Abstract
The interaction of activators with mediator has been proposed to stimulate the assembly of RNA polymerase II (Pol II) preinitiation complexes, but there have been few tests of this model. The finding that the major adenovirus E1A and mitogen-activated protein kinase-phosphorylated Elk1 activation domains bind to Sur2 uniquely among the metazoan mediator subunits and the development of transcriptionally active nuclear extracts from WT and sur2–/– embryonic stem cells, reported here, allowed a direct test of the model. We found that whereas VP16, E1A, and phosphorylated Elk1 activation domains each stimulate binding of mediator, Pol II, and general transcription factors to promoter DNA in extracts from WT cells, only VP16 stimulated their binding in extracts from sur2–/– cells. This stimulation of mediator, Pol II, and general transcription factor binding to promoter DNA correlated with transcriptional activation by these activators in WT and mutant extracts. Because the mutant mediator was active in reactions with the VP16 activation domain, the lack of activity in response to the E1A and Elk1 activation domains was not due to loss of a generalized mediator function, but rather the inability of the mutant mediator to be bound by E1A and Elk1. These results directly demonstrate that the interaction of activation domains with mediator stimulates preinitiation complex assembly on promoter DNA.
Eukaryotic activators regulate transcription by two general mechanisms. First, they interact with multiprotein coactivator complexes that modify chromatin structure to give general transcription factors (GTFs) and RNA polymerase II (Pol II) access to promoter DNA (1–5). Second, they promote the assembly of a preinitiation complex composed of GTFs and Pol II on promoter DNA and stimulate initiation by Pol II (6, 7). Mediator complexes are required for this second aspect of transcriptional activation (8–13) and may also assist in maintaining chromatin in a hyperacetylated, open conformation (14). Several activation domains have been shown to bind directly to mediator complexes, but thus far there is little understanding of how such activation domain–mediator interactions stimulate transcription.
In Saccharomyces cerevisiae, Pol II, GTFs, and mediator are each required for efficient assembly of preinitiation complexes at promoters in vitro (15) and in vivo (16, 17) in what appears to be a highly cooperative process involving a network of protein–protein and protein–DNA interactions. But the relative contributions of interactions between activation domains and mediator vs. interactions between activation domains and other components of the complex transcriptional machinery have been difficult to decipher because the activators studied make multiple interactions that might each contribute to preinitiation complex assembly. Similarly, intensively studied metazoan and viral activation domains, such as VP16, have been reported to interact with GTFs as well as mediator (18–22).
In contrast, several observations suggest that the adenovirus E1A activation domain primarily responsible for stimulating early viral transcription (conserved region 3, hereafter referred to simply as E1A) activates transcription principally through its interaction with the Sur2 subunit of mediator complexes. As for other activators analyzed, mediator is required for E1A to activate transcription in vitro (22). E1A binds directly to the Sur2 mediator subunit, forming a complex that is stable in high salt (2 M KCl) (22). E1A also binds to mediator complexes in vivo in adenovirus-infected and transformed cells (23). The 40-aa E1A activation domain includes four cysteine residues that bind a single Zn2+ ion (24) and are essential for activation function (25, 26). In contrast to acidic activation domains such as those of VP16 (27) and GCN4 (28), multiple single conservative amino acid substitutions in E1A greatly diminish its activation function (25, 26). These same mutations, including mutations in the Zn2+-chelating cysteines, inhibit binding to Sur2 (22). These results suggest that the E1A activation domain folds into a compact zinc-finger domain that forms a stable complex with Sur2 through multiple specific side chain–Sur2 interactions. The perfect correlation between E1A mutant activation function and Sur2-binding (22) argues strongly that E1A activates transcription principally through this interaction with Sur2.
Isolation of embryonic stem (ES) cells with engineered knockout mutations in both SUR2 alleles confirmed that an E1A–Sur2 interaction is required for E1A activation (29). Sur2 was the only mediator subunit entirely missing from mediator complexes in sur2–/– cells. Thyroid hormone receptor-associated protein (TRAP)100 and TRAP95 were reduced, in keeping with the observation that both TRAP95 and Sur2 are lost from mediator complexes in cells with a knockout of TRAP100 (30). These results suggest that Sur2, TRAP100, and TRAP95 form a subcomplex or module within the mediator complex. All other mediator subunits were present at comparable levels in mediator from WT and sur2–/– and TRAP100–/– cells (29, 30). The strong E1A activation function was completely defective in sur2–/– cells but could be rescued by expression of the nearly identical human Sur2. Activation by the Elk1 activation domain in response to mitogen-activated protein kinase (MAPK) phosphorylation was also extremely defective and could be rescued by human Sur2. In contrast, multiple other activation domains analyzed, including VP16, activated similarly in sur2–/– and WT ES cells, indicating that the Sur2-deleted mediator functions normally in transcription control by multiple activators. This conclusion is also consistent with the viability of homozygous sur2–/– ES cells and ongoing studies of sur2–/– mouse embryos. Although sur2–/– embryos die at ≈10 days postcoitum, considerable morphogenesis and cellular differentiation are observed in the mutant embryos, indicating that the mutant mediator can support complex transcriptional programs (J.L.S. and A.J.B., unpublished results). This finding contrasts to the knockout of the mouse Srb7 subunit, an essential subunit in S. cerevisiae, that is also required for viability of murine ES cells and early embryos (31). These results indicate that the Sur2 mediator subunit is required for activation by only a highly restricted subset of activation domains including E1A and MAPK-phosphorylated Elk1.
The selective loss of E1A activation in sur2–/– cells is a unique situation among mammalian mediator subunit mutants analyzed so far. Although knockout of TRAP220, a mediator subunit that interacts with the ligand-binding activation domain of many nuclear receptors, results in a considerable decrease in activation by thyroid receptor, significant thyroid receptor activation is retained in the mutant cells, and there is little decrease in activation by the retinoic acid receptor (32). Knockout of TRAP100 leads to a decrease in activation by all activators analyzed (30). Several S. cerevisiae mutants with deletions of nonessential mediator subunits have been isolated and shown to have reduced expression of a subset of genes in vivo (33, 34); however, the specific activators that are defective for activation in these mutants have not been identified. Some in vitro experiments have illustrated the requirements of certain yeast mediator subunits for activated transcription with some but not all activators (34, 35). However, the effect of these mediator subunit deletions on preinitiation complex assembly was not analyzed. Consequently, the development of ES cells with mediator complexes lacking the E1A-interacting subunit, but sufficient for binding and activation by other activators, provides an unusual opportunity to study the consequences of a single activation domain–mediator interaction in this process.
Mediator complexes isolated from nuclear or whole cell extracts of S. cerevisiae are generally associated with Pol II, and, in some cases, GTFs (36–40). In contrast, mediator complexes in nuclear extracts of mammalian cells prepared at 0.3 M KCl (41) are readily separated from Pol II and GTFs (20, 22, 42–47). When HeLa nuclear extract was directly fractionated by gel filtration without exposure to high salt or ion-exchange chromatography, a single ≈1.5-MDa size class of mediator complexes was observed well separated from Pol II and GTFs (48). We have made similar observations with nuclear extracts prepared from ES cells (G. Wang, G.T.C., and A.J.B., unpublished results). Consequently, in contrast to the situation in S. cerevisiae, mediator complexes are not tightly associated with Pol II or GTFs in nuclear extracts from mammalian cells.
To analyze the mechanism by which the E1A–Sur2 interaction stimulates transcription, we prepared transcriptionally active nuclear extracts from WT and sur2–/– ES cells and studied the interaction of mediator, GTFs, and Pol II with matrix-bound (tethered) template DNA. Similar experiments also were performed with the MAPK-phosphorylated Elk1 and VP16 activation domains. Our results indicate that the interactions of the E1A and Elk1 activation domains with the Sur2 mediator subunit stimulate assembly of a preinitiation complex on promoter DNA.
Materials and Methods
Cell Culture and Nuclear Extract Preparation. WT and sur2–/– murine ES cells were cultured as described (29). Nuclear extract was prepared as described (41) and dialyzed into 0.1 M KCl in D buffer [20 mM Hepes, pH 7.9 (NaOH)/0.2 mM EDTA/20% (vol/vol) glycerol/0.5 mM DTT] made 0.5 mM in PMSF.
Protein Purification. Gal4 DNA-binding domain (residues 1–147) fusions to the activation domains of VP16 or E1A (48) were expressed in Escherichia coli and purified as for Gal4-Elk1. Gal4-Elk1 [containing the Elk1 activation domain from residues 307–428 (C terminus)] was expressed in E. coli BL21 (DE3) by using the pET8C expression vector (Novagen). One liter in 2× TYE (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter) grown to OD600 ≈ 0.6 was induced by adding 0.4 mM isopropyl β-d-thiogalactoside, incubated 3 h at 37°C, and centrifuged. Pelleted cells were resuspended in 20 ml of 0.2 M KCl/0.5 mM PMSF/0.5 mg/ml lysozyme in buffer Z (20 mM Hepes, pH 7.9/10 μM ZnCl2/1 mM DTT) and sonicated to disrupt cells. Nonidet P-40 was added to 0.05%, and the sonicate was mixed for 20 min at 4°C and then centrifuged 15 min at 4°C at 15,000 rpm in a Sorvall SA600 rotor (23,000 × g). The supernatant was subjected to ammonium sulfate precipitation at 15% and 40% saturation, and the 15–40% cut was dissolved in 5 ml of 0.1 M KCl and 0.5 mM PMSF in buffer Z plus 20% glycerol (Z+) and dialyzed in the same buffer. Dialyzed protein was loaded on a 1 ml of HiTrapQ column (Amersham Biosciences), equilibrated with 0.1 M KCl in buffer Z+, and eluted with a linear gradient of 0.1–1 M KCl in buffer Z+. Eluted fractions were analyzed by SDS/PAGE and Western blotting using an antibody to Gal4 DNA-binding domain. Gal4–Elk1-containing fractions were pooled and dialyzed in 0.1 M KCl in buffer Z+ and stored at –70°C in small aliquots.
Plasmids and Immobilized Template Preparation. pG5TATA contains five 17-bp Gal4-binding sites each separated by 2 bp, 14 bp upstream of the adenovirus 2 major late promoter from –53 to +2 followed by a 233-bp G-less cassette inserted between HindIII and EcoRI in pB (SK) (+) (Stratagene). pG5ΔTATA is identical to pG5TATA except for deletion of the major late promoter from –53 to +11. Biotinylated G5TATA was 799 bp and was prepared by PCR using pG5TATA as template with one primer biotinylated at the 5′ end complementary to the sequence 238 bp upstream from the first Gal4-binding site and the second primer complementary to pB (SK) (+) 120 bp downstream from the G-less cassette. Biotinylated G5ΔTATA was 735 bp and was prepared as above by using pG5ΔTATA as template. Biotinylated G-less was 464 bp and was prepared by PCR using pG5TATA as template with one primer biotinylated at the 5′ end complementary to +18 of the G-less cassette and the other complementary to the sequence of pB (SK) (+) 247bp downstream of the G-less cassette.
All PCR products were purified from primers by using a QIAquick PCR purification kit (Qiagen, Valencia, CA). Approximately 1.5 pmol of each biotinylated template was bound per 100 μg of Dynabeads M-280 Streptavidin (Dynal, Great Neck, NY) according to the manufacturer's instructions. Immobilized templates were concentrated by using a magnetic particle concentrator (Dynal) and resuspended in 30 μl of blocking buffer (0.1 M KCl in buffer D plus 8 mM MgCl2/10 μM ZnCl2/2.5 mM DTT/0.01% Triton X-100/50 mg/ml BSA). After 5 min at room temperature, saturating amounts of recombinant Gal4-VP16, Gal4-E1A, or Gal4-Elk1, or buffer Z+ alone were added and incubated 20 min at room temperature. Except for templates bound by Gal4-Elk1, beads were then washed once with blocking buffer before the addition of nuclear extract. For the phosphorylation of Gal4-Elk1 by ERK2, immobilized templates with bound Gal4-Elk1 were washed once with kinase buffer [25 mM Hepes, pH 7.9 (NaOH)/25 mM β-glycerophosphate/25 mM MgCl2/0.2 mM ATP/0.05% Nonidet P-40/0.5 mM DTT/0.5 mg/ml BSA/10% glycerol/0.5 mM PMSF] followed by incubation in kinase buffer with or without 1 unit/μl activated extracellular signal-regulated kinase 2 (ERK2; New England Biolabs) for 30 min at 30°C. Immobilized templates were then washed once with kinase buffer before the addition of nuclear extract.
In Vitro Transcription. In vitro transcription was carried out on 0.25 pmol of immobilized biotinylated G5TATA or G5ΔTATA without bound Gal4-fusion proteins or with saturating Gal4-VP16, Gal4-E1A, Gal4-Elk1, or Gal4-Elk1-P phosphorylated in vitro with recombinant ERK2. Templates were concentrated by using the magnetic particle concentrator, the supernatant was decanted, and 20 μg of WT or sur2–/– nuclear extract protein was added in 35 μl of 60 mM KCl/30 mM Hepes, pH 7.9 (NaOH)/8 mM MgCl2/10 μM ZnCl2/0.2 mM EDTA/12% glycerol/0.5 mM PMSF/40 units of RNasin (Promega)/100 μM ATP/100 μM CTP/3 μM UTP/10 μCi of [α-32P]UTP (3,000 Ci/mmol; Amersham Biosciences; 1 μCi = 37 kBq). Transcription was carried out for 1 h at 30°C. Reactions were processed by extraction with phenol/chloroform and analyzed by gel electrophoresis and autoradiography as described (48).
Factor-Binding Assay. Incubations were carried out for 30 min at room temperature with 2 pmol of immobilized templates, prepared as described above. Templates were concentrated, supernatants were decanted, and 130 μg of WT or sur2–/– ES cell nuclear extract protein in 140 μl of 0.12 M KCl/8 mM MgCl2/10 μM ZnCl2/30 mM Hepes, pH 7.9 (NaOH)/0.1 mM EDTA/20% glycerol/3.0 mM DTT/0.03% Triton X-100/5 μg/ml heparin (Sigma, H-3125)/5 mM NaF/0.1 μg/ml okadaic acid (Sigma). Immobilized templates were washed three times with the aid of the magnetic particle concentrator in 0.2 M KCl/8 mM MgCl2/10 μM ZnCl2/2.5 mM DTT/20 mM Hepes, pH 7.9 (NaOH)/0.2 mM EDTA/20% glycerol/0.06% Triton X-100/5 μg/ml heparin/0.5 mg/ml BSA. Bound protein was eluted with Laemmli buffer and subjected to SDS/10% PAGE followed by Western blotting using antibodies against the Pol II subunit RPB1 [mAb 8WG16 (49)], Sur2 (BD Biosciences no. 550429), and cyclin-dependent kinase 8 (CDK8), TATA box-binding protein (TBP), transcription factor (TF)IIE (p56), TFIIH (CDK7), TFIIB, phosphorylated Elk1 (Elk1-P), Elk1, and G4DBD (Santa Cruz Biotechnology, sc-1521, sc-421, sc-237, sc-7344, sc-225, sc-8406, sc-355, sc-577, respectively).
Results
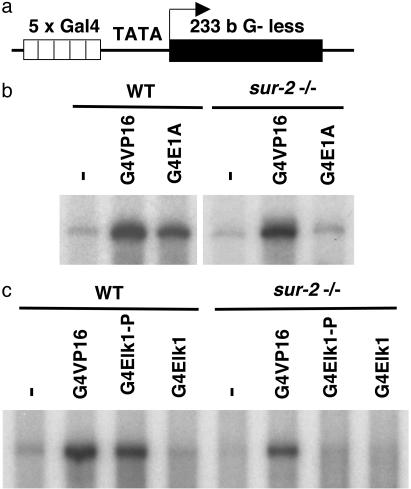
Nuclear extract was prepared from WT and sur2–/– ES cells and used to transcribe tethered templates bound to magnetic streptavidin beads through biotin incorporated at the upstream end of the template, ≈240 bp from the first Gal4 site. The template, G5TATA, contained five Gal4-binding sites upstream of the adenovirus 2 major late promoter TATA box followed by an ≈230-bp G-less cassette (Fig. 1a). The extract from WT ES cells supported strong activation by Gal4-VP16 and substantial, but somewhat weaker, activation by Gal4-E1A (Fig. 1b) and Gal4-Elk1 phosphorylated by ERK2 MAPK (Fig. 1c), consistent with the weaker activation by these Gal4 fusions in vivo (29). In contrast, whereas the extract from sur2–/– cells supported strong activation by Gal4-VP16, little stimulation above that observed in the absence of an added Gal4-fusion protein was observed in reactions with the same concentration of Gal4-E1A or Gal4-Elk-P.
Fig. 1.
Transcriptional activation by E1A and Elk1 in vitro requires the Sur2 mediator subunit. In vitro transcription was carried out by using a biotinylated linear DNA template immobilized on streptavidin-coated magnetic beads. (a) The immobilized template, G5TATA, contains five Gal4-binding sites upstream of a TATA-containing promoter driving transcription of a 233-bp G-less cassette. (b and c) Transcription was carried out on immobilized templates either without activator or with saturating amounts of Gal4 DNA-binding domain fused to the activation domain of VP16 (G4VP16), E1ACR3 (G4E1A), or the activation domain of Elk1 (G4Elk1). G4Elk1 bound to immobilized templates was either phosphorylated by the MAPK ERK2 or left unaltered. Nuclear extracts prepared from either WT or sur2–/– murine ES cells were used in the reactions, as indicated.
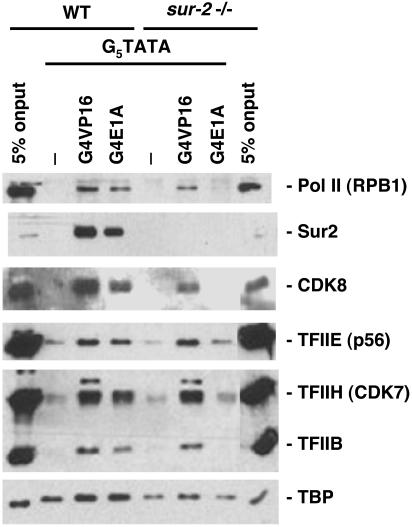
Earlier results showed that Gal4-E1A and Gal4-Elk-P bound to this template bind mediator in nuclear extract from WT, but not sur2–/–, ES cells, whereas Gal4-VP16 bound mediator in both WT and mutant extracts (29). To determine whether these activation domains also influence the binding of Pol II or GTFs to promoter DNA, the binding of these proteins to tethered templates was also assayed. Gal4-VP16, Gal4-E1A, or Gal4-Elk-P was prebound to the tethered templates under conditions that saturated the Gal4-binding sites. After removing excess unbound Gal4-fusion proteins by washing, the templates were incubated in nuclear extract from WT or mutant cells at near physiological salt concentration in the absence of nucleoside triphosphates. After a 30-min incubation, the tethered templates were washed under conditions that removed most nonspecifically bound proteins. Factors that remained stably associated with the templates were eluted with SDS and assayed by Western blotting (Figs. 2 and 3). TBP binding to the high-affinity major late promoter TATA box was observed in the absence of added Gal4-fusion proteins in both the WT and sur2–/– nuclear extract. The Gal4 activators caused only a very modest increase in TBP binding. In contrast, as observed earlier (29), Gal4-VP16, Gal4-E1A, and Gal4-Elk1-P, but not unphosphorylated Gal4-Elk1, greatly stimulated binding of mediator in the WT nuclear extract (as observed by binding of CDK8 and Sur2), whereas only Gal4-VP16 stimulated binding of mediator (observed by the binding of CDK8) when the nuclear extract from sur2–/– cells was used. Although CDK8 and its cyclin C partner appear to be present in only a subset of mediator complexes (48), E1A and the phosphorylated Elk1 activation domains bind mediator complexes with and without CDK8 equally well in vitro, because binding of CDK8 was comparable to binding of other mediator subunits (29).
Fig. 2.
Requirement for the Sur2 mediator subunit in the E1A-induced binding of Pol II and GTFs to promoter DNA. Immobilized G5TATA DNA either without bound activator or with saturating amounts of recombinant Gal4-VP16 or -E1A was incubated with nuclear extract from WT or sur2–/– ES cells. Immobilized templates were then washed, and bound protein was eluted, resolved by SDS/PAGE, and analyzed by Western blotting using antibodies to the indicated proteins.
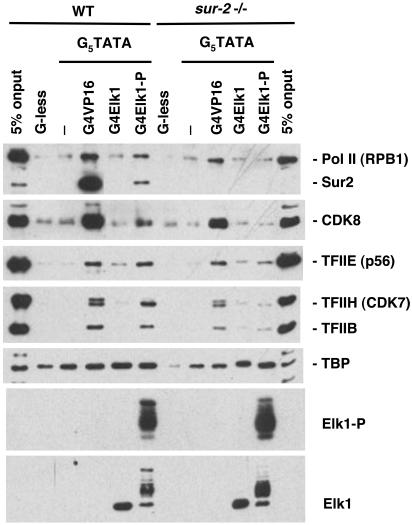
Fig. 3.
Requirement of the Sur2 mediator subunit for the Elk1-P-induced binding of Pol II and GTFs to promoter DNA. Nuclear extracts from WT and sur2–/– ES cells were incubated with the immobilized templates G5TATA or G-less (a DNA fragment lacking the five Gal4-binding sites and the core promoter). Immobilized G5TATA contained either no activator or saturating amounts of Gal4-VP16, -Elk1, or -Elk1 phosphorylated in vitro by recombinant ERK2 (G4Elk1-P). Binding was analyzed as in Fig. 2. Additional antibodies were used that recognize Elk1 and phosphorylated Elk1 (Elk1-P).
Significantly, in the experiments with WT nuclear extract, Gal4-VP16, Gal4-E1A, and Gal4-Elk1-P, but not unphosphorylated Gal4-Elk1, also greatly stimulated the binding of Pol II and GTFs TFIIB, TFIIE, and TFIIH (Figs. 2 and 3). Stable TFIIF binding was not observed. It may have been removed during the washing steps required to remove proteins that bound nonspecifically under these conditions. In contrast to the results observed with nuclear extract from WT ES cells, Gal4-E1A and Gal4-Elk-P did not stimulate the binding of Pol II or GTFs in binding experiments with sur2–/– nuclear extract. Pol II, GTFs, and mediator in the sur2–/– nuclear extract were all functional in that they supported in vitro transcription in response to Gal4-VP16 (Fig. 1) and could be bound to tethered templates in the presence of Gal4-VP16 (Figs. 2 and 3). Consequently, it appeared that elimination of the Sur2 mediator subunit specifically interfered with the ability of the E1A and Elk1-P activation domains, but not the VP16 activation domain, to stimulate Pol II, TFIIB, TFIIE, and TFIIH binding to promoter DNA, as well as transcription.
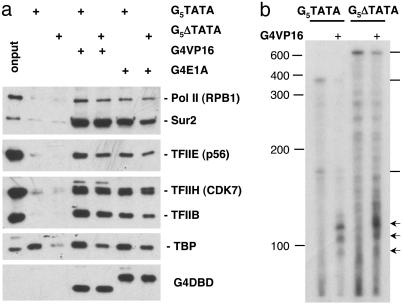
We next determined the significance of the TATA box for binding Pol II and GTFs to tethered templates. The TATA box and cap site region were deleted from the G5TATA template (G5ΔTATA). The G5ΔTATA template was then compared to G5TATA in tethered template binding (Fig. 4a) and in vitro transcription experiments (data not shown) using WT ES nuclear extract. In incubations containing either Gal4-VP16 or Gal4-E1A bound to G5ΔTATA, the binding of Pol II, TFIIB, TFIIE, TFIIH, TBP, and mediator was only slightly reduced compared with G5TATA. The observation that deletion of the TATA box had only a modest effect on TBP binding is in keeping with the limited DNA sequence specificity of TBP binding (50). Transcription of the G-less cassette from the G5ΔTATA template was eliminated by deletion of the TATA box and cap site region, as expected. The observation of only a mild reduction in binding of the general transcription machinery, but a severe loss in transcription of the template strand due to deletion of the TATA box has been reported in previous studies with yeast and human nuclear extracts (15, 51). However, RNase protection assay of transcripts synthesized in vitro revealed that Gal4-VP16 stimulated a low level of transcription from the opposite strand starting from a region near the Gal4 sites (Fig. 4b). Thus, although transcription initiated at a very low rate, Gal4-VP16 likely stimulated the assembly of preinitiation complexes on the G5ΔTATA template despite the deletion of the canonical TATA box.
Fig. 4.
Stimulation of Pol II and GTF binding occurs in the absence of a canonical TATA box. (a)G5TATA and G5ΔTATA immobilized templates, with or without bound Gal4-VP16 or Gal4-E1A, were incubated with WT ES nuclear extract. Bound protein was washed, eluted, and subjected to SDS/PAGE and Western blotting as in Figs. 2 and 3. (b) RNase protection assay of in vitro transcripts transcribed from the strand complementary to the G-less cassette template strand. Transcription was from the G5TATA (Left) or G5ΔTATA (Right) templates in the presence (+) or absence of Gal4-VP16. The probe was transcribed by T7 RNA polymerase from the G5ΔTATA template cut with EcoRI. Arrows indicate RNase-protected fragments corresponding to transcripts with 5′ ends in the region of the Gal4 sites. Lines at the right indicate RNase-protected fragments corresponding to transcripts complementary to the full length of the probe. The positions of single-stranded DNA markers containing the indicated number of nucleotides are shown at the left.
Discussion
Because multiple activation domains have been shown to bind to mediator complexes (9–13) and mediator complexes bind directly to Pol II (37, 45, 52), it has been suggested that activation domain–mediator interactions stimulate preinitiation complex formation by acting as a molecular bridge between enhancerbound activators and the Pol II general machinery, a version of the recruitment model for activated transcription (53). However, there have been few direct tests of this model.
Experiments in S. cerevisiae with mediator subunit mutants have shown that activators stimulate Pol II and GTF binding to promoter DNA both in vitro (15) and in vivo (16, 17) by a mechanism requiring functional mediator. However, it was not clear whether the mutant mediators in these studies were defective because they failed to bind to activation domains or because of a general defect in a mediator activity required for activated transcription. The observations that mediator complexes stimulate basal Pol II transcription in the absence of activators (37, 51, 54–57) raise the possibility that the mediator requirement for Pol II and GTF binding in earlier experiments (15–17) might be for a function that is not dependent on its interaction with activators. In these earlier studies, the mutant mediators were not shown to support activation by control activators.
The findings that the major E1A activation domain binds to Sur2 uniquely among the mediator subunits (22, 29), that the Sur2 subunit is required for E1A to activate transcription in vivo (29), that multiple other activation domains function normally in sur2–/– cells, and that the development of transcriptionally active nuclear extracts from WT and sur2–/– ES cells (reported here) made it possible to test directly whether the interaction between an activation domain and a mediator stimulates preinitiation complex assembly, as has been widely proposed. Comparable experiments were also performed with the MAPK-phosphorylated Elk1 activation domain that is largely dependent on Sur2 for its ability to activate transcription (29). There are few other examples of a mutation in a mediator subunit that blocks activation and mediator binding by a specific activation domain without inhibiting transcription generally (34, 35, 58).
We observed that interactions between the E1A and MAPK-phosphorylated Elk1 activation domains and the Sur2 subunit of the mediator complex stimulated binding of Pol II, TFIIB, TFIIE, and TFIIH to promoter DNA (Figs. 2 and 3). In the absence of this interaction because of omission of an activation domain linked to the Gal4 DNA-binding domain, or because of the absence of the interacting Sur2 subunit in nuclear extract from sur2–/– cells, much less Pol II and these GTFs bound to promoter DNA. This result was not due to a defect in a general function of mediator required for activated transcription because Gal4-VP16 stimulated Pol II and GTF binding in the sur2–/– extract to a similar level as in the WT extract. The ability of activators to stimulate Pol II and GTF binding to promoter DNA in the WT and sur2–/– nuclear extracts correlated with their ability to activate transcription. These results imply that activation by E1A and Elk1-P is due, at least in part, to their ability to stimulate the assembly of preinitiation complexes on promoter DNA through their interaction with mediator complexes by means of the Sur2 subunit. This ability of an activation domain–mediator interaction to stimulate preinitiation complex formation is consistent with earlier studies showing that tethering of some mediator subunits to a promoter by expressing them as fusions to a DNA-binding domain activates transcription in vivo (59).
The network of interactions known to occur in preinitiation complex formation during activation by E1A and Elk1 is summarized in Fig. 5. Among the GTFs, protein–protein interactions occur between Pol II and TFIIB, TFIIE and TFIIF, and TFIIE and TFIIH (60). TFIIB interacts with the TBP subunit of TFIID and promoter DNA (61), with TBP establishing the binding site on a TATA box promoter. TFIIF, TFIIH, and Pol II also interact extensively with promoter DNA (62–64). Mediator interacts directly with Pol II (37, 45, 52), TFIIE (65), and TFIID (51). As observed here, the key regulatory interaction is the binding of the E1A or Elk1-P activation domain to Sur2. In the absence of this interaction, there is only a low rate of transcription and a low level of Pol II, TFIIB, TFIIE, and TFIIH bound to promoter DNA.
Fig. 5.
Model of the known protein–protein and protein–DNA interactions during Gal4-E1A- and Gal4-Elk1-P-activated transcription. DNA binding by the Gal4-activation domain fusions and TBP to the template studied here was independent of the activation domain–Sur2 interaction. But all other interactions shown were stabilized by interactions of these activation domains with Sur2. DBD, DNA-binding domain; TAFs, TBP-associated factors; Med, mediator.
The E1A activation domain probably interacts with Sur2 alone to stimulate Pol II and GTF binding and activate transcription because a perfect correlation was observed between the ability of multiple E1A conservative single amino acid substitutions to bind Sur2 and to activate transcription (22). Elk1 has not been analyzed as extensively as E1A in this regard. Consequently, there is little data to indicate whether Elk1 interacts with other components of the preinitiation complex in addition to mediator to stimulate preinitiation complex assembly and activate transcription. But any other interactions are not sufficient to promote Pol II, TFIIB, TFIIE, and TFIIH binding (Fig. 3), indicating that the Elk1-P interaction with Sur2 is a critical regulatory step.
It was not possible for us to determine what fraction of the Pol II and GTFs that bound to the tethered templates were assembled into functional preinitiation complexes. Some of the bound factors might have been in partial complexes, e.g., complexes of only the activator, mediator, and Pol II. However, the correlation between the ability of Gal4-VP16, -E1A, and -Elk1-P to activate transcription and to stimulate Pol II and GTF binding in the WT and sur2–/– nuclear extracts suggests strongly that some of the observed stably bound factors and Pol II were assembled into functional preinitiation complexes before the templates were removed from the nuclear extract and washed. Taken together, these results provide strong evidence in favor of the widely held model that activation domain binding to mediator stimulates preinitiation complex assembly on promoter DNA and that this is an important aspect of the mechanism of transcriptional activation. Nonetheless, these results do not rule out the possibility that interactions of other activators with additional components of the preinitiation complex may contribute to activation. Moreover, activation domain–mediator interactions may also stimulate transcription initiation at a postrecruitment step subsequent to Pol II and GTF binding.
Acknowledgments
We thank Carol Eng for technical support. This work was supported by National Institutes of Health Grant CA25235. G.T.C. was partially supported by U.S. Public Health Service National Research Service Award GM07185.
Abbreviations: ES, embryonic stem; MAPK, mitogen-activated protein kinase; Pol II, RNA polymerase II; GTF, general transcription factor; TBP, TATA box-binding protein; CDK, cyclin-dependent kinase; TRAP, thyroid hormone receptor-associated protein; TF, transcription factor; ERK2, extracellular signal-regulated kinase 2.
References
- 1.Berger, S. L. (2002) Curr. Opin. Genet. Dev. 12, 142–148. [DOI] [PubMed] [Google Scholar]
- 2.Becker, P. B. & Horz, W. (2002) Annu. Rev. Biochem. 71, 247–273. [DOI] [PubMed] [Google Scholar]
- 3.Horn, P. J. & Peterson, C. L. (2002) Science 297, 1824–1827. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 5.Neely, K. E. & Workman, J. L. (2002) Mol. Genet. Metab. 76, 1–5. [DOI] [PubMed] [Google Scholar]
- 6.Roeder, R. G. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 201–218. [DOI] [PubMed] [Google Scholar]
- 7.Woychik, N. A. & Hampsey, M. (2002) Cell 108, 453–463. [DOI] [PubMed] [Google Scholar]
- 8.Hampsey, M. & Reinberg, D. (1999) Curr. Opin. Genet. Dev. 9, 132–139. [DOI] [PubMed] [Google Scholar]
- 9.Lee, T. I. & Young, R. A. (2000) Annu. Rev. Genet. 34, 77–137. [DOI] [PubMed] [Google Scholar]
- 10.Malik, S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277–283. [DOI] [PubMed] [Google Scholar]
- 11.Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729–749. [DOI] [PubMed] [Google Scholar]
- 12.Naar, A. M., Lemon, B. D. & Tjian, R. (2001) Annu. Rev. Biochem. 70, 475–501. [DOI] [PubMed] [Google Scholar]
- 13.Rachez, C. & Freedman, L. P. (2001) Curr. Opin. Cell Biol. 13, 274–280. [DOI] [PubMed] [Google Scholar]
- 14.Lorch, Y., Beve, J., Gustafsson, C. M., Myers, L. C. & Kornberg, R. D. (2000) Mol. Cell 6, 197–201. [DOI] [PubMed] [Google Scholar]
- 15.Ranish, J. A., Yudkovsky, N. & Hahn, S. (1999) Genes Dev. 13, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuras, L. & Struhl, K. (1999) Nature 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 17.Li, X. Y., Virbasius, A., Zhu, X. & Green, M. R. (1999) Nature 399, 605–609. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman, P. M. & Berk, A. J. (1994) Genes Dev. 8, 995–1006. [DOI] [PubMed] [Google Scholar]
- 19.Uesugi, M., Nyanguile, O., Lu, H., Levine, A. J. & Verdine, G. L. (1997) Science 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 20.Naar, A. M., Beaurang, P. A., Zhou, S., Abraham, S., Solomon, W. & Tjian, R. (1999) Nature 398, 828–832. [DOI] [PubMed] [Google Scholar]
- 21.Hall, D. B. & Struhl, K. (2002) J. Biol. Chem. 277, 46043–46050. [DOI] [PubMed] [Google Scholar]
- 22.Boyer, T. G., Martin, M. E., Lees, E., Ricciardi, R. P. & Berk, A. J. (1999) Nature 399, 276–279. [DOI] [PubMed] [Google Scholar]
- 23.Wang, G. & Berk, A. J. (2002) J. Virol. 76, 9186–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culp, J. S., Webster, L. C., Friedman, D. J., Smith, C. L., Huang, W. J., Wu, F. Y., Rosenberg, M. & Ricciardi, R. P. (1988) Proc. Natl. Acad. Sci. USA 85, 6450–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster, L. C. & Ricciardi, R. P. (1991) Mol. Cell. Biol. 11, 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, K. J., Lillie, J. W. & Green, M. R. (1990) Nature 346, 147–152. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan, S. M., Horn, P. J., Olson, V. A., Koop, A. H., Niu, W., Ebright, R. H. & Triezenberg, S. J. (1998) Nucleic Acids Res. 26, 4487–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan, K., Jackson, B. M., Zhou, H., Winston, F. & Hinnebusch, A. G. (1999) Mol. Cell 4, 657–664. [DOI] [PubMed] [Google Scholar]
- 29.Stevens, J. L., Cantin, G. T., Wang, G., Shevchenko, A., Shevchenko, A. & Berk, A. J. (2002) Science 296, 755–758. [DOI] [PubMed] [Google Scholar]
- 30.Ito, M., Okano, H. J., Darnell, R. B. & Roeder, R. G. (2002) EMBO J. 21, 3464–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tudor, M., Murray, P. J., Onufryk, C., Jaenisch, R. & Young, R. A. (1999) Genes Dev. 13, 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B. & Roeder, R. G. (2000) Mol. Cell 5, 683–693. [DOI] [PubMed] [Google Scholar]
- 33.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 34.Myers, L. C., Gustafsson, C. M., Hayashibara, K. C., Brown, P. O. & Kornberg, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, Y. C., Park, J. M., Min, S., Han, S. J. & Kim, Y. J. (1999) Mol. Cell. Biol. 19, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koleske, A. J. & Young, R. A. (1995) Trends Biochem. Sci. 20, 113–116. [DOI] [PubMed] [Google Scholar]
- 37.Kim, Y. J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. (1994) Cell 77, 599–608. [DOI] [PubMed] [Google Scholar]
- 38.Gustafsson, C. M., Myers, L. C., Beve, J., Spahr, H., Lui, M., Erdjument-Bromage, H., Tempst, P. & Kornberg, R. D. (1998) J. Biol. Chem. 273, 30851–30854. [DOI] [PubMed] [Google Scholar]
- 39.Han, S. J., Lee, Y. C., Gim, B. S., Ryu, G. H., Park, S. J., Lane, W. S. & Kim, Y. J. (1999) Mol. Cell. Biol. 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y., Ranish, J. A., Aebersold, R. & Hahn, S. (2001) J. Biol. Chem. 276, 7169–7175. [PubMed] [Google Scholar]
- 41.Dignam, J. D., Martin, P. L., Shastry, B. S. & Roeder, R. G. (1983) Methods Enzymol. 101, 582–598. [DOI] [PubMed] [Google Scholar]
- 42.Jiang, Y. W., Veschambre, P., Erdjument-Bromage, H., Tempst, P., Conaway, J. W., Conaway, R. C. & Kornberg, R. D. (1998) Proc. Natl. Acad. Sci. USA 95, 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fondell, J. D., Ge, H. & Roeder, R. G. (1996) Proc. Natl. Acad. Sci. USA 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito, M., Yuan, C. X., Malik, S., Gu, W., Fondell, J. D., Yamamura, S., Fu, Z. Y., Zhang, X., Qin, J. & Roeder, R. G. (1999) Mol. Cell 3, 361–370. [DOI] [PubMed] [Google Scholar]
- 45.Sun, X., Zhang, Y., Cho, H., Rickert, P., Lees, E., Lane, W. & Reinberg, D. (1998) Mol. Cell 2, 213–222. [DOI] [PubMed] [Google Scholar]
- 46.Rachez, C., Lemon, B. D., Suldan, Z., Bromleigh, V., Gamble, M., Naar, A. M., Erdjument-Bromage, H., Tempst, P. & Freedman, L. P. (1999) Nature 398, 824–828. [DOI] [PubMed] [Google Scholar]
- 47.Ryu, S. & Tjian, R. (1999) Proc. Natl. Acad. Sci. USA 96, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, G., Cantin, G. T., Stevens, J. L. & Berk, A. J. (2001) Mol. Cell. Biol. 21, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, N. E., Aronson, D. B. & Burgess, R. R. (1990) J. Biol. Chem. 265, 7069–7077. [PubMed] [Google Scholar]
- 50.Hahn, S., Buratowski. S. & Sharp, P. A. (1989) Proc. Natl. Acad. Sci. USA 86, 5718–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson, K. M., Wang, J., Smallwood, A., Arayata, C. & Carey, M. (2002) Genes Dev. 16, 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao, D. M., Gadbois, E. L., Murray, P. J., Anderson, S. F., Sonu, M. S., Parvin, J. D. & Young, R. A. (1996) Nature 380, 82–85. [DOI] [PubMed] [Google Scholar]
- 53.Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 54.Guermah, M., Tao, Y. & Roeder, R. G. (2001) Mol. Cell. Biol. 21, 6882–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittler, G., Kremmer, E., Timmers, H. T. & Meisterernst, M. (2001) EMBO Rep. 2, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baek, H. J., Malik, S., Qin, J. & Roeder, R. G. (2002) Mol. Cell. Biol. 22, 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reeves, W. M. & Hahn, S. (2003) Mol. Cell. Biol. 23, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge, K., Guermah, M., Yuan, C. X., Ito, M., Wallberg, A. E., Spiegelman, B. M. & Roeder, R. G. (2002) Nature 417, 563–567. [DOI] [PubMed] [Google Scholar]
- 59.Balciunas, D., Hallberg, M., Bjorklund, S. & Ronne, H. (2003) J. Biol. Chem. 278, 3831–3839. [DOI] [PubMed] [Google Scholar]
- 60.Bushnell, D. A., Bamdad, C. & Kornberg, R. D. (1996) J. Biol. Chem. 271, 20170–20174. [DOI] [PubMed] [Google Scholar]
- 61.Nikolov, D. B., Chen, H., Halay, E. D., Usheva, A. A., Hisatake, K., Lee, D. K., Roeder, R. G. & Burley, S. K. (1995) Nature 377, 119–128. [DOI] [PubMed] [Google Scholar]
- 62.Coulombe, B., Li, J. & Greenblatt, J. (1994) J. Biol. Chem. 269, 19962–19967. [PubMed] [Google Scholar]
- 63.Kim, T. K., Ebright, R. H. & Reinberg, D. (2000) Science 288, 1418–1422. [DOI] [PubMed] [Google Scholar]
- 64.Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 65.Sakurai, H. & Fukasawa, T. (2000) J. Biol. Chem. 275, 37251–37256. [DOI] [PubMed] [Google Scholar]