Abstract
Notch signaling has been shown to play a pivotal role in inducing T lineage commitment. However, T cell progenitors are known to retain other lineage potential long after the first point at which Notch signaling is required. Thus, additional requirements for Notch signals and the timing of these events relative to intrathymic differentiation remain unknown. Here, we address this issue by culturing subsets of CD4 CD8 double negative (DN) thymocytes on control stromal cells or stromal cells expressing Delta-like 1 (Dll1). All DN subsets were found to require Notch signals to differentiate into CD4+ CD8+ T cells. Using clonal analyses, we show that CD44+ CD25+ (DN2) cells, which appeared committed to the T cell lineage when cultured on Dll1-expressing stromal cells, nonetheless gave rise to natural killer cells with a progenitor frequency similar to that of CD44+ CD25− (DN1) thymocytes when Notch signaling was absent. These data, together with the observation that Dll1 is expressed on stromal cells throughout the thymic cortex, indicates that Notch receptor–ligand interactions are necessary for induction and maintenance of T cell lineage specification at both the DN1 and DN2 stages of T cell development, suggesting that the Notch-induced repression of the B cell fate is temporally separate from Notch-induced commitment to the T lineage.
Keywords: thymus, T cell development, notch ligands, lymphopoiesis and lineage commitment, stromal cell lines
Introduction
A number of studies have demonstrated an essential role for Notch signaling during the commitment of multipotent hematopoietic progenitor cells (HPCs) to the T cell lineage (1–4). Notably, bone marrow cells transduced to express the constitutively active intracellular domain of Notch1 fail to develop into B cells when injected into recipient mice, and instead undergo ectopic T cell development in the bone marrow (1). In contrast, mice with a conditional deletion of Notch1 lack T cells, and B cells develop in the thymus of these Notch1-deficient animals (2). In addition, several studies have implicated the Notch signaling pathway in differentiation events occurring after T cell lineage commitment, including the regulation of TCR-β gene rearrangement (5) and the αβ versus γδ (6), or CD4 versus CD8 (7–10), lineage decisions. Taken together, these findings support the notion that Notch signaling is essential for T lineage commitment and can influence multiple stages of subsequent T cell development. However, the role that Notch plays at the earliest stages of thymocyte development leading up to T cell commitment has not been addressed.
During ontogeny, lymphoid precursors that colonize the thymic rudiment at day 12 of gestation resemble lymphoid progenitors present in the bone marrow and show little evidence of Notch activation (11), suggesting that B cell lineage potential is lost immediately after entry into the thymus. However, several studies have reported that T/NK cell bipotent progenitors can be found in the fetal liver (12–14) and fetal blood (15, 16), indicating that Notch signaling resulting in the initial loss of B cell potential may occur extrathymically. Furthermore, a number of studies have demonstrated a substantial NK cell lineage potential from early progenitor thymocytes (17–22), supporting the notion that T cells develop through a T/NK bipotent intermediate. In this regard, the temporal regulation of Notch signals that result in T cell commitment from bipotent and multipotent progenitors remains unclear.
The earliest populations of progenitor thymocytes lack cell surface expression of CD4 and CD8, and are therefore referred to as double negative (DN) cells (23). The DN population can be further subdivided by surface expression of CD117 (c-kit), CD44, and CD25 (IL-2Rα; reference 24). The CD117+ CD44+ CD25− (DN1) population of progenitor thymocytes is thought to contain multipotent lymphoid progenitors (25), as well as T- and NK cell–restricted bipotent, and perhaps unipotent, progenitors (19, 26). In contrast, CD117+ CD44+ CD25+ (DN2) thymocytes give rise only to T cells when transferred to fetal thymic organ culture or injected into recipient mice (27, 28), and therefore this population is generally thought to represent a population of T lineage–committed precursors. However, it has also been suggested that the DN2 population contains progenitors that are specified to the T cell lineage due to the expression of T lineage–specific genes (29), but that are not yet fully committed, as several groups have reported limited NK and dendritic cell potential from DN2 cells (19, 30–32). CD117− CD44− CD25+ (DN3) cells appear to be fully committed to the T cell lineage, as evidenced by their ongoing process of TCR gene rearrangement (33, 34). CD117− CD44− CD25− (pre–double positive [DP]) cells are the product of DN3 cells that have undergone β selection after successful TCR-β gene rearrangement (35), and have already initiated the process of differentiating to the DP stage (36, 37).
We have recently demonstrated that ectopic expression of a Notch ligand, Delta-like 1 (Dll1), on the bone marrow stromal cell line OP9 allows for the efficient differentiation of fetal liver–derived HPCs into CD4 and CD8 DP T cells, accompanied by the generation of a small population of functionally mature CD8 single positive T cells (38). Furthermore, OP9 cells expressing Dll1 (OP9-DL1) can support the differentiation and extensive proliferation of a single progenitor cell with T or NK cell potential, whereas the control OP9 cell line has a similar capacity to support cells with B or NK cell potential, thus providing an ideal system to specifically address the effect of Notch signaling on the developmental potential of individual cells within the DN subsets.
In this work, we have analyzed the developmental potential and Notch signaling requirements of these DN thymocyte subsets. For each of the DN subsets (DN1-3), the program of T cell differentiation is arrested when cultured on OP9-control cells, but is able to progress normally to the DP stage when cultured on OP9-DL1 cells, indicating that Notch receptor–ligand interactions are required throughout DN T cell development. We confirmed that the DN1 population contains a substantial proportion of cells that are bipotent for the T and NK cell lineages, whereas only a relatively low frequency of DN1 cells possess B cell lineage potential. Furthermore, we demonstrate that DN2 thymocytes have not undergone irreversible commitment to the T cell lineage, but can instead adopt an NK cell fate when cultured on OP9-control cells. Using limiting dilution and clonal assays, we show that DN1 thymocytes are more likely to adopt an NK cell fate in the absence of Notch receptor–ligand interactions, but that DN2 cells can only adopt the NK cell lineage when cultured in the absence of Dll1.
Consistent with this dynamic view of Notch signaling during T cell development, thymic stromal cells throughout the cortex were found to express Dll1, highlighting the requirement for continued and/or recurrent Notch receptor–ligand interactions during all the initial stages of T cell development. Finally, we show that pharmacological inhibition of Dll1-induced Notch signals delayed T cell development from HPCs, and resulted in a substantial increase in the generation of NK cells with increasing concentrations of inhibitor. Together, these findings indicate that Notch signaling is necessary both for initial T cell specification at the DN1 stage, and for T cell commitment at the DN2 stage. In addition, they suggest that Notch signals sequentially restrict the lineage potential of progenitor cells, first by extinguishing B lineage potential, and then by inducing T/NK bipotent progenitors to adopt the T cell lineage.
Materials and Methods
Mice.
C57BL/6 mice and timed-pregnant Swiss.NIH mice were obtained from the National Cancer Institute, Frederick Cancer Research and Development Center.
Cell Lines.
The OP9-control and OP9-DL1 cell lines were generated, as described previously (38), by infecting the bone marrow stromal cell line OP9 (39) with the empty MigR1 retroviral vector (1), or with the MigR1 retroviral vector engineered to express the Dll1 gene (40) 5′ of the internal ribosomal entry site, allowing the bicistronic expression of Dll1 and GFP. OP9 cells, OP9-control cells, and OP9-DL1 cells were cultured as a monolayer in OP9 media (αMEM supplemented with 20% FCS [HyClone] and 2.2 g/liter sodium bicarbonate), to which IL-6, IL-7, IL-15, and Flt3 ligand (all from PeproTech) were added to enhance the growth of T, B, and NK cells.
Isolation of Fetal Cells.
Day 14 fetal thymus and fetal liver were harvested, washed in 5 ml OP9 medium, and disrupted through 40-μm nylon mesh using a syringe plunger. CD24low/Lin− fetal liver cells were obtained by antibody/complement-mediated lysis, as described previously (16). In brief, 2 ml anti-CD24 (J11d.2) culture supernatant and a 1:10 dilution of Low-Tox rabbit complement (Cedar Lane) were added to single cell suspensions in 10 ml complete medium, and cells were incubated at 37°C for 30 min. After incubation, viable cells were recovered by discontinuous density gradient centrifugation over Lympholyte-Mammal (Cedar Lane) and washed before flow cytometric cell sorting.
Flow Cytometry and Cell Sorting.
Flow cytometry was performed using a FACSCalibur™ (BD Biosciences) instrument, as described previously (16). FITC-, PE-, biotin-, and APC-conjugated mAbs and streptavidin-APC were purchased from BD Biosciences. For analysis, live cells were gated based on forward and side scatter, and lack of propidium iodide uptake. Cells were sorted using a FACSDiVa™ (BD Biosciences). Bulk-sorted cells were ≥99% pure, as determined by postsort analysis.
OP9 Cell Cocultures.
Fetal thymus DN subsets were sorted based on the differential surface expression of CD117, CD44, and CD25, as illustrated in Fig. 1 a. Sorted DN subsets were seeded at 103 cells/well into 24-well tissue culture plates containing either OP9-control or OP9-DL1 cells, to which 1 ng/ml IL-6, 1 ng/ml IL-7, 25 ng/ml IL-15, and 5 ng/ml Flt3 ligand were added to enhance the growth of T, B, and NK cells. The cells were cultured for 7 d, and then analyzed by flow cytometry. CD24low/Lin− fetal liver progenitor cells were sorted for CD117+ Sca-1hi HPCs and cultured on either OP9-control or OP9-DL1 cells. Various concentrations of the presenilin Inhibitor X (Calbiochem) were added to the culture wells, as indicated (see Fig. 4). The culture medium containing equal volumes of DMSO alone or presenilin inhibitor (diluted in DMSO) was changed on the fourth day of culture.
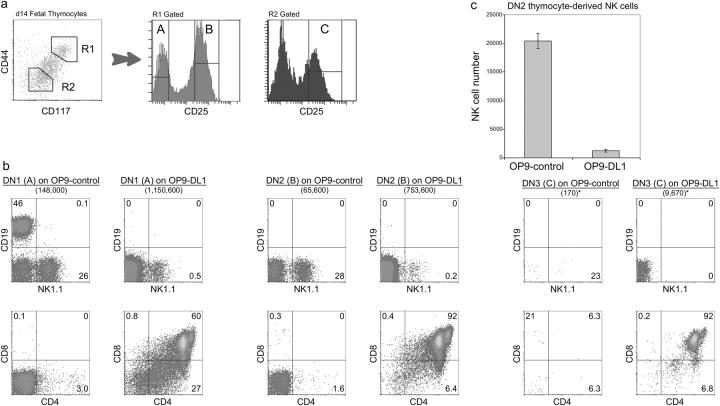
Figure 1.
Developmental potential of early DN thymocytes cultured on OP9 bone marrow stromal cells in the presence (OP9-DL1) or absence (OP9-control) of Dll1 expression. (a) Day 14 fetal thymocytes were sorted for the following populations: DN1 (R1-gated region A, CD117+ CD44+ CD25−), DN2 (R1-gated region B, CD117+ CD44+ CD25+), and DN3 (R2-gated region C, CD117− CD44− CD25+). (b) Thymocyte populations, sorted as indicated, were cultured (1,000 cells/well) on OP9-control cells or OP9-DL1 cells for 7 d, and then analyzed for surface expression of the indicated lineage markers by flow cytometry. Total cellularity is indicated in parentheses below each label. *, samples that could not be accurately counted by hemocytometer, and for which cell counts were determined flow cytometrically. (c) Total number of DN2-derived NK cells generated from DN2 thymocytes cultured on OP9-control and OP9-DL1 stromal cell lines.
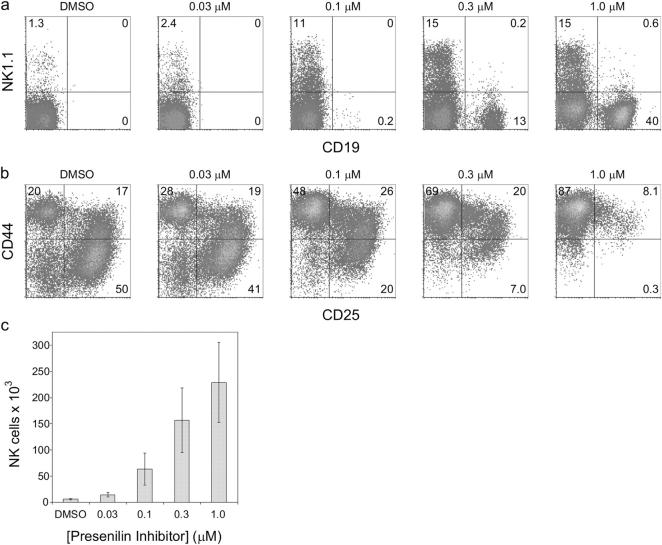
Figure 4.
Inhibition of Notch signaling blocks T cell development and promotes NK cell development. CD117+ Sca-1hi day 14 fetal liver HPCs were cultured on OP9-DL1 cells for 7 d with DMSO alone, or in the presence of Inhibitor X, a presenilin 1/2 inhibitor, at the indicated concentrations. (a) Cells from these cultures were analyzed by flow cytometry for the presence of CD19+ NK1.1− B cells and CD19− NK1.1+ NK cells. (b) CD19− cells from these cultures were electronically gated and analyzed for the expression of CD44 and CD25. (c) Total number of NK cells generated from 1.5 × 103 HPCs cultured on OP9-DL1 stromal cell lines for 7 d in the absence (DMSO alone) or presence of Inhibitor X at the indicated concentrations. The data are representative of at least three independent trials.
Precursor Frequency Analysis.
Limiting dilution analysis was performed using the Clonecyte option of the FACSDiVa™ cell sorter. As mentioned above, IL-6, IL-7, IL-15, and Flt3 ligand were added to these assays to enhance the growth of T, B, and NK cells. Precisely 1, 3, 10, 20, 30, or 100 DN1 and DN2 thymocytes were deposited into 96-well plates containing OP9 cell monolayers, with 56 replicate wells for each sample group of DN1 cells and 53 replicate wells for each sample group of DN2 cells. Limiting dilution analysis was also performed using OP9-DL1 cells by sorting 1, 3, 10, or 30 DN1 and DN2 thymocytes into wells of 96-well plates containing OP9-DL1 cell monolayers, with 24 replicate wells for each sample group. After 12–14 d, the cells were harvested from individual wells and analyzed by flow cytometry. The presence of CD45+ NK1.1+ CD19− NK cells, CD45+ NK1.1− CD19+ B cells, and CD45+ NK1.1− CD3+ T cells was scored and the progenitor frequency was determined by the method of maximum likelihood applied to the Poisson model (41). Clonal assays were performed by depositing single DN1 or DN2 thymocytes into individual wells of 96-well plates containing OP9-DL1 cells by flow cytometric cell sorting. The percentage of T cell progenitors, NK cell progenitors, and T/NK bipotent progenitors within the DN1 and DN2 thymocyte populations was determined flow cytometrically by analyzing the phenotype of cell populations derived from single cells after 11–14 d of culture (NK cell progenitors gave rise only to CD3− NK1.1+ NK cells, T cell progenitors gave rise only to CD3+ T cells, and T/NK bipotent progenitors gave rise to both CD3− NK1.1+ NK cells and CD3+ T cells).
PCR and RT-PCR.
Genomic DNA was purified using the EasyDNA kit (Invitrogen) from total day 14 fetal thymus, ex vivo DN2 thymocytes, T cells derived from DN2 thymocytes cultured on OP9-DL1, CD45+ NK1.1+ CD19− NK cells sorted from DN2 cells cultured on OP9 cells for 14 d, and from embryonic fibroblasts. 100 ng of each DNA sample was amplified using a PTC-225 Peltier Thermal Cycler (MJ Research). Primers used for the TCR D-Jβ rearrangement analysis have been described previously (42). Products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. All PCR products shown correspond to expected molecular sizes.
Immunofluorescent Staining of Thymus Sections.
Transverse frozen sections of 5–6-wk-old C57BL/6 mouse thymus (5-μm thickness) were prepared by embedding the tissue in OCT (Fisher Scientific) followed by cryostatic sectioning onto glass slides. Sections were fixed in ice-cold acetone, dried, and rehydrated in PBS. Endogenous avidin binding activity was blocked using Avidin/Biotin Blocking Kit (Vector Laboratories). Nonspecific protein binding was blocked using PBS containing 5% FBS. This buffer was also used as diluent for all subsequent steps. Next, sections were stained with primary antibody, either goat polyclonal IgG recognizing Dll1 (catalog no. SC9932; Santa Cruz Biotechnology, Inc.), or a matching concentration of nonimmune goat IgG (Jackson ImmunoResearch Laboratories). Sections were washed extensively and stained with biotinylated donkey antiserum raised against goat IgG (Jackson ImmunoResearch Laboratories), followed by blocking of excess secondary antibody binding sites using goat IgG. Sections were then stained with a combination of streptavidin-conjugated Alexa 594 (Molecular Probes) and FITC-conjugated anti-pancytokeratin (catalog no. F3410; Sigma-Aldrich). After washing, nuclei were counterstained by mounting in Prolong solution (Molecular Probes) containing DAPI (Molecular Probes). Single color images (DAPI, FITC, and Alexa 594) were acquired on a fluorescent microscope. Exposure times for each fluorochrome were kept the same for both DL1 and control (nonspecific) primary antibodies. After acquisition, single color images were merged to generate RGB composites.
Online Supplemental Material.
Inhibition of Notch signaling does not affect B or NK cell development on OP9-control cells. See Fig. S1, which is available at http://www.jem.org/cgi/content/full/jem.20040394/DC1.
Results
Requirement for Notch Receptor–Dll1 Interactions throughout Early T Cell Development.
To characterize the requirement for Notch receptor–ligand interactions during early thymocyte development, we sorted each of the three DN thymocyte subsets before β selection and cultured them (1,000 cells/well) on either OP9-control or OP9-DL1 cells for 7 d in conditions that effectively support T, B, and NK cell growth (Fig. 1). As described previously, DN1 cells contain multipotent lymphoid progenitors (27, 43), which, when cultured on OP9-control cells, gave rise to B and NK cells, but not T cells. In contrast, DN1 thymocytes cultured on OP9-DL1 cells gave rise to both T and NK cells, but not B cells (Fig. 1 b, left). DN1 cells proliferated on both stromal cell lines; however, in the presence of Dll1, this subset proliferated to a greater extent (148- vs. 1,150-fold expansion on OP9-control vs. OP9-DL1 cells, respectively; Fig. 1 b).
Consistent with the notion that DN2 progenitors are committed to the T cell lineage, this population failed to give rise to B cells when cultured on OP9-control cells, and T cells were observed almost exclusively when DN2 thymocytes were cultured on OP9-DL1 cells (Fig. 1 b, middle). However, DN2 progenitors gave rise to a substantial number of NK cells when cultured on OP9-control cells, whereas only a few NK cells were generated from DN2 cells cultured on OP9-DL1 cells (∼15-fold more NK cells on OP9-control vs. OP9-DL1 cells, respectively; Fig. 1 c). Therefore, our data supports the view that DN2 thymocytes may not be fully committed to the T cell lineage, and that these cells retain some plasticity toward the NK cell lineage when cultured in an environment that does not support further T cell commitment.
In contrast, DN3 cells, which bear irreversible hallmarks of T lineage commitment in the form of TCR-β gene rearrangements (34), were no longer capable of adopting an NK cell fate, failed to differentiate or expand on OP9-control cells (Fig. 1 b, right), and instead underwent cell death (not depicted). On the other hand, DN3 cells cultured on OP9-DL1 gave rise exclusively to T cells that progressed normally to the DP stage (Fig. 1 b, bottom right). These data show that a requirement for Notch signaling persists at the DN3 stage, even though these cells are already committed to the T cell lineage, implying a role for Notch signaling in functions other than lineage commitment (44).
Requirement for Notch Receptor–Dll1 Interactions during T Cell Lineage Commitment by DN2 Thymocytes.
The results in Fig. 1 b show that DN2 thymocytes do not appear to be fully committed to the T cell lineage, as this population retained the potential to give rise to an alternate cell fate under conditions that do not support T cell development. To determine the frequency of DN2 thymocytes that can differentiate into NK cells, we performed a limiting dilution analysis with DN2 and DN1 thymocytes cultured on OP9 cells. The results presented in Table I indicate that the NK cell progenitor frequency within the DN1 population (1 in 8.8 DN1 cells) is similar to that of the DN2 population (1 in 13.6 DN2 cells). This analysis also revealed that the B cell progenitor frequency within the DN1 subset appears to be surprisingly small (1 in 188), and as demonstrated in Fig. 1 b, DN2 cells do not contain any B cell lineage potential. Taken together, these data suggest that the majority of cells within the DN1 population are either committed T cell progenitors or bipotent T/NK cell progenitors, and that although the transition from the DN1 to DN2 stage results in a loss of B cell potential, it does not result in a substantial loss of NK cell lineage potential. This, in turn, suggests that continued Notch signals specifying T cell lineage fate are required at the DN2 stage.
Table I.
Progenitor Frequency Analysis for DN1 and DN2 Thymocytes Cultured on OP9 Cells
| Subset → lineage analyzeda | Progenitor Frequency−1[95% confidence limits]b |
|---|---|
| DN1 → B cells | 188 [133–266] |
| DN1 → NK cells | 8.8 [7.2–10.7] |
| DN2 → NK cells | 13.6 [11.0–16.7] |
Individual wells (n = 56 wells with DN1 cells or n = 53 wells with DN2 cells for each of the titrations, 100, 30, 20, 10, 3, and 1 cell/well) were analyzed for the generation of B cells (CD19+) and NK cells (NK1.1+; see Fig. 1).
Statistical analysis was performed using the method of maximum likelihood applied to the Poisson model.
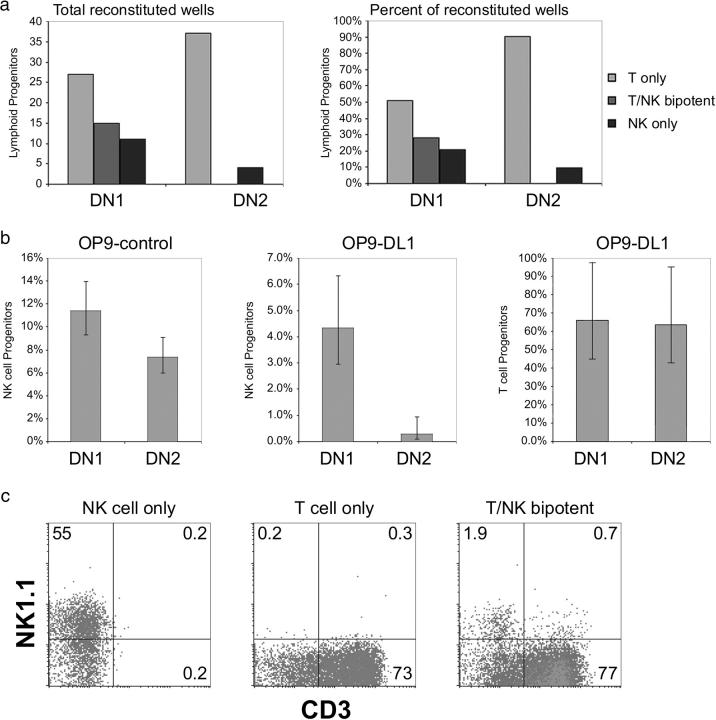
To determine the relative frequency of committed T cell progenitors, NK cell progenitors, and bipotent T/NK cell progenitors within the early DN thymocyte subsets, we performed a clonal assay, in which single DN1 or DN2 thymocytes were placed into individual wells containing OP9-DL1 cells (Fig. 2). T cell lineage potential was determined by CD3 expression, and NK cell lineage potential was determined by the expression of NK1.1 in the absence of CD3 (Fig. 2 c). The frequency of T cell progenitors, NK cell progenitors, and bipotent T/NK cell progenitors within the DN1 and DN2 populations is shown in Fig. 2 a. These results indicate that the DN1 and DN2 subsets both contain a relatively high frequency of progenitors that give rise exclusively to T cells (27 out of 192 wells or ∼51% of the total number of reconstituted wells for DN1; 37 out of 192 wells or ∼90% of reconstituted wells for DN2), whereas the DN1 subset contained a greater percentage of NK cell lineage–only progenitors than the DN2 subset (11 out of 192 wells or ∼21% of reconstituted for DN1; 4 out of 192 wells or ∼10% of reconstituted for DN2). Notably, bipotent T/NK cell progenitors were only observed in wells seeded with DN1 thymocytes (15 out of 192 wells or ∼28% of reconstituted), whereas bipotent T/NK cell progenitors were not detected within the DN2 thymocyte subset (Fig. 2 a).
Figure 2.
Clonal analysis of T and/or NK cell progenitor potential from DN1 and DN2 thymocytes cultured on OP9 bone marrow stromal cells in the presence (OP9-DL1) or absence (OP9-control) of DL1 expression. (a) Single DN1 (CD117+ CD44+ CD25−) and DN2 (CD117+ CD44+ CD25+) thymocytes were sorted into 96-well plates containing OP9-DL1 cells, and then cultured for up to 14 d. Clonal populations were analyzed by flow cytometry to determine the developmental potential of the individually sorted cells (as shown in c). Wells that contained only CD3− NK1.1+ NK cells were scored as NK restricted, wells that contained only CD3+ T cells were scored as T cell restricted, and wells that contained both CD3− NK1.1+ NK cells and CD3+ T cells were scored as T/NK cell bipotent. 192 wells, each containing a single DN1 or DN2 cell, were analyzed for each population, and the total number of wells scoring positive for the presence of T, NK, or both T and NK cells are illustrated (left). The frequency of each progenitor type is also presented as a percentage of the total number of wells that successfully reconstituted (right). (b) A comparison of the NK cell progenitor frequency observed for DN1 and DN2 cells when cultured on either OP9-control (left) or OP9-DL1 cells (middle), and the T cell progenitor frequency observed for DN1 and DN2 cells when cultured on OP9-DL1 cells (right). The data shown were obtained from the limiting dilution analysis presented in Tables I and II. (c) Representative flow cytometric analysis of individual wells seeded with a single cell with NK cell (left), T cell (middle), and T/NK bipotent (right) potential.
These results appear in contrast to the limiting dilution data presented in Table I, which clearly demonstrated a similar progenitor frequency for NK cells (either bipotent or NK cell lineage committed) within the first two DN thymocyte subsets. To determine whether the presence of Dll1 influences the ability of DN2 thymocytes to adopt the NK cell fate, an additional limiting dilution analysis was performed using OP9-DL1 cells to determine the progenitor frequency for T and NK cells within the DN1 and DN2 subsets (Table II).
Table II.
Progenitor Frequency Analysis for DN1 and DN2 Thymocytes Cultured on OP9-DL1 Cells
| Subset → lineage analyzeda | Progenitor frequency−1[95% confidence limits]b |
|---|---|
| DN1 → NK cells | 23.1 [15.8–33.8] |
| DN2 → NK cells | 340 [107–1,080] |
| DN1 → T cells | 1.51 [1.03–2.21] |
| DN2 → T cells | 1.56 [1.07–2.28] |
Individual wells (n = 24 wells for each of the titrations, 30, 10, 3, and 1 cell/well) were analyzed for the generation of T cells (CD3+) and NK cells (CD3− NK1.1+; see Fig. 2 c).
Statistical analysis was performed using the method of maximum likelihood applied to the Poisson model.
The data shown in Fig. 2 b provides a comparison of the NK cell progenitor frequencies from the limiting dilution analysis of each population (from Tables I and II) when cultured on either control-OP9 cells (Fig. 2 b, left) or OP9-DL1 cells (Fig. 2 b, middle), as well as the T cell progenitor frequency for each population (Fig. 2 b, right). This analysis shows that the NK cell progenitor frequency was only slightly decreased for DN2 thymocytes as compared with DN1 thymocytes when cultured on OP9-control cells (11.4 to 7.4%, equivalent to only a ∼1.5-fold higher NK cell progenitor frequency in the DN1 subset), whereas there was a striking loss of NK cell progenitor frequency between the two thymocyte subsets when assayed on OP9-DL1 (4.3 to 0.3%, equivalent to a ∼15-fold higher NK cell progenitor frequency in the DN1 subset). As demonstrated in Fig. 2 a, the only NK cell potential detectable from single DN2 cells cultured on OP9-DL1 cells was derived from a small number of contaminating NK cell progenitors, suggesting that a proportion of committed NK cell progenitors in the thymus may express CD25, and thus cannot be distinguished from DN2 thymocytes. Therefore, the relatively high NK cell progenitor frequency observed from the DN2 population when cultured on control OP9 cells is likely derived from T cell progenitors, which can adopt an NK cell fate in the absence of continued Notch receptor–ligand interactions. In contrast, the progenitor frequency for T cells is not significantly different between the DN1 and DN2 subsets (66 and 64%, respectively) when cultured on OP9-DL1 cells (Fig. 2 b, right).
Notably, the NK cell progenitor frequency of the DN2 population is ∼25-fold higher when assayed on OP9-control versus OP9-DL1 cells. It is also important to note that the measured frequency of NK cell progenitors within the DN1 population is ∼2.5-fold higher when cultured on OP9-control versus OP9-DL1 cells, indicating that although T/NK cell bipotent progenitors are readily detected from DN1 cells cultured on OP9-DL1 (Fig. 2 a), Notch signaling acts at the DN1 stage to promote T lineage specification at the expense of NK cell development. Taken together, these data indicate that Notch receptor–ligand interactions direct T cell development throughout the DN1 and DN2 stages, and that the DN2 population contains progenitors that are specified to the T cell lineage, and thus require continuous Notch signaling both to maintain T lineage specification and to undergo final lineage commitment.
Status of the TCR-β Gene Rearrangement in NK Cells Derived from DN2 Cells.
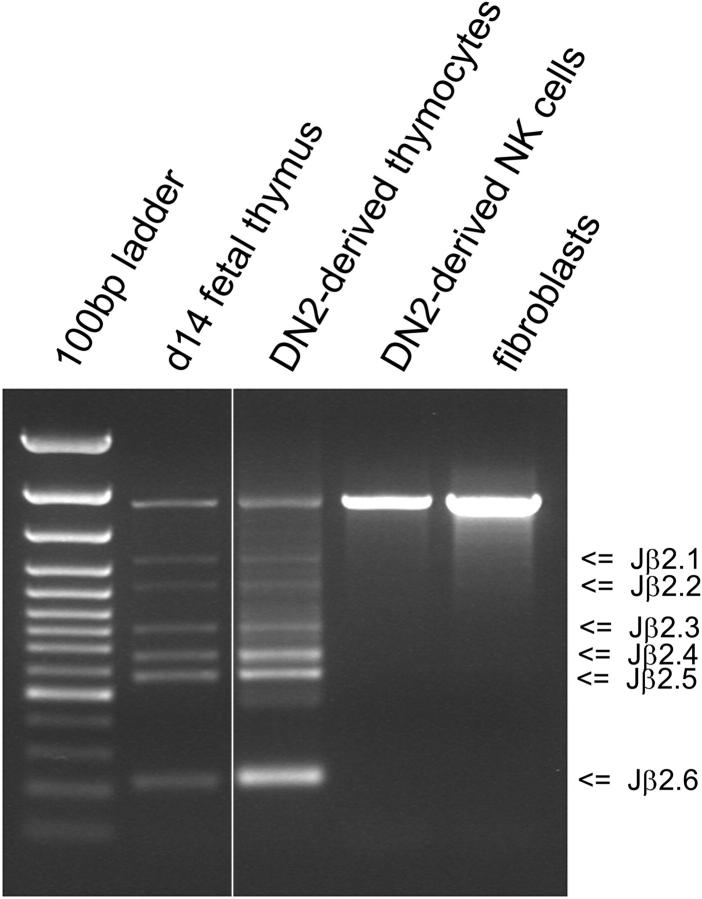
Progenitor thymocytes begin the process of TCR gene rearrangement during the DN2 to DN3 transition (33, 34, 45, 46). Once thymocytes rearrange their TCR genes, they are considered irreversibly committed to the T cell lineage. The TCR-β gene locus of DN2 cells is in the germline configuration, although a small percentage of these cells have initiated the rearrangement process and display TCR D-Jβ rearrangements (33, 34, 45, 46). To determine which DN2 cells are capable of giving rise to NK cells when cultured on OP9 cells lacking Dll1 expression, DN2 thymocytes were cultured for 2 wk on OP9 cells, and the resulting NK cells were analyzed for D-Jβ rearrangement by PCR (Fig. 3). Multiple D-Jβ rearrangements were observed from fetal thymus–derived DNA, and from DNA derived from DN2 thymocytes cultured on OP9-DL1. In contrast, the genomic DNA from DN2-derived NK cells produced a single PCR product corresponding to the expected size of the germline locus, like that observed from the DNA of fibroblasts (Fig. 3). These results indicate that DN2 thymocytes that are removed from a Notch ligand–containing environment are unable to undergo TCR gene rearrangement and furthermore, DN2 thymocytes that have begun TCR gene rearrangement may no longer be capable of adopting an NK cell fate when transferred to an environment lacking Notch receptor–ligand interactions.
Figure 3.
PCR analysis of DN2-derived NK cells for TCR-β gene rearrangements. DN2 (CD117+ CD44+ CD25+) thymocytes were cultured on OP9-control or OP9-DL1 cells for 14 d. DNA was purified from total DN2-derived T cells (from OP9-DL1 cultures) and from DN2-derived NK cells (from OP9-control cultures). The rearrangement status of the TCR-β gene locus was determined by PCR using Dβ2-Jβ2 specific primers. PCR reactions containing DNA from day 14 fetal thymus and fibroblast DNA were included as TCR-rearranged and germline controls, respectively.
Different Thresholds of Notch Signaling Are Required during T Cell Lineage Commitment and Development.
To further elucidate the effect of Notch signaling during the initial stages of T cell lineage commitment, we cultured CD117+ Sca-1hi fetal liver–derived HPCs on OP9-DL1 cells in the presence of increasing concentrations of a presenilin 1 and 2 inhibitor. A presenilin-dependent γ secretase activity is required for the cleavage of the intracellular transactivating domain of all four mammalian Notch receptors in response to engagement by ligand (47). Fig. 4 shows that HPCs cultured on OP9-DL1 cells in the absence of the presenilin 1/2 inhibitor (DMSO alone) failed to give rise to B lineage CD19+ cells, while efficiently generating primarily DN3 and pre-DP/DP T lineage cells and a small percentage of NK1.1+ NK cells after 7 d of culture. However, in the presence of increasing concentrations of the inhibitor there was a striking increase in the proportion of NK cells generated on OP9-DL1 cells (Fig. 4 a), which also correlated with a substantial increase in the absolute number of NK cells in the cultures (Fig. 4 c). Importantly, the effect on T versus NK cell development occurred at lower concentrations of inhibitor than was necessary to allow B cells to develop (Fig. 4 a). Furthermore, progression through the DN stages of T cell development was also impaired or delayed in the presence of low levels of inhibitor (Fig. 4 b). These results suggest that progenitors require relatively high levels of Notch signaling to initiate commitment to the T cell lineage and progress through the DN stages of development, such that at even low inhibitor concentrations, an increase in the generation of NK cells becomes apparent. On the other hand, it appears that the adoption of the B cell lineage by HPCs is sensitive to even low levels of Notch signaling, such that B cells only develop in the presence of the highest concentrations of the presenilin 1/2 inhibitor. The addition of presenilin 1/2 inhibitor to HPCs cultured on OP9-control cells had no effect on NK or B cell development (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20040394/DC1).
The finding that continuous Notch signaling is required for T cell commitment and during the initial DN stages of T cell development suggests that a Notch ligand must be expressed throughout the thymic cortex, where early T cell development occurs. Therefore, we analyzed transverse sections of adult thymus for the expression of Dll1 (Fig. 5). Cortical epithelial cells, which are present throughout the thymic cortex, were found to express Dll1, particularly near the cortico-medullary junction, as staining for Dll1 and cytokeratin colocalized on the same stromal cells. These findings are consistent with those reported by Harman et al. (48), in which transcripts for the different Notch ligands, including Dll1, were shown to be expressed by thymic stromal cells. Because cortical stromal cells provide the matrix for adhesion and migration of DN cells across the cortex (49), this pattern of Dll1 expression shows that Dll1 is in the position to provide the necessary Notch signals that induce and support the commitment and early development of T cells within the thymus.
Figure 5.
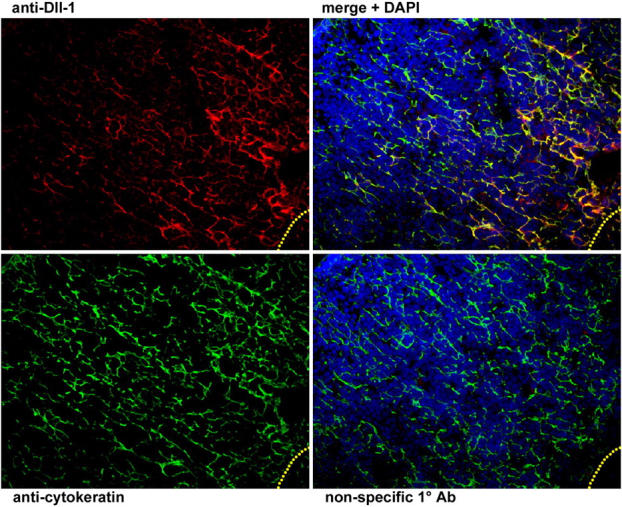
Analysis of Dll1 expression by thymic epithelial cells. Immunofluorescent staining of thymus sections reveals that Dll1 (top left, red) is expressed in a reticular pattern in the thymic cortex (in all panels, the capsule is in the extreme top left corner and the medulla begins in the bottom right corner, with the cortico-medullary junction indicated by the yellow dotted line). This pattern of expression is mirrored by cortical stromal cells that express cytokeratin (bottom left, green), as can be seen when the Dll1 and cytokeratin images are merged (top right, counterstained with DAPI, blue). Expression of Dll1 generally appears to be strongest in the deep cortex, but is present throughout the cortex on most cytokeratin+ stromal cells. The bottom right panel shows a serial section of the same thymus stained with a nonspecific control antibody of the same isotype and concentration as the Dll1 antibody.
Discussion
In this work, we demonstrate that Notch signals are required at each of the early DN stages of thymocyte development because the DN1-3 subsets could progress to subsequent T cell developmental stages only when cultured on stromal cells expressing Dll1, but not when transferred to OP9 cells that lack Dll1 expression. The DN2 population is generally thought to represent the earliest thymocyte population that is committed to the T cell lineage (50). Therefore, we were surprised to find that a robust population of NK cells developed from DN2 cells cultured on OP9-control cells. This observation suggested that DN2 cells retain some plasticity under certain conditions. In fact, previous studies have also noted that DN2 thymocytes may possess some potential for NK cell development (19, 27, 51). However, the contribution of Notch signals to the NK cell progenitor frequency from early DN thymocytes was not addressed.
As opposed to previously reported assay systems (19), the OP9/OP9-DL1 cell lines represent a simple, well-defined system that is ideally suited to address such questions (52). Collectively, these cell lines can support the differentiation of all lymphocyte lineages coupled with robust cellular expansion, allowing limiting dilution analysis and powerful clonal assays to be performed. Using this approach, we were surprised to find that both DN1 cells and DN2 cells displayed a similar, relatively high NK cell progenitor frequency when cultured in the absence of Dll1-induced signals. Furthermore, we observed a very low B cell progenitor frequency (∼0.5%) from the DN1 population, indicating that the purported common lymphoid progenitor within the DN1 population must be relatively rare or may not exist within the fetal thymus (53).
To determine whether the NK cell potential within the DN1 and DN2 populations was derived from T/NK cell bipotent or NK cell lineage–committed progenitors, a clonal assay using OP9-DL1 cells was used. Although our results revealed that NK cell–committed precursors could be found in both populations, we found that DN1 cells have twice the frequency of NK cell–committed progenitors than that observed for DN2 cells (Fig. 2). However, although ∼60% of the total NK cell progenitor activity from DN1 cells was derived from T/NK cell bipotent progenitors, a striking absence of bipotent progenitors was noted from the DN2 subset, such that the only NK cell potential was derived from a small number of likely contaminating NK cell–committed progenitors. These results indicate that DN2 cells, which were shown to possess substantial NK cell lineage potential when cultured on OP9-control cells (Fig. 1 b and Table I), are unable to effectively adopt an NK cell fate when cultured in the presence of Dll1.
Limiting dilution analyses of DN1 and DN2 thymocytes cultured on OP9-DL1 cells revealed a decrease in the observed NK cell progenitor frequency for both the DN1 and DN2 subsets than that observed on OP9-control cells. These results are consistent with the notion that Notch acts to direct T cell fate throughout early thymocyte development. However, Notch receptor–ligand interactions resulted in a dramatic 25-fold reduction in the frequency of NK cell lineage potential from DN2 thymocytes, whereas the effect of Notch was less pronounced at the DN1 stage, resulting in only an ∼2.5-fold decrease in the frequency of NK cell lineage potential in the presence of Dll1. Taken together, these findings point to a mechanism whereby Notch receptor–ligand interactions maintain T cell specification by suppressing the remaining potential of DN2 thymocytes to adopt an NK cell fate.
Our data suggests that the DN1 subset contains bipotent T/NK cell progenitors that undergo a stochastic cell fate choice, which might be biased toward the T cell lineage in the presence of Notch signaling. On the other hand, specified DN2 thymocytes can only adopt the NK cell lineage when Notch receptor–ligand interactions are interrupted, such as by transfer to OP9 cells that lack Dll1 expression. These findings shed light on earlier studies in which DN2 cells cultured in fetal thymic organ culture developed predominantly into T cells (27), or in which a low frequency of NK cell lineage potential could be detected from DN2 cells using a modified (yet still able to strongly induce Notch signals) fetal thymic organ culture system (19). As demonstrated by Wolfer et al. (5), Notch1 signaling is also essential for TCR-β gene rearrangement, which likely coincides with T cell lineage commitment. This notion is supported by our findings that the TCR-β gene locus of DN2-derived NK cells is in the germline configuration (Fig. 3), indicating that DN2 thymocytes with TCR-D-Jβ rearrangements may not readily adopt an NK cell fate.
Changes in the strength and duration of Notch signals can influence cell fate choices during development (54). We demonstrate that low levels of pharmacological inhibition of Notch signaling during fetal liver–derived HPC differentiation on OP9-DL1 cells results in a partial block of the T cell lineage developmental pathway, and preferentially promotes the development of NK cells (Fig. 4). In contrast, B cells develop on OP9-DL1 cells only in the presence of high concentrations of the presenilin 1/2 inhibitor. Importantly, lymphoid progenitors in the fetal liver express a level of Notch1 comparable to fetal DN1 and DN2 thymocytes (11). Thus, it is likely that the observed differences in sensitivity to the presenilin 1/2 inhibitor correlates with the sensitivity to Notch signaling at different developmental stages.
In contrast to the established role that Notch plays in T versus B cell fate determination, the subsequent effect that Notch signaling may play in T versus NK cell differentiation has been overlooked. However, a number of studies have demonstrated the presence of lymphoid progenitors that are restricted to the T and NK cell lineages at various extrathymic sites of hematopoietic development (12–16, 55). The notion that these progenitors require Notch signaling to develop is supported by the well-established role of Notch in blocking B cell potential, while directing the differentiation of multipotent progenitors toward the T lineage (4). However, Harman et al. (11) have recently demonstrated that IL-7Rα+ Lin− lymphoid progenitors from the fetal liver exhibit only low levels of Notch activation, and migrant lymphoid precursors in the perithymic mesenchyme of day 12 embryos lack clear evidence of Notch activation before thymic entry. Thus, it remains unclear when T cell progenitors that seed the thymus lose B cell lineage potential.
Our results show that inhibition of B lymphopoiesis requires low levels of Notch signaling, whereas the induction of T lymphopoiesis requires high levels of Notch signaling. Therefore, entry into the thymus may not be required to extinguish B cell potential. Rather, HPCs may lose B cell lineage potential in response to low levels of Notch activation in the fetal liver or in the adult bone marrow, but are unable to undergo further T lineage differentiation before thymic entry. Thus, several days may separate the Notch signal that results in the loss of B cell potential and the sustained Notch signaling that leads to T lineage commitment. These distinct and temporally separable Notch signaling events are consistent with our new understanding of the time period (up to 10 d) required for DN1 cells to transit to the DN2 stage within the adult thymus (56).
The requirement for high levels of Notch ligand–induced signaling for the maintenance of T cell development within the thymus is confirmed by our results showing that Dll1 is expressed throughout the thymic cortex and, in particular, found to be expressed at higher levels near the cortico-medullary junction (Fig. 5), which is the site where incoming progenitors would first encounter the thymic stroma (57). Our data does not rule out that other unknown factors, including other Delta-like family members (such as Delta-like 4), may also play an important role in the induction and maintenance of T cell development in the thymus. Nevertheless, our findings suggest that thymocytes require constant contact with Notch ligand–bearing thymic epithelial cells to progress through early T cell differentiation. In keeping with this, it has recently been shown that thymocytes use cortical stromal cells as a matrix for transmigration across the cortex during differentiation, and thus maintain intimate contact with such stromal cells throughout their differentiation (49). Because these are the same stromal cells that we now find to express Dll1, it appears that one critical reason for using a stromal cell matrix for migration rather than an extracellular one, is to enforce continuous Notch signaling throughout the differentiation process.
Acknowledgments
We thank S. Palencia (Memorial Sloan-Kettering Cancer Center) for the immunofluorescent staining of thymic sections and G. Knowles for her expert assistance in flow cytometry and cell sorting.
J.C. Zúñiga-Pflücker is supported by an Investigator Award from the Canadian Institute of Health Research (CIHR) and M. Ciofani is supported by a Studentship Award from the CIHR. This work was supported by funds from the CIHR.
The authors have no conflicting financial interests.
Abbreviations used in this paper: Dll1, Delta-like 1; DN, double negative; DP, double positive; HPC, hematopoietic progenitor cell.
References
- 1.Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnell, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, et al. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. [DOI] [PubMed] [Google Scholar]
- 2.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 3.Jaleco, A.C., H. Neves, E. Hooijberg, P. Gameiro, N. Clode, M. Haury, D. Henrique, and L. Parreira. 2001. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J. Exp. Med. 194:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pear, W.S., and F. Radtke. 2003. Notch signaling in lymphopoiesis. Semin. Immunol. 15:69–79. [DOI] [PubMed] [Google Scholar]
- 5.Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 16:869–879. [DOI] [PubMed] [Google Scholar]
- 6.Washburn, T., E. Schweighoffer, T. Gridley, D. Chang, B.J. Fowlkes, D. Cado, and E. Robey. 1997. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 88:833–843. [DOI] [PubMed] [Google Scholar]
- 7.Fowlkes, B.J., and E.A. Robey. 2002. A reassessment of the effect of activated Notch1 on CD4 and CD8 T cell development. J. Immunol. 169:1817–1821. [DOI] [PubMed] [Google Scholar]
- 8.Izon, D.J., J.A. Punt, L. Xu, F.G. Karnell, D. Allman, P.S. Myung, N.J. Boerth, J.C. Pui, G.A. Koretzky, and W.S. Pear. 2001. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 14:253–264. [DOI] [PubMed] [Google Scholar]
- 9.Robey, E., D. Chang, A. Itano, D. Cado, H. Alexander, D. Lans, G. Weinmaster, and P. Salmon. 1996. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 87:483–492. [DOI] [PubMed] [Google Scholar]
- 10.Deftos, M.L., and M.J. Bevan. 2000. Notch signaling in T cell development. Curr. Opin. Immunol. 12:166–172. [DOI] [PubMed] [Google Scholar]
- 11.Harman, B.C., E.J. Jenkinson, and G. Anderson. 2003. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J. Immunol. 170:1299–1303. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto, H., T. Ikawa, K. Ohmura, S. Fujimoto, and Y. Katsura. 2000. T cell progenitors emerge earlier than B cell progenitors in the murine fetal liver. Immunity. 12:441–450. [DOI] [PubMed] [Google Scholar]
- 13.Douagi, I., F. Colucci, J.P. Di Santo, and A. Cumano. 2002. Identification of the earliest prethymic bipotent T/NK progenitor in murine fetal liver. Blood. 99:463–471. [DOI] [PubMed] [Google Scholar]
- 14.Jaleco, A.C., B. Blom, P. Res, K. Weijer, L.L. Lanier, J.H. Phillips, and H. Spits. 1997. Fetal liver contains committed NK progenitors, but is not a site for development of CD34+ cells into T cells. J. Immunol. 159:694–702. [PubMed] [Google Scholar]
- 15.Ikawa, T., K. Masuda, M. Lu, N. Minato, Y. Katsura, and H. Kawamoto. 2004. Identification of the earliest prethymic T-cell progenitors in murine fetal blood. Blood. 103:530–537. [DOI] [PubMed] [Google Scholar]
- 16.Carlyle, J.R., and J.C. Zúñiga-Pflücker. 1998. Requirement for the thymus in αβ T lymphocyte lineage commitment. Immunity. 9:187–197. [DOI] [PubMed] [Google Scholar]
- 17.Carlyle, J.R., A.M. Michie, C. Furlonger, T. Nakano, M.J. Lenardo, C.J. Paige, and J.C. Zúñiga-Pflücker. 1997. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J. Exp. Med. 186:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodewald, H.R., P. Moingeon, J.L. Lucich, C. Dosiou, P. Lopez, and E.L. Reinherz. 1992. A population of early fetal thymocytes expressing FcγRII/III contains precursors of T lymphocytes and natural killer cells. Cell. 69:139–150. [DOI] [PubMed] [Google Scholar]
- 19.Ikawa, T., H. Kawamoto, S. Fujimoto, and Y. Katsura. 1999. Commitment of common T/natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J. Exp. Med. 190:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez, M.J., H. Spits, L.L. Lanier, and J.H. Phillips. 1993. Human natural killer cell committed thymocytes and their relation to the T cell lineage. J. Exp. Med. 178:1857–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Res, P., E. Martinez-Caceres, A. Cristina Jaleco, F. Staal, E. Noteboom, K. Weijer, and H. Spits. 1996. CD34+ CD38dim cells in the human thymus can differentiate into T, natural killer, and dendritic cells but are distinct from pluripotent stem cells. Blood. 87:5196–5206. [PubMed] [Google Scholar]
- 22.Blom, B., P. Res, E. Noteboom, K. Weijer, and H. Spits. 1997. Prethymic CD34+ progenitors capable of developing into T cells are not committed to the T cell lineage. J. Immunol. 158:3571–3577. [PubMed] [Google Scholar]
- 23.Anderson, G., N.C. Moore, J.J. Owen, and E.J. Jenkinson. 1996. Cellular interactions in thymocyte development. Annu. Rev. Immunol. 14:73–99. [DOI] [PubMed] [Google Scholar]
- 24.Godfrey, D.I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244–4252. [PubMed] [Google Scholar]
- 25.Allman, D., A. Sambandam, S. Kim, J.P. Miller, A. Pagan, D. Well, A. Meraz, and A. Bhandoola. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174. [DOI] [PubMed] [Google Scholar]
- 26.Michie, A.M., J.R. Carlyle, T.M. Schmitt, B. Ljutic, S.K. Cho, Q. Fong, and J.C. Zúñiga-Pflücker. 2000. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J. Immunol. 164:1730–1733. [DOI] [PubMed] [Google Scholar]
- 27.Moore, T.A., and A. Zlotnik. 1995. T-cell lineage commitment and cytokine response of thymic progenitors. Blood. 86:1850–1860. [PubMed] [Google Scholar]
- 28.Zúñiga-Pflücker, J.C., J. Di, and M.J. Lenardo. 1995. Requirement for TNF-α and IL-1α in fetal thymocyte commitment and differentiation. Science. 268:1906–1909. [DOI] [PubMed] [Google Scholar]
- 29.Anderson, M.K., G. Hernandez-Hoyos, R.A. Diamond, and E.V. Rothenberg. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 126:3131–3148. [DOI] [PubMed] [Google Scholar]
- 30.Wu, L., C.L. Li, and K. Shortman. 1996. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 184:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardavin, C., L. Wu, L. Chung-Leung, and K. Shortman. 1993. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 362:761–763. [DOI] [PubMed] [Google Scholar]
- 32.Shen, H.Q., M. Lu, T. Ikawa, K. Masuda, K. Ohmura, N. Minato, Y. Katsura, and H. Kawamoto. 2003. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J. Immunol. 171:3401–3406. [DOI] [PubMed] [Google Scholar]
- 33.Godfrey, D.I., J. Kennedy, P. Mombaerts, S. Tonegawa, and A. Zlotnik. 1994. Onset of TCR-β rearrangement and role of TCR-β expression during CD3−CD4−CD8− thymocyte differentiation. J. Immunol. 152:4783–4792. [PubMed] [Google Scholar]
- 34.Petrie, H.T., F. Livak, D. Burtrum, and S. Mazel. 1995. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J. Exp. Med. 182:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudley, E.C., H.T. Petrie, L.M. Shah, M.J. Owen, and A.C. Hayday. 1994. T cell receptor β chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1:83–93. [DOI] [PubMed] [Google Scholar]
- 36.Petrie, H.T., P. Hugo, R. Scollay, and K. Shortman. 1990. Lineage relationships and developmental kinetics of immature thymocytes: CD3, CD4, and CD8 acquisition in vivo and in vitro. J. Exp. Med. 172:1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolic-Zugic, J., M.W. Moore, and M.J. Bevan. 1989. Characterization of the subset of immature thymocytes which can undergo rapid in vitro differentiation. Eur. J. Immunol. 19:649–653. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, T.M., and J.C. Zúñiga-Pflücker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 39.Kodama, H., M. Nose, S. Niida, and S. Nishikawa. 1994. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp. Hematol. 22:979–984. [PubMed] [Google Scholar]
- 40.Kuroda, K., S. Tani, K. Tamura, S. Minoguchi, H. Kurooka, and T. Honjo. 1999. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274:7238–7244. [DOI] [PubMed] [Google Scholar]
- 41.Fazekas de St. Groth, S. 1982. The evaluation of limiting dilution assays. J. Immunol. Methods. 49:R11–R23. [DOI] [PubMed] [Google Scholar]
- 42.Rodewald, H.-R., K. Kretzschmar, S. Takeda, C. Hohl, and M. Dessing. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zúñiga-Pflücker, J.C., and M.J. Lenardo. 1996. Regulation of thymocyte development from immature progenitors. Curr. Opin. Immunol. 8:215–224. [DOI] [PubMed] [Google Scholar]
- 44.Ciofani, M., T.M. Schmitt, A. Ciofani, A.M. Michie, N. Çuburu, A. Aublin, J.L. Maryanski, and J.C. Zúñiga-Pflücker. 2004. Obligatory role for cooperative signaling by pre-T cell receptor and Notch during thymocyte differentiation. J. Immunol. 172:5230–5239. [DOI] [PubMed] [Google Scholar]
- 45.Livak, F., M. Tourigny, D.G. Schatz, and H.T. Petrie. 1999. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 162:2575–2580. [PubMed] [Google Scholar]
- 46.Capone, M., R.D. Hockett, Jr., and A. Zlotnik. 1998. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 95:12522–12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena, M.T., E.H. Schroeter, J.S. Mumm, and R. Kopan. 2001. Murine notch homologs (N1-4) undergo presenilin-dependent proteolysis. J. Biol. Chem. 276:40268–40273. [DOI] [PubMed] [Google Scholar]
- 48.Harman, B.C., E.J. Jenkinson, and G. Anderson. 2003. Microenvironmental regulation of Notch signalling in T cell development. Semin. Immunol. 15:91–97. [DOI] [PubMed] [Google Scholar]
- 49.Prockop, S.E., S. Palencia, C.M. Ryan, K. Gordon, D. Gray, and H.T. Petrie. 2002. Stromal cells provide the matrix for migration of early lymphoid progenitors through the thymic cortex. J. Immunol. 169:4354–4361. [DOI] [PubMed] [Google Scholar]
- 50.Shortman, K., and L. Wu. 1996. Early T lymphocyte progenitors. Annu. Rev. Immunol. 14:29–47. [DOI] [PubMed] [Google Scholar]
- 51.Diamond, R.A., S.B. Ward, K. Owada-Makabe, H. Wang, and E.V. Rothenberg. 1997. Different developmental arrest points in RAG-2−/− and SCID thymocytes on two genetic backgrounds: developmental choices and cell death mechanisms before TCR gene rearrangement. J. Immunol. 158:4052–4064. [PubMed] [Google Scholar]
- 52.Zúñiga-Pflücker, J.C. 2004. T-cell development made simple. Nat. Rev. Immunol. 4:67–72. [DOI] [PubMed] [Google Scholar]
- 53.Katsura, Y. 2002. Redefinition of lymphoid progenitors. Nat. Rev. Immunol. 2:127–132. [DOI] [PubMed] [Google Scholar]
- 54.Lai, E.C. 2004. Notch signaling: control of cell communication and cell fate. Development. 131:965–973. [DOI] [PubMed] [Google Scholar]
- 55.Rodewald, H.R., K. Kretzschmar, S. Takeda, C. Hohl, and M. Dessing. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porritt, H.E., K. Gordon, and H.T. Petrie. 2003. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J. Exp. Med. 198:957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lind, E.F., S.E. Prockop, H.E. Porritt, and H.T. Petrie. 2001. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J. Exp. Med. 194:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]