Abstract
Cytokines, such as tumor necrosis factor-α (TNFα), potently inhibit the differentiation of mesenchymal cells and down-regulate the expression of Sox9 and MyoD, transcription factors required for chondrocyte and myocyte development. Previously, we demonstrated that NF-κB controls TNFα-mediated suppression of myogenesis through a mechanism involving MyoD mRNA down-regulation. Here, we show that NF-κB also suppresses chondrogenesis and destabilizes Sox9 mRNA levels. Multiple copies of an mRNA cis-regulatory motif (5′-ACUACAG-3′) are necessary and sufficient for NF-κB-mediated Sox9 and MyoD down-regulation. Thus, in response to cytokine signaling, NF-κB modulates the differentiation of mesenchymal-derived cell lineages via RNA sequence-dependent, posttranscriptional down-regulation of key developmental regulators.
Keywords: NF-κB, differentiation, mesenchymal, muscle, chondrocyte, posttranscriptional control
In multicellular organisms, control of proliferation, differentiation, and apoptosis is essential for survival. Defects in cell differentiation can lead to aberrations in other cellular processes, resulting in diseases such as cancer (Scott 1997). Mesenchymal progenitor cells give rise to the skeletal cells, including myocytes, chondrocytes, osteoblasts, and adipocytes (Pittenger et al. 1999; Caplan and Bruder 2001). Notably, cytokines such as tumor necrosis factor-α (TNFα) can inhibit the differentiation of mesenchymal stem cells (Filipak et al. 1988; Gilbert et al. 2000) and suppress the expression of genes important for their differentiation (Guttridge et al. 2000; Murakami et al. 2000; Gilbert et al. 2002; Ruan et al. 2002). However, the molecular mechanism underlying this suppression is not known.
Nuclear factor κ B (NF-κB) complexes are dimeric, sequence-specific transcription factors that regulate the expression of a large number of genes in response to a variety of cellular conditions, including infection and inflammation (Silverman and Maniatis 2001; Ghosh and Karin 2002; Li and Verma 2002). There are five distinct NF-κB proteins, p65/RelA, c-Rel, RelB, NF-κB2/p52 and NF-κB1/p50, each containing a Rel homology domain that mediates DNA binding and dimerization. In the absence of stimuli, NF-κB is sequestered predominantly in the cytoplasm by inhibitory IκB proteins. Stimulation by effectors such as TNFα, interleukin-1 (IL-1), viral proteins, and double-stranded RNA, triggers IκB phosphorylation and degradation, NF-κB nuclear translocation, and transcriptional regulation of target genes (Silverman and Maniatis 2001; Ghosh and Karin 2002; Li and Verma 2002). Generally, NF-κB is described as a transcriptional activator, and there is ample data demonstrating that NF-κB positively regulates transcription. However, there is also some evidence that NF-κB can repress transcription. For example, the p50 NF-κB subunit, which binds DNA but lacks transcriptional activation domains, may inhibit the transcription of certain genes through interaction with transcriptional corepressors such as HDAC-1 (Zhong et al. 2002).
Recently, we demonstrated that NF-κB can inhibit the differentiation of mesenchymally derived skeletal muscle cells (Guttridge et al. 2000). TNFα-mediated activation of NF-κB in C2C12 myoblasts led to a dramatic decrease in endogenous MyoD mRNA levels. Expression of the p65 subunit of NF-κB induced the loss of coexpressed MyoD message, and a fragment of the MyoD-coding sequence fused to a heterologous gene conferred NF-κB-dependent loss of that chimeric mRNA. These data suggested that the mechanism underlying MyoD mRNA down-regulation and inhibition of muscle-cell differentiation involves a unique posttranscriptional silencing mechanism. Cartilage, another mesenchymal-derived cell lineage, is a highly specialized connective tissue that is important for skeletal development (de Crombrugghe et al. 2001; Shum and Nuckolls 2002). The transcription factor Sox9 is required for cartilage development (Bi et al. 1999; Panda et al. 2001), and loss of Sox9 causes campomelic dysplasia, a dominant skeletal disease (Foster 1996; Schafer et al. 1996). Sox9 expression has been shown to be down-regulated by cytokines such as TNFα and IL-1 (Murakami et al. 2000), which may explain the deleterious role of inflammatory cytokines in diseases such as rheumatoid arthritis and osteoporosis (Fujita et al. 1990; Chen and Goeddel 2002). Cytokine-induced NF-κB inhibits Sox9 expression and activity (Murakami et al. 2000), however, it is not known whether this occurs at the transcriptional or posttranscriptional level. Additionally, functional characterization of the Sox9 promoter (Kanai and Koopman 1999) did not identify binding sites for NF-κB, suggesting that posttranscriptional regulatory pathways may regulate NF-κB-mediated inhibition of Sox9.
Here, we report that NF-κB suppresses chondrogenesis and controls the loss of Sox9 mRNA. Furthermore, NF-κB-mediated loss of both MyoD and Sox9 mRNAs requires a 5′-ACUACAG-3′ motif present multiple times in the mRNAs. Our data can explain, at least partly, the ability of inflammatory cytokines, such as TNFα, to suppress differentiation of mesenchymal-derived cells, and they demonstrate a novel mechanism by which NF-κB can silence gene expression.
Results and Discussion
NF-κB inhibits chondrocytic differentiation and mediates posttranscriptional down-regulation of Sox9 mRNA
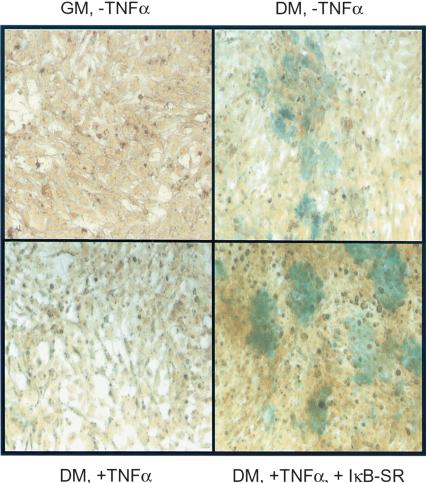
Cytokines, such as TNFα and IL-1 can potently inhibit the differentiation of chondrocytes, the precursors of cartilage (Murakami et al. 2000). To determine whether TNFα-mediated inhibition of differentiation of chondrocytes was mediated by or required NF-κB, we performed differentiation experiments in the chondrocytic cell line MC615-TRexI, which stably expresses a Tet-promoter inducible IκBα super-repressor (IκBα-SR) construct. We first determined that doxycycline (dox) induction of IκBα-SR in MC615-TrexI cells blocked TNFα-mediated NF-κB DNA-binding activity in electromobility shift assays (data not shown). Confluent cells were driven toward chondrocytic maturation by culturing in differentiation medium (DM), and we examined the formation of a cartilage-like matrix containing sulfated glycosaminoglycans by histochemical staining with Alcian blue. Cells grown in DM showed increased Alcian blue staining compared with cells grown in rich growth medium (GM). As expected, TNFα-treated cells grown in DM showed decreased staining, indicating inhibition of chondrocytic differentiation (Fig. 1). In contrast, dox-induction of IκBα-SR promoted chondrocytic differentiation in both untreated and TNFα-treated MC615 cells grown in DM. These results demonstrate a definitive role for NF-κB in modulating the differentiation of chondrocytic cells.
Figure 1.
NF-κB inhibits mesenchymal chondrocytic differentiation in vivo. MC615 cells expressing Tet-IκB-SR were cultured in growth medium (GM) or differentiation medium (DM) and grown in the absence or presence of TNFα and doxycycline to induce IκBα-SR. Cells were stained with alcian blue to determine extent of matrix formation containing sulfated glycosaminoglycans, an indicator of chondrocytic differentiation. Increased alcian blue staining is seen in cells grown in DM in the absence TNFα, as well as in the presence of TNFα and dox-induced IκBα-SR.
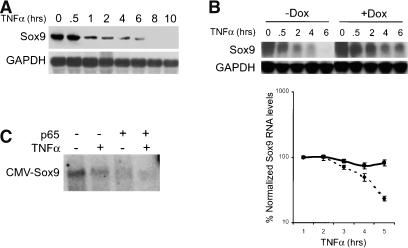
It has been reported that TNFα treatment of MC615 chondrocytic cells leads to the NF-κB-dependent loss of Sox9 (Murakami et al. 2000), a transcription factor that is required for cartilage differentiation. However, the mechanism underlying this process is not known. We sought to determine whether NF-κB-mediated down-regulation of Sox9 mRNA occurs through a mechanism similar to the loss of MyoD in TNFα-induced suppression of myocyte differentiation. To address whether TNFα-mediated regulation of Sox9 is dependent on NF-κB, we first examined endogenous Sox9 expression in MC615 chondrocytic cells by Northern analysis. After reaching 80%-90% confluency in growth medium, MC615 cells were switched to differentiation medium for 2-3 d, and then subject to TNFα treatments. All cells were cultured for the same total time. Total RNA from untreated or TNFα-treated cells were harvested and hybridized with a Sox9 cDNA probe (Fig. 2A). Sox9 mRNA expression was potently inhibited by TNFα treatment starting at 1 h, with complete inhibition seen at 8 h. Interestingly, destabilization of Sox appears to be biphasic, with an initial decrease in message at 1 h and a second decrease at 8 h. The kinetics of RNA loss is similar to that observed in previously published work describing inhibition of Sox9 mRNA by TNFα and IL-1 (Murakami et al. 2000). To determine whether TNFα-induced loss of Sox9 transcript was mediated by NF-κB, we examined Sox9 expression by Northern analysis in MC615-TRexI chondrocytes stably expressing IκBα-SR. We observed that Sox9 expression is inhibited with TNFα treatment and dox-induction of IκBα-SR resulted in loss of sensitivity to TNFα. These results, which are quantitated in the corresponding graph, demonstrate that NF-κB mediates down-regulation of Sox9 (Fig. 2B). The incomplete stabilization of Sox9 mRNA may be due to incomplete inhibition of NF-κB activity by IκBα-SR or to an NF-κB-independent down-regulation of Sox9 mRNA.
Figure 2.
NF-κB regulates Sox9 expression in chondrocytic cells. (A) TNF regulates Sox9 expression in chondrocytic cells. Total RNA from TNFα-treated MC615 chondrocytic cells was harvested at the indicated time points and hybridized with a Sox9 cDNA probe. Loss of Sox9 mRNA is seen with TNFα treatment. (B) TNF-induced loss of Sox9 mRNA requires NF-κB activity. Sox9 mRNA levels in MC615-TRexI chondrocytes stably expressing Tet-inducible IκBα-SR were examined by Northern analysis. A representative blot shows stabilization of Sox9 message in dox-treated cells. Graph shows quantitation of Sox9 transcripts relative to the zero time point (=value 100). (C) NF-κB p65 subunit mediates regulation of Sox9 expression and does not require the Sox9 promoter. 10T½ fibroblasts were transfected with a Sox9 expression vector and either pcDNA3.1 vector or CMV-p65, ±TNFα treatment. Northern analysis was performed to detect Sox9 expression.
We next asked whether NF-κB-mediated regulation of Sox9 occurred transcriptionally through a promoter-dependent mechanism and examined whether expression of a Sox9 construct under the control of the CMV promoter was affected by the p65 subunit of NF-κB. Northern analysis revealed that CMV-Sox9 expression was inhibited by either p65 or TNFα compared with vector control (Fig. 2C). The presence of both TNFα and p65 resulted in even greater loss of Sox9 mRNA. These results demonstrate that NF-κB p65-dependent down-regulation of Sox9 is not dependent on its promoter, but occurs posttranscriptionally (see below).
NF-κB mediates posttranscriptional down-regulation of MyoD
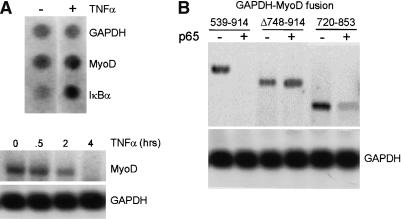
Similar to the cytokine-mediated down-regulation of Sox9, TNFα can down-regulate MyoD expression in an NF-κB-dependent manner (Guttridge et al. 2000). MyoD down-regulation appears to occur through a posttranscriptional mechanism on the basis of the observation that the MyoD promoter itself was not required for regulation by NF-κB. To determine whether NF-κB can regulate the transcription of the endogenous MyoD gene, nuclear run-on assays were performed. Results from these experiments demonstrated that transcription of MyoD was largely unaffected after 4 h of TNFα treatment, despite the virtually complete loss of endogenous MyoD mRNA assayed by Northern analysis (Fig. 3A). IκBα exhibited increased TNFα-dependent transcription, as expected for an NF-κB-regulated gene, and GAPDH remained unchanged. Together, these results clearly establish that TNFα-induced down-regulation of MyoD mRNA occurs posttranscriptionally.
Figure 3.
NF-κB down-regulates MyoD expression via a posttranscriptional mechanism. (A) Nuclear run-on and Northern blot assays were performed to analyze MyoD expression in untreated and TNFα-treated C2C12 cells. Transcription of GAPDH and IκBα were measured as negative and positive controls for NF-κB activation, respectively. TNFα (4 h) treatment does not result in significantly altered MyoD transcription, although MyoD mRNA levels are decreased by Northern assay. GAPDH expression was unaffected and IκBα mRNA levels were increased by TNFα treatment. (B) Expression analysis of different GAPDH-MyoD fusion constructs were performed by Northern analysis of RNA from transiently transfected 10T½ cells. The MyoD (539-914)-GAPDH fusion is down-regulated by p65 (cf. lanes 1 and 2). A construct containing a deletion of a region containing the 9-nucleotide motifs, MyoD (D748-D914), is insensitive to regulation by NF-κB. A 133-nucleotide fragment containing both motifs, MyoD (720-853)-GAPDH is down-regulated in a p65-dependent manner.
We next sought to identify the cis-regulatory elements required for NF-κB-dependent down-regulation of MyoD. A fragment encompassing sequence elements from 539 to 914 of MyoD fused to the GADPH gene conferred NF-κB-dependent down-regulation of the chimeric RNA (Guttridge et al. 2000). To further map a minimal cis-regulatory region in the MyoD mRNA required for regulation by NF-κB, we created two smaller deletions within the 539-914 region and tested their ability to confer down-regulation on the heterologous GAPDH mRNA. Northern analysis of transiently transfected 10T½ cells showed that MyoD Δ748-914-GAPDH is resistant to NF-κB regulation (Fig. 3B). In contrast, the MyoD 720-853-GAPDH construct retained sensitivity to NF-κB, thus defining a 130-nucleotide region within the MyoD mRNA as a crucial cis-regulatory element.
Conserved cis-regulatory elements are required for NF-κB-dependent, posttranscriptional down-regulation of Sox9 and MyoD mRNAs
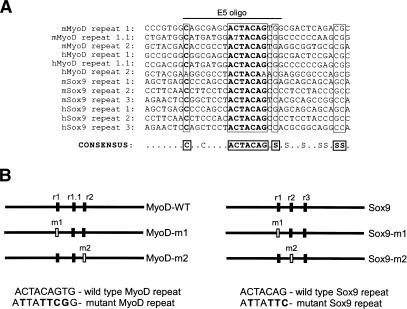
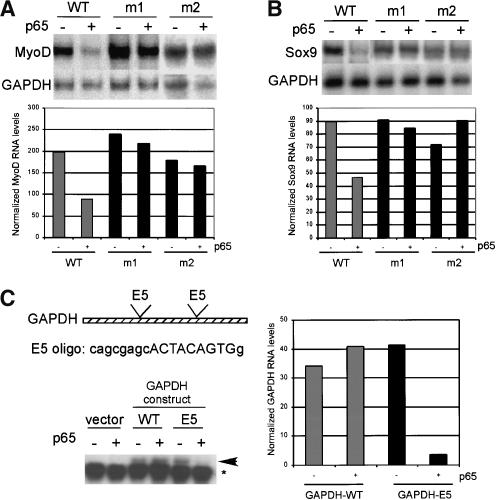
Using a Pustell DNA matrix analysis to identify regions of similarity within MyoD 720-853 (MacVector Software), we initially discovered two 9-nucleotide motifs located near the boundaries of that fragment of the sequence 5′-ACTACAGTG-3′ (written here and below as DNA sequence; Fig. 4). Another similar sequence, 5′-AtTACAGcG-3′ (repeat 1.1), was found between these 9-nucleotide repeat elements. The deletion construct that eliminated the most 5′ of these motifs (MyoDΔ748-914) was resistant to p65-dependent down-regulation (Fig. 3B), suggesting that the first ACTA CAGTG motif plays a significant role in this process. To investigate the potential function of the 9-nucleotide motifs, we introduced point mutations within each motif in the context of the full-length MyoD sequence and tested for NF-κB-dependent degradation in transiently transfected 10T½ cells (Fig. 5A). Northern blot analysis revealed that point mutations generated in either motif (ACTACAGTG → ATTATTCGG) abrogated p65-dependent destabilization of MyoD mRNA, suggesting that both intact motifs are required for full NF-κB-mediated regulation of MyoD expression.
Figure 4.
The coding sequences of MyoD and Sox9 contain multiple ACTCAG motifs. (A) Sequence alignment of repeat motifs in MyoD and Sox9 mouse and human sequences. Conserved nucleotides indicated with bold type. Srepresents C or G. The E5 oligo used in the experiment shown in Figure 5B is indicated. (B) Position and spacing of wild-type and mutant repeat motifs within MyoD and Sox9. Sequence of wild-type and mutant repeats is shown with mutated nucleotides in bold.
Figure 5.
The ACTACAG motifs are required for NF-κB-dependent mRNA destabilization. Representative Northern blots are shown with data quantitated in corresponding graphs. (A) MyoD mutants were generated within the context of the full-length coding sequence (see Fig. 4B). 10T½ cells were transfected with wild-type or mutant MyoD constructs and with or without p65. MyoD constructs with mutations in either motif (m1 or m2) are largely resistant to p65-mediated down-regulation compared with wild-type MyoD. (B) A 7-nucleotide motif in Sox9 is similar to the 9-nucleotide MyoD motif and required for efficient down-regulation by NF-κB. Mutations in Sox9 were generated within the context of the full-length coding sequence. The Sox9 m1 and m2 mutants were insensitive to down-regulation by p65 compared with wild-type Sox9. (C) A short oligonucleotide containing the ACTACAG motif is sufficient to confer p65-dependent destabilization of GAPDH-E5. The sequence and position of the E5 oligo is shown. Transfected constructs are indicated by an arrow. Endogenous GAPDH expression is indicated with an asterisk.
Interestingly, a core 7-nucleotide sequence (ACTA CAG), as well as other homologous nucleotides, are highly conserved in mouse and human MyoD sequences (Fig. 4A). Scanning of Sox9 for distinctive motifs revealed a 7-nucleotide repeat (ACTACAG) that is present three times within a 250-nucleotide region in the middle of the mRNA-coding sequence (Fig. 4A). This motif in Sox9 is identical to the first 7 nucleotides of the ACTAC AGTG repeat found in MyoD, and is also conserved between mouse and human Sox9 sequences, suggesting that down-regulation of Sox9 and MyoD involves similar mechanisms. Given the similarity of the motifs present in both MyoD and Sox9, we investigated whether the 7-nucleotide motifs present in Sox9 were required for regulation by NF-κB. We introduced mutations into the first repeat sequence (ACTACAG→ATTATTC, see Fig. 4B) and analyzed Sox9 expression in the absence or presence of p65. Whereas the wild-type Sox9 sequence is down-regulated in the presence of p65, mutation of either the first and second ACTACAG motif (Sox9-m1) stabilized the Sox9 mRNA (Fig. 5B).
To determine whether the ACTACAG motifs could specify down-regulation by NF-κB, we introduced an oligonucleotide containing a single motif into two positions within GAPDH and assayed their effect on the stability of the chimeric GAPDH mRNA (Figs. 4A, 5C). The oligos were spaced 125 nucleotides apart within the central coding region of GAPDH, similar to the spacing of the motifs within MyoD. After transient transfection of 10T½ cells, we examined the expression of wild-type and chimeric GAPDH transcript (E5) by Northern analysis. The expression of GAPDH-E5 was similar to wild-type GAPDH (Fig. 5C, lanes 3,5). We observed almost complete destabilization of the GAPDH-E5 transcript in the presence of p65, whereas both the wild-type construct, as well as endogenous GAPDH, remained unaffected by p65 expression (Fig. 5C, lanes 4,6). The ability of the E5 oligo to confer p65-mediated down-regulation of GAPDH, a transcript that is not typically regulated by NF-κB, demonstrates that this short sequence containing the ACTACAG motifs is sufficient to generate an NF-κB response. These results suggest that the ACTACAG motifs represent, presumably indirect, NF-κB-responsive elements in the MyoD and Sox9 mRNAs (also see below). In sum, our results indicate that the conserved ACTACAG motifs are both necessary and sufficient for NF-κB-mediated Sox9 and MyoD down-regulation, demonstrating a novel mechanism by which NF-κB regulates gene expression and modulates cell differentiation.
Conclusions
Our data demonstrate that NF-κB can modulate the differentiation status of mesenchymal-derived cell lineages in response to cytokine signaling, and that NF-κB-dependent inhibition of chondrocytic and myocytic differentiation occurs through the posttranscriptional down-regulation of the key developmental regulators, Sox9 and MyoD. Using mutagenesis experiments, we have established that this down-regulation requires a novel ACTACAG motif found multiple times in the coding region of both mRNAs, and demonstrated that a short oligo containing this motif is sufficient to confer NF-κB-dependent destabilization of a heterologous mRNA. Interestingly, the ACTACAG heptamer, as well as some additional nucleotides close to each motif, is conserved in both mouse and human MyoD and Sox9 sequences (Fig. 4). The significance of the additional conserved nucleotides remains to be determined. It is also not clear whether the localization or spacing of the repeat elements within the coding regions of Sox9 and MyoD is significant. The ACTACAG motif may form a binding site for a factor that regulates the stability of Sox9 and MyoD mRNAs. Preliminary secondary structure analysis using Mfold 3.0 (M. Zuker, Washington University, and the NRCC) suggests that the ACTACAG motif may form part of a stem-loop structure. However, because NF-κB transcriptional activity is required for down-regulation of MyoD (Guttridge et al. 2000), it is likely that NF-κB does not directly interact with this motif, but rather, induces the expression of a factor that regulates RNA stability. This hypothetical factor could directly promote RNA degradation, or inhibit RNA stability.
Several studies indicate that NF-κB can inhibit gene expression in other mesenchymal cell lineages. For example, it has been shown that TNFα stimulation of adipocytes suppressed expression of adipocytic genes, including PPARγ, as well as induced pre-adipocytic genes, in an NF-κB-dependent manner (Xing et al. 1997; Ruan et al. 2002). It has also been reported that TNFα-mediated down-regulation of the transcription factor Runx2/CbfaI, which is required for osteoblast differentiation (Karsenty 2001; Komori 2002), occurs at multiple levels, including RNA destabilization and transcriptional repression (Gilbert et al. 2002). Interestingly, the Runx2/Cbfa1 mRNA-coding sequence contains similar, but not perfect, AC TACAG motifs. Thus, it seems likely that both transcriptional and posttranscriptional mechanisms may be used by NF-κB to regulate the differentiation of other mesenchymal lineages and/or the expression of other genes. It will be important to determine the breadth of the NF-κB-mediated, posttranscriptional down-regulation of mRNAs through this RNA sequence-dependent mechanism in order to relate this process to inflammatory disorders and oncogenesis.
Materials and methods
Constructs
NF-κB p65, IκBα, IκBα-SR, GAPDH, full-length MyoD (pcDNA3-MyoD), and MyoD (539-914) and MyoD 539-914-GAPDH fusion constructs were described previously (Guttridge et al. 2000). MyoD Δ748-914 was created by excising a 75-nucleotide fragment in pcDNA-GAPDH-MyoD 539-914 with NarI and MluI, filling in and religating the plasmid. The deletion maintains an ORF within the mouse MyoD fragment. MyoD 720-853 fragment was PCR amplified from pcDNA3-MyoD using the following oligos: 5′-CTGCAGAACCAAAGGCTTGGCAGCGAGCAC TACAGT-3′ and 5′-ATGCATCCAAAGGCTTTGCACTGTAGTAGG CGGTGT-3′, digested with BstXI and subcloned into the GAPDH BstXI site. Full-length mouse Sox9 was amplified by reverse transcriptase PCR (RT-PCR) from MC615 total RNA, with the following oligos: 5′-CGAATTCCGTATGAATCTCCTGGACCC-3′ and 5′-CGGGATCCAT CATCTCGGCCATCGTCG-3′, and subcloned into pcDNA3.1 vector (Invitrogen). pcDNA3-MyoD and Sox9 mutant constructs were made using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the primers for MyoD (mMD-1/5′, CCCCCGGGCCGTGGCAGCGAG CATTATTCGGGCGACTCAGACGCG; mMD-1/3′, CGCGTCTGAGT CGCCCGAATAATGCTCGCTGCCACGGCCCGGGG; mMD-2/5′, GG CTACGACACCGCCTATTATTCGAGGCGGTTCGCGAGTCC; mMD-2/3′, GGACTCGCGAACCGCCTCCGAATAATAGGCGGTGTCGTA GCC) and for Sox9. Mutated nucleotides are indicated in bold. Tetinducible IκBα-SR (TRex-IκBα-SR) was constructed by subcloning a HindIII/SmaI human IκBα-SR fragment into the HindIII/SmaI of pcDNA/TO/Myc/HisA (Invitrogen). For GAPDH-E5, first oligos 5′-CATG GCAGCGAGCACTACAGTGGA-3′ and 5′-CATGTCCACTGTAGTG CTCGCTGC-3′ were annealed and subcloned into GAPDH NcoI site, then oligos 5′-CCAGCGAGCACTACAGTGGATGCA-3′ and 5′-TCCACTGTAGTGCTCGCTGGTGCA-3′ were annealed and subcloned into GAPDH NsiI site.
Cell culture and histochemical analysis
C2C12 and 10T½ cells were obtained from ATTC. Dr. B. Olsen kindly provided the chondrocytic cell line, MC615 (Mallein-Gerin and Olsen 1993). C2C12 and MC615 cells were maintained in growth medium (GM) containing DMEM-H, 20% fetal bovine serum, 100 μg/mL penicillin and 100 μg/mL streptomycin (Sigma). For C2C12 myogenic differentiation, cells were changed to DMEM containing 2% horse serum, 5 μg/mL insulin and pen/strep. For MC615 chondrocytic differentiation, cells were grown to 80%-90% confluency and changed to differentiation medium [DM: DMEM-H 5% FCScontaining 50 μg/mL ascorbic acid (Invitrogen)], and 10 mM β-glycerophosphate (Sigma) for 2-3 d. NF-κB activation was induced with 20 ng/mL rhTNFα (Promega). Transient transfections were carried out using Fugene6 (Roche) according to the manufacturer's instructions.
MC615 stably expressing IκB-SR (MC615-TRexI) were generated with the TRex system (Invitrogen). Briefly, MC615 cells were cotransfected with TRex-IκB-SR and pcDNA6/TR, which expresses the Tet repressor under the control of the human CMV promoter. Stable integrants were selected with Blasticidin (5 μg/mL) and Zeocin (250 μg/mL) and individual clones were tested for tetracycline/doxycycline induction of IκBα-SR expression by Western blot (data not shown).
Chondrogenic differentiation was determined by staining of sulfated glycosaminoglycans with Alcian Blue 8GX (pH 1; Sigma), as described (Asahina et al. 1996). Briefly, cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min, and stained with 0.5% Alcian Blue 8GX in 0.1 N HCl overnight.
Nuclear run-on assays
For each condition, 4 × 106 cells were harvested and incubated on ice for 3 min in lysis buffer (10 mM Tris-HCL at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40). Nuclei were pelleted by centrifugation at 500g, resuspended in 100 μL Nuclear Freezing buffer (50 mM Tris-HCl at pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA at pH 8) and either frozen at -80°C or used immediately. For the run-on reaction, 30 μL of 5× Run-on buffer (25 mM Tris-HCl at pH 8; 12.5 mM MgCl2; 750 mM KCl; 1.25 mM each of ATP, GTP, and CTP; and 100 μCi [α32P]UTP) and 0.06% Sarkosyl was added to 100 μL of nuclei on ice. Reactions were allowed to proceed at 30°C for 30 min, after which DNase I (Promega) was added and incubation proceeded for another 15 min. at 30°C. RNA was isolated from nuclei with Trizol (Invitrogen). Plasmid fragments were electrophoresed and dot-blotted on nylon membrane. Membranes were hybridized with 3 × 106 cpm of transcribed RNA for 16-24 h, then washed three to four times with 2× SSC, and analyzed by autoradiography.
Northern blot assays
Total RNA was isolated from cells from a confluent 100-mm plate using Trizol (Invitrogen). 5-20 μg of RNA was electrophoresed on a denaturing formaldehyde gel and transferred onto a nylon membrane (HyBond-NX, Amersham). RNAs were UV-cross-linked to the membrane and hybridized using ExpressHyb (Clontech) according to the manufacturer's instructions. Northern blot data was quantitated using Scion Image (NIH), normalized to endogenous GAPDH levels and multiplied by a factor of 100 for presentation purposes. Experiments were repeated at least three times.
Acknowledgments
We are grateful to Denis Guttridge for MyoD constructs and interesting scientific discussions and Ahmad Amin for assistance in generating the MC615-TRexI cells. We thank Hubert Amrein and Kris Steinbrecher for critical reading of the manuscript. This work was funded by NIH grants AI35098 and CA73756 to A.S.B. R.S. is funded by a Cancer Research Institute postdoctoral fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1114503.
References
- Asahina I., Sampath, T.K., Hauschka, P.V. 1996. Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp. Cell. Res. 222: 38-47. [DOI] [PubMed] [Google Scholar]
- Bi W., Deng, J.M., Zhang, Z., Behringer, R.R., and de Crombrugghe, B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85-89. [DOI] [PubMed] [Google Scholar]
- Caplan A.I. and Bruder, S.P. 2001. Mesenchymal stem cells: Building blocks for molecular medicine in the 21st century. Trends Mol. Med. 7: 259-264. [DOI] [PubMed] [Google Scholar]
- Chen G. and Goeddel, D.V. 2002. TNF-R1 signaling: A beautiful pathway. Science 296: 1634-1635. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Lefebvre, V., and Nakashima, K. 2001. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell. Biol. 13: 721-727. [DOI] [PubMed] [Google Scholar]
- Filipak M., Sparks, R.L., Tzen, C.Y., and Scott, R.E. 1988. Tumor necrosis factor inhibits the terminal event in mesenchymal stem cell differentiation. J. Cell. Physiol. 137: 367-373. [DOI] [PubMed] [Google Scholar]
- Foster J.W. 1996. Mutations in SOX9 cause both autosomal sex reversal and campomelic dysplasia. Acta Pediatr. Jpn. 38: 405-411. [DOI] [PubMed] [Google Scholar]
- Fujita T., Matsui, T., Nakao, Y., Shiozawa, S., and Imai, Y. 1990. Cytokines and osteoporosis. Ann. NY Acad. Sci. 587: 371-375. [DOI] [PubMed] [Google Scholar]
- Ghosh S. and Karin, M. 2002. Missing pieces in the NF-κB puzzle. Cell 109: S81-S96. [DOI] [PubMed] [Google Scholar]
- Gilbert L., He, X., Farmer, P., Boden, S., Kozlowski, M., Rubin, J., and Nanes, M.S. 2000. Inhibition of osteoblast differentiation by tumor necrosis factor-α. Endocrinology 141: 3956-3964. [DOI] [PubMed] [Google Scholar]
- Gilbert L., He, X., Farmer, P., Rubin, J., Drissi, H., van Wijnen, A.J., Lian, J.B., Stein, G.S., and Nanes, M.S. 2002. Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2α A) is inhibited by tumor necrosis factor-α. J. Biol. Chem. 277: 2695-2701. [DOI] [PubMed] [Google Scholar]
- Guttridge D.C., Mayo, M.W., Madrid, L.V., Wang, C.Y., and Baldwin Jr., A.S. 2000. NF-κB-induced loss of MyoD messenger RNA: Possible role in muscle decay and cachexia. Science 289: 2363-2366. [DOI] [PubMed] [Google Scholar]
- Kanai Y. and Koopman, P. 1999. Structural and functional characterization of the mouse Sox9 promoter: Implications for campomelic dysplasia. Hum. Mol. Genet. 8: 691-696. [DOI] [PubMed] [Google Scholar]
- Karsenty G. 2001. Minireview: Transcriptional control of osteoblast differentiation. Endocrinology 142: 2731-2733. [DOI] [PubMed] [Google Scholar]
- Komori T. 2002. Runx2, a multifunctional transcription factor in skeletal development. J. Cell. Biochem. 87: 1-8. [DOI] [PubMed] [Google Scholar]
- Li Q. and Verma, I.M. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2: 725-734. [DOI] [PubMed] [Google Scholar]
- Mallein-Gerin F. and Olsen, B.R. 1993. Expression of simian virus 40 large T (tumor) oncogene in mouse chondrocytes induces cell proliferation without loss of the differentiated phenotype. Proc. Natl. Acad. Sci. 90: 3289-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Lefebvre, V., and de Crombrugghe, B. 2000. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-α. J. Biol. Chem. 275: 3687-3692. [DOI] [PubMed] [Google Scholar]
- Panda D.K., Miao, D., Lefebvre, V., Hendy, G.N., and Goltzman, D. 2001. The transcription factor SOX9 regulates cell cycle and differentiation genes in chondrocytic CFK2 cells. J. Biol. Chem. 276: 41229-41236. [DOI] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay, A.M., Beck, S.C., Jaiswal, R.K., Douglas, R., Mosca, J.D., Moorman, M.A., Simonetti, D.W., Craig, S., and Marshak, D.R. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284: 143-147. [DOI] [PubMed] [Google Scholar]
- Ruan H., Hacohen, N., Golub, T.R., Van Parijs, L., and Lodish, H.F. 2002. Tumor necrosis factor-α suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: Nuclear factor-κB activation by TNF-α is obligatory. Diabetes 51: 1319-1336. [DOI] [PubMed] [Google Scholar]
- Schafer A.J., Foster, J.W., Kwok, C., Weller, P.A., Guioli, S., and Goodfellow, P.N. 1996. Campomelic dysplasia with XY sex reversal: Diverse phenotypes resulting from mutations in a single gene. Ann. NY Acad. Sci. 785: 137-149. [DOI] [PubMed] [Google Scholar]
- Scott R.E. 1997. Differentiation, differentiation/gene therapy and cancer. Pharmacol. Ther. 73: 51-65. [DOI] [PubMed] [Google Scholar]
- Shum L. and Nuckolls, G. 2002. The life cycle of chondrocytes in the developing skeleton. Arthritis Res. 4: 94-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N. and Maniatis, T. 2001. NF-κB signaling pathways in mammalian and insect innate immunity. Genes & Dev. 15: 2321-2342. [DOI] [PubMed] [Google Scholar]
- Xing H., Northrop, J.P., Grove, J.R., Kilpatrick, K.E., Su, J.L., and Ringold, G.M. 1997. TNF α-mediated inhibition and reversal of adipocyte differentiation is accompanied by suppressed expression of PPARγ without effects on Pref-1 expression. Endocrinology 138: 2776-2783. [DOI] [PubMed] [Google Scholar]
- Zhong H., May, M.J., Jimi, E., and Ghosh, S. 2002. The phosphorylation status of nuclear NF-κ B determines its association with CBP/p300 or HDAC-1. Mol. Cell 9: 625-636. [DOI] [PubMed] [Google Scholar]