 2002. Cellular identity and lineage choice. Nat. Rev. Immunol. 2:977–982. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33:245–254. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein, T., and C.D. Allis. 2001. Translating the histone code. Science. 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 4.Brown, K.E., S.S. Guest, S.T. Smale, K. Hahm, M. Merkenschlager, and A.G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. [DOI] [PubMed] [Google Scholar]
- 5.Gasser, S.M. 2001. Positions of potential: nuclear organization and gene expression. Cell. 104:639–642. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K.E., J. Baxter, D. Graf, M. Merkenschlager, and A.G. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 3:207–217. [DOI] [PubMed] [Google Scholar]
- 7.Su, R.-C., K.E. Brown, S. Saaber, A.G. Fisher, M. Merkenschlager, and S.T. Smale. 2004. Assembly of silent chromatin at a developmentally regulated gene. Nat. Genet. 36:502–506. [DOI] [PubMed] [Google Scholar]
- 8.Dillon, N., and R. Festenstein. 2002. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18:252–258. [DOI] [PubMed] [Google Scholar]
- 9.Csink, A.K., and S. Henikoff. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 381:529–531. [DOI] [PubMed] [Google Scholar]
- 10.Dernburg, A.F., K.W. Broman, J.C. Fung, W.F. Marshall, J. Philips, D.A. Agard, and J.W. Sedat. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 85:745–759. [DOI] [PubMed] [Google Scholar]
- 11.Kisielow, P., and H. von Boehmer. 1995. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58:87–209. [DOI] [PubMed] [Google Scholar]
- 12.Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. [DOI] [PubMed] [Google Scholar]
- 13.Lundberg, K., W. Heath, F. Kontgen, F.R. Carbone, and K. Shortman. 1995. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4−8+ TCRhi thymocytes. J. Exp. Med. 181:1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki, H., J.A. Punt, L.G. Granger, and A. Singer. 1995. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 2:413–425. [DOI] [PubMed] [Google Scholar]
- 15.Benveniste, P., G. Knowles, and A. Cohen. 1996. CD8/CD4 lineage commitment occurs by an instructional/default process followed by positive selection. Eur. J. Immunol. 26:461–471. [DOI] [PubMed] [Google Scholar]
- 16.Lucas, B., and R.N. Germain. 1996. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 5:461–477. [DOI] [PubMed] [Google Scholar]
- 17.Barthlott, T., H. Kohler, H. Pircher, and K. Eichmann. 1997. Differentiation of CD4(high) CD8(low) coreceptor-skewed thymocytes into mature CD8 single-positive cells independent of MHC class I recognition. Eur. J. Immunol. 27:2024–2032. [DOI] [PubMed] [Google Scholar]
- 18.Cibotti, R., A. Bhandoola, T.I. Guinter, S.O. Sharrow, and A. Singer. 2000. CD8 coreceptor extinction in signaled CD4(+)CD8(+) thymocytes: coordinate roles for both transcriptional and posttranscriptional regulatory mechanisms in developing thymocytes. Mol. Cell. Biol. 20:3852–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Y.H., D.L. Li, A. Winoto, and E.A. Robey. 2004. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc. Natl. Acad. Sci. USA. 101:4936–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogquist, K.A., S.C. Jameson, W.R. Heath, J.L. Howard, M.J. Bevan, and F.R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. [DOI] [PubMed] [Google Scholar]
- 21.Kaye, J., M.L. Hsu, M.E. Sauron, S.C. Jameson, N.R. Gascoigne, and S.M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. [DOI] [PubMed] [Google Scholar]
- 22.Spanopoulou, E., P. Cortes, C. Shih, C.M. Huang, D.P. Silver, P. Svec, and D. Baltimore. 1995. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 3:715–726. [DOI] [PubMed] [Google Scholar]
- 23.Cosgrove, D., D. Gray, A. Dierich, J. Kaufman, M. Lemeur, C. Benoist, and D. Mathis. 1991. Mice lacking MHC class II molecules. Cell. 66:1051–1066. [DOI] [PubMed] [Google Scholar]
- 24.Koller, B.H., P. Marrack, J.W. Kappler, and O. Smithies. 1990. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science. 248:1227–1230. [DOI] [PubMed] [Google Scholar]
- 25.van Kaer, L., P.G. Ashton-Rickardt, H.L. Ploegh, and S. Tonegawa. 1992. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4−8+ T cells. Cell. 71:1205–1214. [DOI] [PubMed] [Google Scholar]
- 26.Merkenschlager, M., D. Graf, M. Lovatt, U. Bommhardt, R. Zamoyska, and A.G. Fisher. 1997. How many thymocytes audition for selection? J. Exp. Med. 186:1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostelny, S.A., M.S. Cole, and J.Y. Tso. 1992. Formation of a bispecific antibody by the use of leucine zippers. J. Immunol. 148:1547–1553. [PubMed] [Google Scholar]
- 28.Georgopoulos, K. 2002. Haematopoietic cell-fate decisions, chromatin regulation and Ikaros. Nat. Rev. Immunol. 2:162–174. [DOI] [PubMed] [Google Scholar]
- 29.Basson, M.A., U. Bommhardt, P.J. Mee, V.L. Tybulewicz, and R. Zamoyska. 1998. Molecular requirements for lineage commitment in the thymus–antibody-mediated receptor engagements reveal a central role for lck in lineage decisions. Immunol. Rev. 165:181–194. [DOI] [PubMed] [Google Scholar]
- 30.Nawijn, M.C., R. Ferreira, G.M. Dingjan, O. Kahre, D. Drabek, A. Karis, F. Grosveld, and R.W. Hendriks. 2001. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J. Immunol. 167:715–723. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez-Hoyos, G., M.K. Anderson, C. Wang, E.V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. [DOI] [PubMed] [Google Scholar]
- 32.Pai, S.Y., M.L. Truitt, C.N. Ting, J.M. Leiden, L.H. Glimcher, and I.C. Ho. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., and R. Bosselut. 2004. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nat. Immunol. 5:280–288. [DOI] [PubMed] [Google Scholar]
- 34.Yasutomo, K., C. Doyle, L. Miele, C. Fuchs, and R.N. Germain. 2000. The duration of antigen receptor signaling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 404:506–510. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez-Hoyos, G., S.J. Sohn, E.V. Rothenberg, and J. Alberola-Ila. 2000. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 12:313–322. [DOI] [PubMed] [Google Scholar]