Abstract
A massively parallel pyro-sequencing technology commercialized by 454 Life Sciences Corporation was used to sequence the transcriptomes of shoot apical meristems isolated from two inbred lines of maize using laser capture microdissection (LCM). A computational pipeline that uses the POLYBAYES polymorphism detection system was adapted for 454 ESTs and used to detect SNPs (single nucleotide polymorphisms) between the two inbred lines. Putative SNPs were computationally identified using 260 000 and 280 000 454 ESTs from the B73 and Mo17 inbred lines, respectively. Over 36 000 putative SNPs were detected within 9980 unique B73 genomic anchor sequences (MAGIs). Stringent post-processing reduced this number to > 7000 putative SNPs. Over 85% (94/110) of a sample of these putative SNPs were successfully validated by Sanger sequencing. Based on this validation rate, this pilot experiment conservatively identified > 4900 valid SNPs within > 2400 maize genes. These results demonstrate that 454-based transcriptome sequencing is an excellent method for the high-throughput acquisition of gene-associated SNPs.
Keywords: SNPs, ESTs, maize, 454 sequencing, markers
Introduction
SNPs (single nucleotide polymorphisms) are single base differences between haplotypes. Once discovered, SNPs can be converted into genetic markers that can be inexpensively assayed in a high-throughput manner (Gut, 2001; Kwok, 2001). Due to their abundance, it is possible to use SNP-based markers to generate very dense genetic maps (Rafalski, 2002). Such maps can be used to conduct marker-assisted selection (MAS) programs, construct the specific genotypes required for quantitative genetic studies, and to enhance our understanding of genome organization and function and address fundamental questions relating to evolution and meiotic recombination. SNPs can also be used for genome-wide linkage disequilibrium and association studies that assign genes to specific functions or traits. Furthermore, transcript-associated SNPs can be used to develop allele-specific assays for the examination of cis-regulatory variation within a species (Bray et al., 2003; Cowles et al., 2002; Guo et al., 2003; Pastinen et al., 2004; Stupar and Springer, 2006).
Although SNPs can be identified by sequencing candidate genes from a set of individuals that represent diversity in the species of interest, this is neither high-throughput nor inexpensive. Alternative approaches used during construction of the human SNP map included identifying sequence polymorphisms within overlapping BAC clones derived from different individuals and shotgun sequencing of genomic fragments (Sachidanandam et al., 2001). However, this approach is not always possible because many genome sequencing projects use DNA extracted from highly similar or inbred individuals. Instead, SNP-based markers are typically mined from whole-genome sequences or expressed sequence tags (ESTs) obtained from genetically diverse individuals. For example, SNPs have been identified by comparing genomic sequences from two or more genetically distinct inbred lines of mouse (Wiltshire et al., 2003), the indica and japonica sub-species of rice (Feltus et al., 2004), the Columbia and Landsberg ecotypes of Arabidopsis (Jander et al., 2002), and different lines of maize (Yamasaki et al., 2005). EST collections from genetically dissimilar individuals have similarly been mined for SNPs in humans (Marth et al., 1999), pine (Dantec et al., 2004), barley (Kota et al., 2001, 2003), cassava (Lopez et al., 2005) and maize (Batley et al., 2003).
The latest maize genetic map (IBM_IDP_bd map, ver4) contains over 3000 gene-based PCR markers distributed across the 2.5 Gbp genome (Fu et al., 2006). Even so, this map is not dense enough to support high-resolution mapping applications and association genetics, particularly given the decay of linkage disequilibrium outside of maize genes (Ching et al., 2002; Tenaillon et al., 2001). Additionally, because the maize inbred B73 line is being hierarchically sequenced, a higher density genetic map would be invaluable for anchoring each sequenced BAC contig to its proper place in the genome. Increasing the marker density of this crop therefore has applications in accurately assembling this highly complex genome and ultimately in improving agricultural traits.
Maize is genetically very diverse; SNP and indel polymorphism frequencies between inbred lines and landraces average one variation per 124 or 28 bases for coding regions (Ching et al., 2002) or all associated regions (Tenaillon et al., 2001), respectively. We were particularly interested in identifying SNPs between B73 and the inbred line Mo17. These two inbred lines represent two of the major heterotic groups, and historically are the parental lines of much of the commercial corn grown in the USA. These inbred lines are also the parents of the IBM RILs (recombinant inbred lines) that were used to develop the maize genetic community’s high-resolution genetic maps.
The size and complexity of the maize genome make it unlikely that a second inbred line will be sequenced in the immediate future. Although there are currently over 650 000 maize EST sequences available in GenBank, nearly all of these were drawn from a small subset of inbred lines, principally B73, W23 and Oh43A. Hence, the identification of B73/Mo17 SNPs requires the development of Mo17 EST sequence resources. Although genome sequencing technology has become progressively more efficient, EST projects require substantial investments in library construction and sequencing efforts to achieve the overall coverage required to locate SNPs.
Recently, 454 Life Sciences (http://www.454.com) reported a highly parallel DNA sequencing system that is 100 times faster than standard sequencing methods and is capable of providing over 20 Mbp of sequence in a single four-hour run (Margulies et al., 2005). Increased throughput comes at the expense of read length (100 bp average length) and the absence of clone pair information, making it less attractive for whole-genome sequencing of complex genomes. However, 454 sequencing of maize cDNAs obtained from shoot apical meristem (SAM) tissue isolated by laser capture microdissection (LCM; reviewed by (Schnable et al., 2004) has recently been shown to be an effective method for tagging tens of thousands of maize genes without cloning and its associated costs (Emrich et al., 2007a). Therefore, 454-based sequencing of the B73 and Mo17 SAM transcriptome was expected to provide a collection of diverse ESTs that could support high-throughput computational identification of gene-associated SNPs. Because 454 reads contain more sequence errors than do reads generated by traditional sequencing technology (Margulies et al., 2005), it was not, however, clear whether 454-based ESTs could be used for SNP discovery.
Here, we describe the generation of over 280 000 Mo17 SAM ESTs using 454 sequencing technology, the development of an efficient computational SNP mining pipeline based on the POLYBAYES sequence polymorphism detection tool, and the subsequent identification of over 7000 putative Mo17/B73 SNPs within expressed sequences, a subset of which has been experimentally validated.
Results
The shoot apical meristem ultimately gives rise to all above-ground tissues. Thus, it is expected that many rare and developmentally important transcripts are present in the SAM transcriptome. Indeed, we have demonstrated that 454 sequencing of maize SAM cDNA captures fragments of thousands of genes, including many that may be expressed only rarely or only in the SAM (Emrich et al., 2007a).
Using 454 sequencing, we previously generated from the B73 inbred line a collection of 260 887 high-quality SAM ESTs with an average length of 101 bp (Emrich et al., 2007a). Using the same methodology, a collection of 454 SAM ESTs was generated from the maize inbred line Mo17 (Experimental procedures). After trimming polyA/T tails, the 287 917 resulting SAM ESTs from Mo17 had an average length of 100 bp, and consisted of 30.7 Mbp in total.
Assignment of Mo17 and B73 SAM ESTs to maize genomic anchor sequences
MAGIs are maize genomic sequence assemblies (Fu et al., 2005) composed of gene-enriched B73 genomic survey sequences (Whitelaw et al., 2003). Because these sequences are highly accurate (1 disagreement per 10 000 bp; Fu et al., 2005) and comprehensive (> 75% of all maize genes are present), they provide an excellent collection of B73 reference sequences for SNP detection. Attempts were made to align each of the 260 887 B73 and 287 917 Mo17 454 ESTs to the MAGI version 3.1 partial maize B73 genome assembly using a two-step approach. The initial pre-processing step uses BLAST to save time and improve accuracy by grouping together individual 454 SAM ESTs that preferentially align to a single MAGI template (Experimental procedures). This analysis assigned 432 431 of the 454 ESTs (207 294 B73 and 225 137 Mo17) to 48 063 MAGIs (Table 1). Of these MAGIs, 20 432 aligned to both B73 (n= 120 662) and Mo17 (n= 135 249) ESTs. An additional 14 496 and 13 135 MAGIs aligned to only Mo17 (n= 89 888) or only B73 (n= 86 632) ESTs, respectively. The MAGI assembly sequences identified above served as templates upon which associated 454 ESTs were multiply aligned by CROSS_MATCH (P. Green, University of Washington, personal communication).
Table 1.
Summary of multiple sequence alignments (MSAs) between MAGI 3.1 anchors and B73 and Mo17 454 ESTs
| Types of ESTs in MSAs | ||||||
|---|---|---|---|---|---|---|
| All ESTs | All B73 ESTs | All Mo17 ESTs | Both B73 and Mo17 ESTs | Only B73 ESTs | Only Mo17 ESTs | |
| Number of MAGIs aligned | 48 063 | 33 567 | 34 928 | 20 432 | 13 135 | 14 496 |
| Bases covered | 8 897 508 | 4 989 045 | 5 798 933 | 1 890 459 | 3 098 586 | 3 908 463 |
| Coverage depth | 1.8 x | 2.3 x | 2.3 x | 8.4 x | 1.3 x | 1.3 x |
For this analysis, 454 sequences were initially mapped to individual MAGIs using BLAST, which later served as the template on which these MSA were computed using CROSS_MATCH (Experimental procedures). Coverage data are presented for all alignments, as well as alignments between individual subsets of ESTs.
Doing so produced 48 063 anchored multiple sequence alignments (MSAs) that covered a total of 8 897 508 MAGI template bases with 454 ESTs. Approximately 5 Million and 5.8 Million anchor template bases were sampled by B73 and Mo17, respectively, while slightly fewer than 1.9 Million bases were sampled by both inbred lines (Table 1). The relative proportions and average sequence depths (coverage) of the 1.9 Mbp MAGI nucleotides sampled by B73 and Mo17 454 ESTs are presented in Table 2. Although it is theoretically possible to identify putative B73/Mo17 SNPs across the entire region of the MAGI 3.1 sequence space that was simultaneously sampled by B73 and Mo17 454 ESTs (approximately 1.9 Mbp), analysis of those regions that contain deeper sequencing coverage for both inbred lines is expected to yield putative SNPs that are more likely to be valid. We therefore defined a high-confidence set of bases on the MAGI anchor that was sampled to ≥ threefold by Mo17 ESTs and to ≥ twofold by B73 ESTs. With the inclusion of the MAGI 3.1 anchor sequence (B73), these bases are sampled a minimum of three times for both inbred lines. This set comprises 42% (606 180) of the simultaneously sampled sequence space (Table 2).
Table 2.
Average coverage of nucleotide sites represented within B73 454 ESTs, Mo17 454 ESTs and MAGI 3.1 anchored multiple sequence alignments
| 454 EST component depths | Number of nucleotides | Average coverage | ||
|---|---|---|---|---|
| Mo17 | B73 | 1 092 570 | 3.2 | |
| 1 x | ≥1 x | |||
| or | ||||
| ≥ 1 x | 1 x | 326 095 | 5.9 | |
| 2 x | ≥ 2 x | |||
| or | ||||
| 2 x | ≥ 2 x | |||
| ≥ 3 x | 2 x | 134 386 | 6.7 | |
| ≥ 3 x | ≥ 3 x | 471 794 | 22 | |
Although the alignment of a single Mo17 EST to a B73-derived MAGI is sufficient to predict a SNP, increased sampling depth is expected to increase the accuracy of SNP calling by filtering out sequencing errors. Depth classes that are grouped together were pooled for analysis.
Polymorphism detection using POLYBAYES
Putative SNPs were identified from the MSA using the POLYBAYES polymorphism software package (Marth et al., 1999). POLYBAYES uses a Bayesian statistical model that considers depth of coverage, sequence quality and an expected polymorphism rate to determine the probability that polymorphic sites within an MSA are SNPs rather than disagreements resulting from either sequencing errors or the alignment of paralogous (rather than allelic) sequences (Marth et al., 1999).
454 sequencing technology is susceptible to indel-type errors (Margulies et al., 2005), and the resulting ESTs exhibit an overall rate of sequencing error of approximately 1.5% (Emrich et al., 2007a). To address the issue of indel-type errors, we used MAGI assemblies as templates on which 454 SAM ESTS were aligned (Experimental procedures). Template-based MSAs such as these are often correct even in the presence of abundantly expressed or alternatively spliced transcripts (Marth et al., 1999), and are therefore more likely to overcome the technical issues associated with 454 ESTs.
POLYBAYES identifies single base substitutions as well as single base insertions and deletions. However, because of the high number of indel errors associated with 454 technology (Margulies et al., 2005), only base substitutions (i.e. SNPs) were considered in the current analysis. Initially, a total of 36 006 putative SNPs (P= 0.5) were detected within 9980 unique MAGI anchor sequences. This number of putative SNPs is expected to over-estimate the diversity present in SAM-expressed genes in the two maize inbred lines. Because Mo17 and B73 are inbred lines, they should be mono-allelic at every base position, with relatively rare exceptions caused by nearly identical paralogs (NIPs) (Emrich et al., 2007b). Hence, the observation that many of the putative SNPs discovered initially are multi-allelic within Mo17, B73 or both, suggests that many are false positives due to sequencing errors. With this in mind, we purposefully set the SNP probability low (P= 0.5) and filtered the putative SNPs using the following rules designed to substantially decrease the rate of false positives within the context of this study:
Polymorphic sites require a minimum of twofold representation in the Mo17 454 ESTs.
All Mo17 base calls at sites that were polymorphic between Mo17 454 ESTs and the B73 MAGI anchors were expected to be identical. This ensures mono-allelism within the Mo17 454 ESTs.
When B73 454 EST sequences also align across polymorphic sites that pass rules 1 and 2, all of the B73 454 ESTs and the MAGI 3.1 anchor base calls must agree. This avoids polymorphisms resulting from incorrect MAGI base calls or NIPs within B73.
To reduce the possibility of an erroneous base in the MAGI anchor mimicking a true SNP, regions of the MAGI assemblies composed of sequences from high-Cot selected clones that are not covered by B73 ESTs were avoided because 40% of high-Cot clones contain cloning artifacts that mimic SNPs (Fu et al., 2004) (see Experimental procedures).
Applying these stringent rules to the raw SNP data returned 7016 putative B73/Mo17 SNPs distributed among 3403 MAGIs. The numbers of 454 ESTs that cover these polymorphic sites range from only two Mo17 454 ESTs to at least three B73 and three Mo17 ESTs (Table 3).
Table 3.
Number of putative SNPs, depth at each SNP site by inbred line, and estimates of the total number of maize genes that contain at least one putative SNP between the B73 and Mo17 inbred lines in this SNP dataset
| 454 EST component depths of MSAs | ||||||
|---|---|---|---|---|---|---|
| Mo17 | B73 | Number of putative SNPs | Number of MAGI 3.1 anchorsa | Additive SNP number | Additive minimum estimate of SNP-containing genesb | |
| 1 x | 1 x | 1762 | 1154 | |||
| or | ||||||
| 1 x | 0 | |||||
| 2 x | ≥ 2 x | 1648 | 1039 | |||
| or | ||||||
| ≥ 2 x | 2 x | |||||
| ≥ 3 x | ≥ 3 x | 1452 | 900 | 1452 | 900 | |
| ≥ 3 x | 2 x | 565 | 404 | 2017 | 1205 | |
| ≥ 3 x | 1 x | 717 | 513 | 2734 | 1570 | |
| 2 x | ≥ 3 x | 537 | 372 | 3271 | 1821 | |
| 2 x | 2 x | 546 | 363 | 3817 | 2053 | |
| 2 x | 1 x | 1045 | 707 | 4862 | 2548 | |
| ≥ 3 x | 0 | 481 | 283 | 5353 | 2775 | |
| 2 x | 0 | 1673 | 830 | 7016 | 3403 | |
Polymorphic bases sampled with low redundancy (rows 1 and 2) were not further analyzed. In contrast, rows 4 and 5 illustrate polymorphic sites with a minimum sampling depth of threefold for both inbred lines, and, as a result, have the highest confidence. The remaining rows summarize alignments that predict SNPs with decreasing confidence levels. Sub-categories that are grouped together were pooled for analysis.
MAGIs are gene-enriched maize genomic sequence assemblies that are likely to contain only a single gene or gene fragment (Emrich et al., 2004; Fu et al., 2005).
Numbers represent a non-redundant collection at each row.
For completeness, Table 3 presents all polymorphism data. The total numbers of polymorphic bases sampled by only one or two Mo17 454 ESTs and/or B73 454 ESTs are displayed in rows 1 and 2, respectively; these were removed from further consideration. The numbers of putative SNPs that pass the above rules and their associated MAGIs are presented in rows 3–12. Rows 3 and 4 illustrate the total number of polymorphic sites sampled simultaneously by a minimum of three Mo17 454 ESTs, two B73 454 ESTs and the B73 MAGI 3.1 anchor. This represents the highest-confidence data set, with a minimum sampling depth of threefold for both inbred lines. Rows 5–12 display putative SNPs at sites with decreasing depths of coverage, which are expected to represent decreasingly confident data sets. This expectation is supported by their corresponding POLYBAYES-assigned SNP probabilities (pSNP) (Supplementary Tables S1 and S2). The number of potential SNPs, the number of their associated MAGI anchors for each B73/Mo17 sampling depth, and the total number (additive) of potential SNPs and the number of unique MAGIs anticipated by systematically including data sets (starting with row 3) is also presented in Table 3. In summary, after single 454 GS-20 sequencing runs of B73 and Mo17 SAM cDNA, our computational polymorphism mining strategy identified over 7000 putative SNPs (Supplementary Table S1).
Validation of SNPs
A set of 110 putative B73/Mo17 SNPs were subjected to validation by sequencing (using Sanger technology) the corresponding alleles that had been PCR-amplified from B73 and Mo17 genomic DNA. Detailed results of these validation experiments are presented in Supplementary Table S2. The overall rate of validation was over 85% (94/110). Most of the SNPs selected for testing represent sites with at least moderate levels of B73/Mo17 coverage. Over 88% (85/96) of SNPs sampled by three or more Mo17 454 ESTs and two or more B73 454 ESTs (Table 3, rows 3 and 4) were validated. Fewer of the lesser-confidence SNPs were assayed; these exhibit a collective validation rate of 64% (9/14). Using the above validation rates, the number of SNPs that could be validated was estimated (Table 4); these data suggest that 4984 computationally identified B73/Mo17 SNPs represent ‘true’ polymorphisms, and that these are distributed within 2472 MAGIs. The average sizes of the MAGI assemblies suggest they contain only one (or a portion of one) maize gene. Because these polymorphisms were mined from cDNA sequences derived from mRNA and conservatively filtered, we estimate that this analysis identified at least 4900 valid SNPs within at least 2400 maize genes.
Table 4.
Number of putative SNPs, depth at each SNP site by inbred line, and estimates of the potential number of polymorphic maize genes adjusted for validation rates
| 454 EST component depths | |||||||
|---|---|---|---|---|---|---|---|
| Mo17 | B73 | Number of putative SNPs (Table 3) | Validation rate | Estimated number of valid SNP sitesa | Number of MAGI 3.1 anchorsab | Number of additive SNPs | Additive minimum estimate of genes impactedc |
| ≥ 3 x | ≥ 2 x | 2017 | 0.885 | 1785 | 1154 | 1785 | 1066 |
| ≥ 2 x | 0–1 x | 3916 | |||||
| 1–2 x | ≥ 2 x | 1083 | 0.64 | 3199 | 1963 | 4984 | 2472 |
Validation of 110 putative B73/Mo17 SNPs divided into two groups was performed by sequencing the corresponding B73 and Mo17 alleles using Sanger technology. Using the validation rates obtained, the number of SNPs that could be validated was estimated. Because many MAGIs correspond to single genes (see Results), the number of non-redundant MAGI anchors was used to generate the estimate of the number of genes impacted. Depths that are grouped together were pooled for analysis.
Numbers are corrected for validation rate (see text).
MAGIs are gene-enriched maize genomic sequence assemblies that are likely to contain only a single gene or gene fragment (Emrich et al., 2004; Fu et al., 2005). These numbers represent a non-redundant collection of MAGIs (see text).
Numbers represent a non-redundant collection See comment above at each row.
Discussion
Once discovered, SNPs have a wide variety of applications in biological research. One means to discover SNPs is to align ESTs from more than one genotype. LCM 454 sequencing enables efficient deep sampling of ESTs obtained from specific cell types (Emrich et al., 2007a), but suffers from the disadvantage of higher error rates than Sanger sequencing. Even so, this study demonstrates that it is possible to use ESTs obtained via LCM 454 sequencing to achieve high-throughput SNP discovery. Over 260 000 Mo17 ESTs were obtained from a single GS-20 sequencer run on cDNA isolated from SAM tissue, and over 7000 putative SNPs were identified relative to B73 genomic and 454 EST sequences. A subset of these SNPs was validated via direct sequencing of PCR products amplified from B73 and Mo17 genomic DNA.
Putative SNPs are identified as mismatches between aligned sequences, and several computational tools for SNP identification are available (Manaster et al., 2005; Marth et al., 1999; Nickerson et al., 1997; Wang and Huang, 2005; Weckx et al., 2005; Zhang et al., 2005). Our SNP discovery pipeline implements POLYBAYES, which has been used to identify SNPs in several studies (Dantec et al., 2004; Marth et al., 1999; Pavy et al., 2006; Useche et al., 2001). We assigned default values to 454 sequences based on an empirical evaluation of the base error rate rather than using the relatively new 454 quality scores. As a result, sequence depth and relative allele proportions have the greatest influence on polymorphism detection, and, based on this observation, potential SNPs were filtered by examining these statistics at each polymorphic site. The highest-confidence polymorphisms are those that are minimally covered by both Mo17 and B73 sequences to threefold. Experimentally, > 88% of these sites could be validated as being polymorphic, and are assigned prior probability scores (pSNP Check nomenclature, cf pSNP used above) of at least 0.997 by POLYBAYES.
POLYBAYES is designed to use template-driven MSAs, in which sequences are scaffolded across a high-quality template sequence that serves as an anchor. In addition to being highly accurate (Marth et al., 1999), this approach eliminates the need to perform de novo assemblies of 454 ESTs, which are complicated by the short lengths of 454 reads. Furthermore, gaps and insertions in this template-driven multiple sequence alignment approach are propagated throughout all members, so 454 semi-random indels can be easily identified and ignored (Figure 1). Finally, the ability of POLYBAYES to use quality scores during SNP detection provides the option of utilizing 454 sequence calls once they are better accepted by the research community, or if Sanger sequences are also used, or if the base accuracy of the template is suspect. In all of these cases, the availability of accurate base quality data could improve the accuracy of SNP detection.
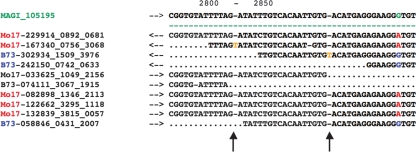
Figure 1.
A portion of the CROSS_MATCH-produced, template-driven, padded alignment between B73 and Mo17 454 EST sequences and the high-quality MAGI_105195 sequence assembly constructed from the B73 maize genomic survey sequence that serves as an alignment template. A G/A polymorphism occurs at position 2846 of the template (green highlight), with the Mo17 allele (A) in red and the B73 allele (G) in blue. Two insertions have occurred (yellow), one within a Mo17 454 EST and the second within a B73 454 EST. Because these insertions are not supported by other sequences, they are easily identified as errors by the POLYBAYSE pipeline and are not called as polymorphisms.
We estimate that our SNP collection contains at least 4984 valid SNPs within 2472 genes (see Results). This estimate is based on an observed validation rate of > 0.88 for polymorphic sites minimally sampled to threefold by each inbred line, and the assumption that all other depth classes of polymorphism have a conservative validation rate of 0.64. A subset of 2017 high-confidence SNPs was detected within B73 genomic sequence that was sampled by a minimum of two B73 ESTS and a minimum of three Mo17 454 ESTs (Table 3). The size of this reduced sequence space is 621 956 bp (Table 2), providing an observed polymorphism rate of at least 1/300. This rate is only about half of that previously reported in maize coding sequence (Ching et al., 2002); however, the published rate was based on only 18 genes and may not be representative of the genome. Furthermore, the conservative parameters used in this study are expected to under-estimate polymorphism rates. Specifically, in the absence of 454 quality information, we required that B73 and Mo17 inbred lines both be mono-allelic at each nucleotide before calling a putative SNP. In fact, 17 671 instances where either inbred line (or both) exhibits bi-allelism were initially ignored to simplify polymorphism detection and subsequent validation. These were further parsed to identify putative SNPs where the B73 and/or Mo17 major allele frequencies are ≥ 0.75, and each major allele is represented at least three times within the MSA. There are 879 such cases (Supplementary Table S1), which, if all were validated, would increase our polymorphic rate to at most 1/214 bp.
All of the polymorphic sites discussed in this study were detected by comparing the sequences obtained from single 454 GS-20 sequencer runs on cDNA obtained from Mo17 and B73 SAM tissue. Additional sequencing runs would be expected to increase the proportion of the transcriptome sequence space covered, and, perhaps most importantly, increase the overall depth of coverage. Consequently, additional sequencing runs would be expected to increase the confidence of at least a fraction of the putative SNPs that are currently poorly supported due to insufficient sampling depth (Table 3). Increased depth would also lend additional support to the identification of NIPs, a process that is particularly dependent on deep sampling.
Maize is a globally important crop and a model system for the study of genome structure, evolution and genetics. Between 5000 and 10 000 years ago, the wild grass teosinte was domesticated to produce modern maize. Domestication resulted in a population bottleneck that reduced allelic diversity in maize relative to teosinte. Over the past decade, analysis of DNA sequence polymorphism data to detect signatures of genes that were involved in domestication and subsequent selection has become a well-established approach (e.g. Wang et al., 1999; Whitt et al., 2002; Tenaillon et al., 2004; Wright et al., 2005; Yamasaki et al., 2005).
The maize genome is composed of approximately 2.5 billion bases and contains an estimated 50 000 genes (Fu et al., 2005). The vast majority of this genome is composed of a small number of highly repetitive retrotransposons (Bennetzen, 1996; Meyers et al., 2001; SanMiguel et al., 1996; Whitelaw et al., 2003). Hence, it has not been economically feasible to conduct whole-genome scans for SNPs by sequencing multiple maize haplotypes. However, the 454 EST-based SNP mining procedure described here, which is focused on a specific transcriptome using LCM, provides the underpinning for a high-throughput SNP discovery platform than could be used to cost-effectively identify genes that exhibit signatures of having been involved in the domestication or improvement of maize and other large-genome crops, and that are therefore potential targets for improving agriculturally relevant traits.
Experimental procedures
Isolation of SAM mRNA and 454 sequencing
Maize SAM cDNA isolation, 454 sequencing and raw sequence processing were performed as previously described (Emrich et al., 2007a). A single GS-20 run produced 260 887 (28.8 Mbp) and 287 917 (30.7 Mbp) B73 and Mo17 SAM ESTs, respectively.
Identification of B73 reference sequences for 454 ESTs
Mo17 454 ESTs were initially mapped to a specific contig or singleton (217 773 total) from the MAGI 3.1 partial genome assembly of the maize inbred line B73 (Fu et al., 2005) using best BLASTN matches (minimum E-value 1e-8). Although ‘best hit’ criteria were used, it is possible that some 454 ESTs align to paralogous genomic fragments, especially given the partial nature of the MAGI assembly. To compensate, we used POLYBAYES (see below), which includes an internal paralog filter and should identify and discard these instances. These ESTs were also aligned to MAGIs using GeneSeqer (http://deepc2.psi.iastate.edu/cgi-bin/gs.cgi) and its maize-specific splice models (Usuka et al., 2000) for display on the MAGI website (http://magi.plantgenomics.iastate.edu). Only alignments consisting of at least 50 bp in length and with identity ≥ 95% over at least 80% of the length of the 454 EST were used to annotate genomic sequences.
Multiple sequence alignments and SNP detection of 454 sequence data
Custom PERL scripts were written to create a pipeline to process MAGI 3.1 anchor sequences and their associated B73 and Mo17 454 EST sequences for detecting SNPs. Anchored MSAs were produced by CROSS_MATCH with the following parameters: -discrep_lists -tags -masklevel 5 -gap_init -1 -gap_ext -1. Low initiation (-gap_init) and gap extension (-gap_ext) were used to increase alignment tolerance between the short 454 ESTs and the unplaced MAGI 3.1 genomic anchors. Sequence polymorphisms were detected by POLYBAYES using the following parameters:
anchorBaseQualityDefault 34 memberBaseQualityDefault 18
maskAmbiguousMatches nofilterParalogs -priorParalog 0.03 -thresholdNative 0.75 -screenSnps -considerAnchor -noconsiderTemplateConsensus -prescreenSnps -priorPoly 0.01 -thresholdSnp 0.5.
Default anchor quality values (34) were based on a previous assessment of sequence error rates within the MAGI 3.1 assembly (Fu et al., 2005). Default quality values of 18 were assigned to the 454 reads. This corresponds to an error rate of approximately 1/65, which over-compensates for the error rate observed for current 454 sequencing (Emrich et al., 2007a; Margulies et al., 2005). Although each base within the 454 sequence reads is given a quality score, these scores are only reliable when confirmed within independent sequences covering the same region. Because CROSS_MATCH aligns each sequence individually to the anchor during MSA construction, and POLYBAYES assesses base quality on an individual basis, use of a stringent default rather than the base quality information provided by 454 Life Sciences is expected to increase the accuracy of polymorphism detection.
SNP parsing
Mo17 and B73 are inbred lines, and thus should be mono-allelic at every base position. Custom PERL scripts were written to parse the POLYBAYES output (see Results). POLYBAYES identifies indel polymorphisms. Because indels are a common form of 454 sequencing error, only base substitutions were considered during this analysis. MAGI 3.1 assemblies contain a low frequency of base substitutions propagated during shotgun sequencing of the high-Cot selected maize genomic DNA (Fu et al., 2004). High-Cot selected maize DNA sequences account for only a portion of the MAGI 3.1 assembly sequence, but unidentified base substitutions within these regions could increase the number of false polymorphisms detected. Strict parsing rules (see Results) ensured that potential MAGI 3.1 sequence errors were avoided when B73 454 EST sequences are present in the multiple alignment. In cases where B73 454 ESTs are not present in the multiple alignment, SNPs called within regions of the MAGI 3.1 assemblies containing high-Cot selected DNA were avoided.
Acknowledgments
We thank Ruth Swanson-Wagner (Iowa State University) for preparing the SAM-specific Mo17 mRNA used for 454 sequencing. This research was supported by grants from the National Science Foundation (DBI-0321595, DBI-0321711 and DBI-0527192), Iowa State University’s Plant Science Institute and Pioneer Hi-Bred International Inc.; additional support was provided by the Hatch Act, State of Iowa, and the Donald Danforth Plant Science Center.
Supplementary material
The following supplementary material is available for this article online:
All raw polymorphism data predicted in this study.
All validated SNPs from this study.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Batley J. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003;132:84–91. doi: 10.1104/pp.102.019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- Bray NJ. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- Ching A. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 2002;3 doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Dantec LL, et al. Automated SNP detection in expressed sequence tags: statistical considerations and application to maritime pine sequences. Plant Mol. Biol. 2004;54:461–470. doi: 10.1023/B:PLAN.0000036376.11710.6f. [DOI] [PubMed] [Google Scholar]
- Emrich SJ. A strategy for assembling the maize (Zea mays L.) genome. Bioinformatics. 2004;20:140–147. doi: 10.1093/bioinformatics/bth017. [DOI] [PubMed] [Google Scholar]
- Emrich SJ. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007a;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich SJ. Nearly identical paralogs (NIPs): implications for maize (Zea mays L.) genome evolution. Genetics. 2007b;175:429–439. doi: 10.1534/genetics.106.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus FA. An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res. 2004;14:1812–1819. doi: 10.1101/gr.2479404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Types and frequencies of sequencing errors in methyl-filtered and high Cot maize genome survey sequences. Plant Physiol. 2004;135:2040–2045. doi: 10.1104/pp.104.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc. Natl Acad. Sci. USA. 2005;102:12282–12287. doi: 10.1073/pnas.0503394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, et al. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics. 2006;175:429–439. doi: 10.1534/genetics.106.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 2003;36:30–44. doi: 10.1046/j.1365-313x.2003.01852.x. [DOI] [PubMed] [Google Scholar]
- Gut IG. Automation in genotyping of single nucleotide polymorphisms. Hum. Mutat. 2001;17:475–492. doi: 10.1002/humu.1131. [DOI] [PubMed] [Google Scholar]
- Jander G. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota R. Generation and comparison of EST-derived SSRs and SNPs in barley (Hordeum vulgare L.) Hereditas. 2001;135:145–151. doi: 10.1111/j.1601-5223.2001.00145.x. [DOI] [PubMed] [Google Scholar]
- Kota R. Snipping polymorphisms from large EST collections in barley (Hordeum vulgare L.) Mol. Genet. Genomics. 2003;270:24–33. doi: 10.1007/s00438-003-0891-6. [DOI] [PubMed] [Google Scholar]
- Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- Lopez C. Using cDNA and genomic sequences as tools to develop SNP strategies in cassava (Manihot esculenta Crantz) Theor. Appl. Genet. 2005;110:425–431. doi: 10.1007/s00122-004-1833-3. [DOI] [PubMed] [Google Scholar]
- Manaster C. InSNP: a tool for automated detection and visualization of SNPs and InDels. Hum. Mutat. 2005;26:11–19. doi: 10.1002/humu.20188. [DOI] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- Meyers BC. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N. Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. Novel genetic mapping tools in plants: SNPs and LD-based approaches. Plant Sci. 2002;162:329–333. [Google Scholar]
- Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schnable PS. Global expression profiling applied to plant development. Curr. Opin. Plant Biol. 2004;7:50–56. doi: 10.1016/j.pbi.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Stupar RM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc. Natl Acad. Sci. USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- Useche FJ. High-throughput identification, database storage and analysis of SNPs in EST sequences. Genome Inform. 2001;12:194–203. [PubMed] [Google Scholar]
- Usuka J. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics. 2000;16:203–211. doi: 10.1093/bioinformatics/16.3.203. [DOI] [PubMed] [Google Scholar]
- Wang J. A method for finding single-nucleotide polymorphisms with allele frequencies in sequences of deep coverage. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Weckx S. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CA, et al. Enrichment of gene-coding sequences in maize by genome filtration. Science. 2003;302:2118–2120. doi: 10.1126/science.1090047. [DOI] [PubMed] [Google Scholar]
- Whitt SR. Genetic diversity and selection in the maize starch pathway. Proc. Natl Acad. Sci. USA. 2002;99:12959–12962. doi: 10.1073/pnas.202476999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire T, et al. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl Acad. Sci. USA. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Yamasaki M. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput. Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All raw polymorphism data predicted in this study.
All validated SNPs from this study.